Cell-Intrinsic Regulation of Axonal Morphogenesis by the Cdh1-APC Target SnoN
|
|
|
- Brook Gordon
- 6 years ago
- Views:
Transcription
1 Neuron 50, , May 4, 2006 ª2006 Elsevier Inc. DOI /j.neuron Cell-Intrinsic Regulation of Axonal Morphogenesis by the Cdh1-APC Target SnoN Judith Stegmüller, 1 Yoshiyuki Konishi, 1,2 Mai Anh Huynh, 1 Zengqiang Yuan, 1 Sara DiBacco, 1 and Azad Bonni 1, * 1 Department of Pathology Harvard Medical School 77 Avenue Louis Pasteur Boston, Massachusetts Summary Axonal growth is fundamental to the establishment of neuronal connectivity in the brain. However, the cell-intrinsic mechanisms that govern axonal morphogenesis remain to be elucidated. The ubiquitin ligase Cdh1-anaphase-promoting complex (Cdh1-APC) suppresses the growth of axons in postmitotic neurons. Here, we report that Cdh1-APC operates in the nucleus to inhibit axonal growth. We also identify the transcriptional corepressor SnoN as a key target of neuronal Cdh1-APC that promotes axonal growth. Cdh1 forms a physical complex with SnoN and stimulates the ubiquitin-dependent proteasomal degradation of SnoN in neurons. Knockdown of SnoN in neurons significantly reduces axonal growth and suppresses Cdh1 RNAi enhancement of axonal growth. In addition, SnoN knockdown in vivo suggests an essential function for SnoN in the development of granule neuron parallel fibers in the cerebellar cortex. These findings define Cdh1-APC and SnoN as components of a cell-intrinsic pathway that orchestrates axonal morphogenesis in a transcription-dependent manner in the mammalian brain. Introduction The growth of axons is critical to the establishment of neuronal connectivity and normal wiring of the developing nervous system. Studies of axonal development have led to the identification of several families of polypeptide growth factors that regulate and guide the growth of axons toward their targets (Dickson, 2002; Tessier-Lavigne and Goodman, 1996). Axon growth and guidance cues act on neurons via cell surface receptors that couple attractive and repulsive extrinsic signals to the cytoskeletal machinery of the axon growth cone (Dent and Gertler, 2003; Henley and Poo, 2004; Huber et al., 2003; Luo et al., 1997). Growing evidence suggests that cell-intrinsic mechanisms also play a crucial role in the control of axonal morphogenesis, but the nature of these mechanisms remains incompletely understood (Goldberg, 2004). A few transcription factors have been implicated in the control of distinct aspects of axonal development including axonogenesis and refinement of axonal projections *Correspondence: azad_bonni@hms.harvard.edu 2 Present address: Molecular Gerontology Group, Mitsubishi Kagaku Institute of Life Science, 11 Minamiooya, Machida-shi, Tokyo , Japan. (Arlotta et al., 2005; Kania et al., 2000; Segawa et al., 2001; Wang et al., 2002; Weimann et al., 1999). However, the transcriptional mechanisms that specifically control axonal elongation and stabilization remain to be identified. In addition to transcriptional control, the ubiquitinproteasome machinery is emerging as a key cell-intrinsic regulator of axonal development (Campbell and Holt, 2001; Konishi et al., 2004; Stegmuller and Bonni, 2005; Watts et al., 2003). Ubiquitin-dependent protein turnover in Xenopus retinal neurons modulates axon growth cone responses to chemotropic signals (Campbell and Holt, 2001). In addition, the ubiquitin-proteasome machinery promotes the local degeneration of axon terminals during development in Drosophila (Watts et al., 2003). While ubiquitination of proteins within the growth cone has been implicated in the regulation of axonal growth, with few exceptions specific ubiquitin ligases and their substrates in these processes remain to be identified (Campbell and Holt, 2001; Watts et al., 2003). We have found that the ubiquitin ligase Cdh1- anaphase-promoting complex (Cdh1-APC) plays a critical role in the control of axonal growth and patterning in the mammalian brain (Konishi et al., 2004). The finding that Cdh1-APC controls axonal growth raises the major question of how Cdh1-APC exerts this function in postmitotic neurons. Cdh1-APC is a multisubunit E3 ubiquitin ligase that promotes the ubiquitination and consequent degradation of B-type cyclins and other proteins in dividing cells and thereby ensures the proper transitions of the cell cycle (Harper et al., 2002; Page and Hieter, 1999; Peters, 2002). The regulatory subunit Cdh1 has the dual function of stimulating the APC ubiquitin ligase activity and targeting APC to its substrates. Substrate recognition by Cdh1 is dependent on specific peptide motifs, including the D box and KEN box, that are present in APC substrates (King et al., 1996; Pfleger and Kirschner, 2000). The ubiquitin ligase activity of Cdh1-APC is required for axon growth inhibition in mammalian neurons (Konishi et al., 2004). Therefore, to determine the mechanisms by which Cdh1-APC controls axonal morphogenesis, it will be crucial to identify the substrates of Cdh1-APC in neurons. In this study, we report a mechanism by which Cdh1- APC controls axonal morphogenesis in the mammalian brain. We have found that Cdh1-APC operates in the nucleus to inhibit the growth of axons. Using a candidate approach, we have discovered that Cdh1-APC controls axonal growth by inhibiting the transcriptional corepressor SnoN. Cdh1 interacts with SnoN and thereby targets SnoN for ubiquitin-dependent proteasomal degradation in primary cerebellar granule neurons. Knockdown of SnoN in primary neurons by RNAi significantly reduces the growth of axons and completely suppresses the ability of Cdh1 RNAi to enhance axonal growth. In addition, expression of a mutant SnoN protein that is resistant to Cdh1-mediated degradation robustly stimulates the growth of axons, an effect that is nonadditive with Cdh1 RNAi-enhanced axonal growth. We also find that the in vivo knockdown of SnoN in the developing rat
2 Neuron 390 cerebellum profoundly impairs the development of granule neuron parallel fibers by inhibiting their elongation and/or stabilization. These findings uncover an essential function for the transcriptional corepressor SnoN in axonal development in the mammalian brain. In addition, our study suggests that, by acting in the nucleus via regulation of SnoN-dependent programs of gene expression, Cdh1-APC may play a pivotal role in the control of axonal morphogenesis. Results Cdh1-APC Acts in the Nucleus to Control Axonal Growth To characterize the mechanism by which Cdh1-APC controls axonal growth, we first determined the subcellular site of action of Cdh1-APC in neurons. Cdh1, the APC core protein Cdc27, and the associated Cdh1- APC ubiquitin ligase activity in neurons are found predominantly but not exclusively in the nucleus in cerebellar granule neurons (Konishi et al., 2004) (Figure S1A in the Supplemental Data available with this article online). Consistent with these results, immunohistochemical analyses of the rat cerebellar cortex revealed Cdh1 immunoreactivity in the nucleus and cytoplasm of granule neurons (Figure S1B). Cdh1-APC is thought to function outside the nucleus in Drosophila neurons in the control of synapse development (van Roessel et al., 2004). Finally, ubiquitination of proteins within the growth cone regulates axonal development (Campbell and Holt, 2001; Watts et al., 2003). Together, these observations raised the question of whether Cdh1-APC operates in the nucleus or cytoplasm in mammalian neurons to control axonal growth. We carried out structure-function analyses of Cdh1 in the inhibition of axon growth. Since the overexpression of Cdh1 in neurons failed to alter axon growth (data not shown), we asked if an RNAi-resistant form of Cdh1 (Cdh1-Res) might rescue the Cdh1 knockdown-induced axonal phenotype in neurons and thus allow Cdh1 structure-function analyses in this background. To perform the rescue experiment, we constructed an expression plasmid encoding a GFP-Cdh1 fusion protein using wild-type Cdh1 cdna and a plasmid encoding an RNAi-resistant form of Cdh1 (GFP-Cdh1-Res; Figure 1A). Immunoblotting revealed that Cdh1 RNAi induced the knockdown of Cdh1, encoded by wild-type Cdh1 cdna, but failed to effectively reduce the expression of Cdh1-Res (Figure 1A). We next transfected cerebellar granule neurons with the Cdh1 RNAi (U6/cdh1) or control U6 plasmid together with a DsRed expression plasmid (Figure 1B). In the background of Cdh1 RNAi, we also expressed GFP-Cdh1 or GFP-Cdh1-Res. We found that GFP-Cdh1-Res, but not GFP-Cdh1, reversed the Cdh1 knockdown-induced enhancement of axonal growth (Figure 1B). These experiments indicate that the axonal phenotype upon Cdh1 RNAi is the result of specific knockdown of Cdh1. To determine if the localization of Cdh1 is of importance for Cdh1-APC function in neurons, we targeted GFP-Cdh1-Res to the nucleus or the cytoplasm (Figure 1C). The GFP-Cdh1-Res expression plasmid was modified by appending a nuclear exclusion sequence (NES) or a nuclear localization sequence (NLS) to the N terminus of Cdh1-Res (Yoneda et al., 1999). The expression of GFP-NES-Cdh1-Res and GFP-NLS- Cdh1-Res was assessed in both 293T cells and granule neurons. The localization of GFP-NLS-Cdh1-Res was predominantly nuclear in 293T cells and in primary neurons. By contrast, GFP-NES-Cdh1-Res was excluded from the nucleus (Figure 1C). We next examined the ability of Cdh1-Rescue mutants to reverse the increase in axonal length triggered by Cdh1 knockdown (Figure 1D). Granule neurons were transfected with the Cdh1 RNAi plasmid together with the control GFP or one of the GFP-Cdh1-Res series of plasmids and the DsRed expression vector. Neurons were analyzed 3 days after transfection. Analysis of axon length revealed that GFP-Cdh1-Res and GFP- NLS-Cdh1-Res behaved in a very similar manner, reversing the Cdh1 RNAi-induced enhancement of axonal growth (Figure 1D). In contrast, GFP-NES-Cdh1-Res failed to reverse the Cdh1 RNAi axonal phenotype, and axons remained at a comparable length as control axons (Figure 1D). In control experiments, appending the NES or NLS sequence to Cdh1 had little or no effect on the ability of Cdh1 to interact with the APC core protein Cdc27 (Figure S2), suggesting that NES- or NLStagged Cdh1 can still function as the regulatory subunit of the APC. Together, these results suggest that the localization of Cdh1 in the nucleus is required for Cdh1- APC inhibition of axon growth. The Cdh1-APC Target SnoN Promotes Axonal Growth The importance of the nuclear localization of Cdh1-APC in the control of axonal growth suggests that the targets of Cdh1-APC reside in the nucleus. Mounting evidence points to a key role for transcriptional mechanisms in the regulation of axonal growth (Goldberg, 2004; Stegmuller and Bonni, 2005). Together, these observations raised the possibility that Cdh1-APC might control axonal growth at a transcriptional level. To identify potential targets of neuronal Cdh1-APC that might regulate axonal growth, we considered targets of APC in proliferating cells that control transcription. Almost all APC substrates in dividing cells have functions in anaphase and DNA replication (Harper et al., 2002; Peters, 2002). Intriguingly, the transcriptional corepressor SnoN represents a substrate of Cdh1- APC in proliferating mammalian cells that directly regulates transcription (Stroschein et al., 2001; Wan et al., 2001). We asked if SnoN is expressed in neurons and whether SnoN might control axonal growth downstream of Cdh1-APC. We characterized the expression of SnoN in granule neurons in the cerebellum. Immunoblotting of lysates from primary cerebellar granule neurons with an antibody to SnoN revealed the expression of two alternatively spliced forms of SnoN (Figure 2A). Immunoprecipitation of lysates from granule neurons followed by immunoblotting with the SnoN antibody demonstrated the specificity of the two bands representing two SnoN isoforms (Figure 2A). SnoN was localized predominantly in the nucleus in granule neurons as determined by immunocytochemical analysis as well as immunoblotting of fractionated lysates of granule neurons (Figures 2B and 2C). SnoN localization remained
3 SnoN Regulation of Axonal Growth 391 Figure 1. Cdh1-APC Acts in the Nucleus to Control Axonal Growth (A) Schematic of GFP-Cdh1-Res and modified versions that contain the nuclear localization (NLS) or nuclear export (NES) sequences. Silent mutations that render Cdh1-Res resistant to RNAi are indicated in red. (Right) Lysates of COS cells transfected with the U6 or U6/cdh1 plasmid together with a plasmid encoding FLAG-Cdh1 or FLAG-Cdh1-Res were immunoblotted using a FLAG or antibody. Cdh1 RNAi induced the knockdown of Cdh1 encoded by wild-type cdna but failed to effectively induce knockdown of Cdh1-Res. (B) Primary cerebellar granule neurons were transfected 8 hr after plating with the U6 or U6/cdh1 plasmid and the GFP, GFP-Cdh1 (wild-type), or GFP-Cdh1-Res expression plasmid together with the DsRed and Bcl-x L expression plasmid. Neurons were kept in media supplemented with insulin. Three days later, cultures were subjected to immunocytochemistry using a polyclonal DsRed antibody. Total axonal length was measured and shown as mean 6 SEM. Cdh1 knockdown in granule neurons significantly increased axon length as compared to control U6- transfected neurons (p < 0.001, ANOVA); in the background of Cdh1 RNAi, GFP-Cdh1-Res but not GFP-Cdh1 (wild-type) significantly reduced axon length when compared to Cdh1 knockdown neurons (p < 0.001, ANOVA). A total of 447 neurons were measured. (C) 293T cells (left panels) and granule neurons (right panels) were transfected with the GFP, GFP-Cdh1-Res, GFP-NES-Cdh1-Res, or GFP-NLS- Cdh1-Res expression plasmid. Cultures were subjected to immunocytochemistry using a monoclonal GFP antibody. (D) Granule neurons were transfected with the U6/cdh1 plasmid together with the GFP, GFP-Cdh1-Res, GFP-NES-Cdh1-Res, or GFP-NLS-Cdh1- Res plasmid and the DsRed and Bcl-x L expression plasmids. Neurons were analyzed as in (B). In the background of Cdh1 RNAi, total axonal length of GFP-Cdh1-NES-Res- but not of GFP-Cdh1-NLS-Res-expressing neurons was significantly greater than in Cdh1Res-expressing neurons (p < 0.001, ANOVA). A total of 519 neurons were measured. nuclear under different culture conditions, including in the presence or absence of serum or membrane depolarization (data not shown). Immunohistochemical analysis of the rat cerebellar cortex revealed SnoN expression in both granule neurons and Purkinje cells (Figure 2D). The pattern of SnoN expression in the cerebellar cortex overlapped that of Cdh1, which was also found to be expressed in both granule neurons and Purkinje cells (Figure S1B). In particular, both Cdh1 and SnoN were found in granule neurons in the internal granule layer (IGL) (Figure 2D; Figure S1B). SnoN expression in IGL neurons persisted from P6 to P13 (Figure 2D), thus correlating temporally with axon growth in these neurons during brain development (Altman and Bayer, 1997). To study the function of SnoN in granule neurons, we used RNAi interference to acutely knock down SnoN (Figure 3A). We used a SnoN RNAi plasmid (U6/snon) that induces the knockdown of mouse SnoN (Sarker et al., 2005). The SnoN hairpin RNAs (hprnas) target a sequence in murine SnoN mrna that is identical in rat SnoN (XM ). Consistent with this observation, expression of the SnoN hprnas in primary rat cerebellar granule neurons led to the efficient knockdown of endogenous SnoN as determined by immunocytochemical analyses (Figure 3A). Whereas 70% of control
4 Neuron 392 Figure 2. SnoN Is Expressed in Granule Neurons in the Developing Cerebellum (A) (Left) Lysates of granule neurons prepared from P6 rat pups and placed in culture for indicated days were immunoblotted using a polyclonal SnoN antibody. (Right) Lysates of granule neurons were subjected to immunoprecipitation with the SnoN antibody or an antibody against actin followed by immunoblotting with the SnoN antibody. (B) Granule neurons were subjected to immunocytochemistry using the SnoN antibody and the DNA dye bisbenzimide (Hoechst 33258). SnoN appeared to be predominantly in the nucleus in all granule neurons. (C) Granule neurons were subjected to subcellular fractionation. The nuclear fraction (NF) and postnuclear supernatant (PNS) were immunoblotted with the SnoN, SP1, or antibody. (D) Sagittal sections of cerebella from postnatal rat pups at indicated ages were subjected to immunohistochemistry using the SnoN antibody. Cell nuclei were stained with the DNA dye bisbenzimide (Hoechst 33258). The external granule layer (EGL), molecular layer (ML), and internal granule layer (IGL) are indicated. Asterisks indicate Purkinje cells. Scale bar, 100 mm. U6-transfected neurons displayed robust SnoN immunoreactivity, only 30% of SnoN hprna-expressing neurons had SnoN immunoreactivity (Figure 3A). SnoN knockdown in granule neurons led to a striking axonal phenotype. Granule neurons in which SnoN RNAi was triggered had significantly shorter axons than control U6-transfected neurons (Figure 3B). The SnoN knockdown and control U6-transfected neurons had comparable expression of proteins enriched in postmitotic granule neurons (Figure S3), suggesting that SnoN RNAi did not alter the general differentiation state of granule neurons. We next determined if the SnoN RNAi-induced axonal phenotype is the result of specific knockdown of SnoN. First, we used a plasmid encoding human SnoN hprnas (U6/snon-h), which contain three nucleotide mismatches with the rodent SnoN hprnas (Sarker et al., 2005). While expression of SnoN hprnas induced the efficient knockdown of endogenous SnoN in the mouse neuronal cell line Neuro2A, human SnoN hprnas failed to induce SnoN knockdown in these cells (Figure S4). In granule neurons, expression of human SnoN hprnas failed to reduce axon length (Figure 3B). Second, we performed a rescue experiment in the background of SnoN RNAi (Figure 3C). In these experiments, we used a plasmid encoding human SnoN harboring additional silent mutations in its cdna designed to render it resistant to RNAi (SnoN- Res) (Sarker et al., 2005). We determined the effect of SnoN-Res expression on axonal growth in the background of SnoN knockdown. We found that the expression of SnoN-Res triggered a significant increase in axonal length, restoring total axonal length to 70% of control U6-transfected granule neurons (Figure 3C). These results indicate that the SnoN RNAi-triggered reduction in axonal length is the result of specific knockdown of SnoN rather than off-target effects of SnoN RNAi or nonspecific activation of the RNAi machinery.
5 SnoN Regulation of Axonal Growth 393 Figure 3. SnoN Promotes Axonal Growth in Cerebellar Granule Neurons (A) Granule neurons were transfected with the SnoN RNAi (U6/snon) or control U6 plasmid together with an expression plasmid encoding farnesylated GFP. Three days later, cultures were subjected to immunocytochemical analysis using the GFP and SnoN antibodies. Arrowhead points to a U6-transfected GFP-positive neuron that is also SnoN positive, and arrow points to a neuron transfected with the U6/snon plasmid as indicated by GFP expression that is SnoN negative. Approximately 70% of the control U6-transfected neurons and only 30% of SnoN hprnas-expressing neurons are SnoN positive. Scale bar, 20 mm. (B) Granule neurons transfected with the control U6, U6/snon (mouse), or U6/snon-h (human) RNAi plasmid together with the GFP and Bcl-x L expression plasmids and cultured in media supplemented with calf serum for 3 days were subjected to immunocytochemistry using the GFP antibody and analyzed as in Figure 1B. Axon length in U6/snon (mouse)-expressing neurons is significantly reduced as compared to control U6-transfected neurons (p < , ANOVA) or U6/snon (human)-expressing neurons (p < 0.002, ANOVA). A total of 378 neurons were measured. (C) Granule neurons transfected with the SnoN RNAi or control U6 plasmid together with an expression plasmid encoding SnoN-Res or its control vector (pcmv5) and the GFP and Bcl-x L expression plasmids were analyzed as in Figure 3B. In the background of SnoN knockdown in granule neurons, expression of SnoN-Res significantly increased axonal length as compared to vector pcmv5-expressing neurons (p < 0.02, ANOVA). A total of 241 neurons were measured. (D) Granule neurons transfected with the SnoN RNAi or control U6 plasmid together with the GFP and Bcl-x L expression plasmids were cultured in BME supplemented with calf serum for indicated days and analyzed as in Figure 3B. SnoN knockdown in granule neurons significantly reduced axonal length at 3 days and subsequent days as compared to control U6-transfected neurons (p < 0.005, ANOVA). A total of 716 neurons were measured. Taken together, our results suggest that SnoN is required for the normal development of axons. To assess the nature of SnoN function in the development of axons, we transfected neurons with the SnoN RNAi or control U6 plasmid at a time when they begin to extend axons and measured the length of axons in cohorts of neurons each day for 4 days, beginning 1 day after transfection (Figure 3D). In control U6-transfected granule neurons, axons increased significantly in length from day 2 to day 5 after transfection. However, upon SnoN knockdown the axonal growth curve was significantly reduced in slope (Figure 3D). Axons were 2-fold shorter in neurons with SnoN knockdown as compared to control U6-transfected neurons at day 5 (Figure 3D). These results suggest that a key function of the transcriptional corepressor SnoN in neurons is to promote the growth of axons. We also performed time-lapse analyses to ascertain whether SnoN promotes axonal growth by stimulating their elongation or by inhibiting axonal retraction. We monitored individual neurons transfected with the SnoN RNAi or control U6 plasmid (Figure 4A). While the majority of control U6-transfected neurons robustly extended axons over the entire period of observation, SnoN knockdown almost completely inhibited axonal extension (Figure 4B). However, SnoN RNAi did not appear to induce retraction of axons. In short-term time-lapse analyses over a period of 3 hr, we found rapid changes and high motility of axonal tips in control U6-transfected neurons, but the axonal tips in SnoN knockdown neurons displayed little or no motility (data not shown). Taken together, these results are consistent with the interpretation that SnoN knockdown impairs the elongation of axons in primary neurons. We next determined if SnoN function in axonal growth is affected by extrinsic culture conditions. We assessed the effect of SnoN knockdown in granule neurons that were exposed to the growth factor insulin, serum, or serum together with membrane depolarization. Under each of these conditions, SnoN knockdown significantly
6 Neuron 394 Figure 4. SnoN Knockdown Impairs Axonal Elongation in Primary Granule Neurons (A) Representative images of neurons transfected with the SnoN RNAi or control U6 plasmid. Starting at 2 DIV, images of neurons were taken every 8 hr over a period of 48 hr. Axons of control U6-transfected neurons increased in length. In contrast, axons of SnoN knockdown neurons did not grow or retract. Arrows indicate axons. (B) (Left) Slope of axonal growth of individual neurons transfected with the SnoN RNAi or control U6 plasmid. (Right) Average slope of axonal growth, presented as mean 6 SEM, was significantly higher in control U6-transfected neurons as compared to SnoN knockdown neurons (p < 0.005, Student s t test). reduced total axonal length in granule neurons (Figures 3B, 5A, and 5B). These results suggest that SnoN represents a cell-intrinsic regulator of axonal growth. Having identified a cell-autonomous function for SnoN in axonal growth in primary dissociated cultures of granule neurons, we next asked if SnoN promotes axonal growth in the context of the cerebellar cortex (Figure 5C). To address this question, we used cerebellar slice overlay assays. We plated P6 neurons that were transfected with a plasmid encoding both SnoN hprnas and GFP bicistronically (U6/snon-cmvGFP) or the control U6- cmvgfp plasmid on top of cerebellar slices prepared from P9 rat pups. Three days later, slices were subjected to immunohistochemistry. Granule neurons in which SnoN RNAi was triggered had a significant reduction in total axonal length as compared to the control U6 plasmid-transfected neurons (Figure 5C). These results suggest that SnoN promotes axonal growth in granule neurons in the tissue environment of the cerebellar cortex. We also assessed if SnoN contributes to axonal growth in neuronal cell types other than cerebellar granule neurons. We found that SnoN is also expressed in cortical and hippocampal neurons (Figure 5D). Importantly, SnoN knockdown significantly reduced axonal growth in cortical neurons (Figure 5E). These data suggest that SnoN function in regulation of axonal morphogenesis may be widespread in the mammalian brain. SnoN Acts Downstream of Neuronal Cdh1-APC in the Control of Axon Growth To determine if Cdh1-APC and its substrate SnoN act in a linear pathway to regulate axonal growth, we performed epistatic analysis of the effects of Cdh1 and SnoN knockdown on axonal length (Figure 6A). Primary granule neurons were transfected with the Cdh1 or SnoN RNAi plasmid or both RNAi plasmids together. Cdh1 knockdown significantly stimulated an increase in total axonal length, while SnoN knockdown significantly inhibited axon growth compared to control neurons
7 SnoN Regulation of Axonal Growth 395 Figure 5. SnoN Knockdown Impairs Axonal Growth under Different Environmental Conditions and in Distinct Populations of Neurons (A and B) Granule neurons transfected with the SnoN RNAi or control U6 plasmid together with the GFP and Bcl-x L expression plasmids were cultured in BME supplemented with insulin (A) or calf serum together with membrane-depolarizing concentrations of KCl (B) and were analyzed as in Figure 3B. Images of representative transfected neurons in BME + insulin are shown. Scale bar, 50 mm. SnoN knockdown in granule neurons in insulin and in serum plus KCl is significantly reduced as compared to the corresponding control U6-transfected neurons (p < and p < , respectively, Student s t test). A total of 110 and 165 neurons were measured, respectively. (C) Granule neurons transfected in suspension with the U6/snon-cmvGFP RNAi or control U6-cmvGFP plasmid together with an expression plasmid encoding Bcl-x L were placed on top of cerebellar slices from P9 rat pups. Slices were fixed 3 days later, subjected to immunohistochemistry with the GFP antibody, and subjected to morphometry. Axonal length of U6/snon-expressing neurons was significantly reduced as compared to control U6-transfected neurons (p < 0.002, Student s t test; values indicate mean 6 SEM). A total of 375 neurons were measured. (D) Hippocampal and cortical neurons were isolated from E18 rat embryos. Neurons were cultured for the indicated time period, and lysates were subjected to immunoblotting with the SnoN antibody. Both hippocampal and cortical neurons express SnoN. Asterisks indicate nonspecific band. (E) Axonal length was measured in cortical neurons transfected with the SnoN RNAi or control U6 plasmid together with the GFP and Bcl-X L expression plasmids. Axon length was significantly reduced in neurons in which SnoN RNAi was induced as compared to control U6-transfected neurons (p < , Student s t test; values indicate mean 6 SEM). A total of 194 neurons were measured. Scale bar, 200 mm. (Figure 6A). The simultaneous induction of Cdh1 and SnoN RNAi led to an axonal phenotype that was identical to that of SnoN knockdown, resulting in significantly shorter axons as compared to Cdh1 hprna-expressing neurons or as compared to control U6-transfected neurons (Figure 6A). These experiments indicate that SnoN knockdown completely suppressed the Cdh1 RNAiinduced axonal growth phenotype. These findings are consistent with the interpretation that Cdh1-APC and SnoN operate in a linear pathway, where SnoN acts downstream of Cdh1-APC in the control of axon growth. To corroborate our results suggesting that SnoN acts downstream of Cdh1-APC in neurons, we carried out structure-function analyses of SnoN (Figure 6B). SnoN contains a conserved Cdh1 recognition destruction (D box) peptide motif (Stroschein et al., 2001; Wan et al., 2001), whose mutation renders SnoN resistant to APCmediated ubiquitination. We measured axonal length in granule neurons in which we expressed wild-type SnoN (SnoN WT) or a SnoN protein with a mutated D box (SnoN DBM). Expression of SnoN in granule neurons had little effect on axon length (Figure 6B). In contrast, neurons in which SnoN DBM was expressed had a significant increase in total axonal length as compared to SnoN-expressing neurons or control vector-transfected neurons (Figure 6B). In experiments in which we measured axonal length for several days after transfection, SnoN DBM but not SnoN significantly enhanced the rate of axonal growth as compared to controltransfected neurons (Figure 6C). Together, these results indicate that mutation of the Cdh1 recognition D box in SnoN unmasks SnoN s ability to promote axonal growth. We next tested the ability of SnoN DBM to promote axonal growth in neurons in which Cdh1 RNAi was induced. While expression of SnoN DBM and Cdh1 knockdown each enhanced axonal growth in granule neurons, the combination of SnoN DBM overexpression and Cdh1 knockdown did not result in an additive effect on axonal length (Figure 6D). These findings corroborate the results of the epistasis analyses in Figure 6A, suggesting that Cdh1 and SnoN act in a shared pathway. Taken together, our results support the conclusion that Cdh1 controls axonal growth by inhibiting SnoN function in neurons. To characterize the mechanism by which neuronal Cdh1-APC inhibits SnoN function, we first asked if SnoN is regulated by the ubiquitin-dependent proteasome machinery in granule neurons. Endogenous SnoN levels increased in granule neurons treated with the proteasome inhibitor lactacystin or MG132 as determined by immunoblotting (Figure 6E and data not shown). In other experiments, endogenous SnoN was found to be conjugated with ubiquitin in neurons as determined by immunoblotting of immunoprecipitated SnoN with antibodies to ubiquitin (Figure 6F). Together, these results suggest that SnoN undergoes ubiquitindependent proteasomal degradation in neurons.
8 Neuron 396 Figure 6. SnoN Acts Downstream of Cdh1- APC in the Control of Axonal Growth (A) Granule neurons transfected with the control U6, U6/cdh1, U6/snon plasmid, or both U6/cdh1 and U6/snon RNAi plasmids together with the GFP and Bcl-x L expression plasmids were analyzed as in Figure 3B. Knockdown of both Cdh1 and SnoN in granule neurons significantly reduced axonal length as compared to control U6- and U6/ cdh1-transfected neurons, respectively (p < and p < 0.05 respectively, ANOVA). A total of 521 neurons were measured. (B) Granule neurons transfected with an expression plasmid encoding wild-type SnoN (WT), mutant D box SnoN (DBM), or the control vector pcmv5 together with the GFP and Bcl-x L expression plasmids were analyzed as in Figure 3B. SnoN DBM expression in granule neurons but not the expression of SnoN WT significantly increased axonal length as compared to control U6-transfected neurons (p < 0.02, ANOVA). A total of 328 neurons were measured. (C) Granule neurons transfected as in Figure 5B were analyzed at the indicated times as in Figure 3B. Expression of SnoN DBM in granule neurons significantly increased axonal growth at day 4 as compared to SnoN expression or control U6-transfected neurons (p < 0.001, ANOVA). A total of 423 neurons were measured. (D) Granule neurons transfected with the Cdh1 RNAi plasmid or SnoN DBM expression plasmid alone or together with their control vectors were analyzed as in Figure 3B. Simultaneous knockdown of Cdh1 and expression of SnoN DBM did not result in additive axonal growth. A total of 314 neurons were measured. (E) Lysates of granule neurons treated with 10 mm lactacystin or vehicle for 10 hr were subjected to immunoblotting with the SnoN or antibody. (F) Lysates of granule neurons were subjected to immunoprecipitation with the HA (ctrl) or SnoN antibody followed by immunoblotting with an antibody to ubiquitin. (G) Lysates of granule neurons were immunoprecipitated with the HA (ctrl) and SnoN antibody followed by immunoblotting with the Cdh1 antibody. (H) Schematic of Renilla-SnoN WT and Renilla-SnoN DBM. Lysates of granule neurons transfected with Renilla-SnoN WT or Renilla-SnoN DBM expression plasmid together with the SV40 firefly luciferase (pgl3 promoter) plasmid, the latter to serve as internal control for transfection efficiency, were subjected to a Dual Luciferase assay (Promega). Renilla-SnoN DBM activity was significantly increased compared to SnoN WT (p < 0.03, Student s t test; n = 4; values indicate mean 6 SEM). (I) Lysates of granule neurons transfected with the Ren-SnoN WT or Ren-SnoN DBM expression plasmid together with the firefly luciferase (pgl3 promoter) plasmid and a Cdh1 RNAi plasmid (psuper/cdh1) or its control vector (psuper). Renilla-SnoN activity was significantly increased upon Cdh1 RNAi compared to control-transfected neurons (p < 0.02, ANOVA; n = 4; values indicate mean 6 SEM). Cdh1 knockdown had little or no effect on activity of renilla-snon DBM. We next assessed the role of Cdh1-APC in the regulation of SnoN protein turnover in neurons. In coimmunoprecipitation experiments, endogenous Cdh1 was found to associate with endogenous SnoN (Figure 6G). To determine if neuronal Cdh1-APC promotes SnoN degradation, we asked if Cdh1 knockdown increases the level of SnoN protein in neurons. First, we established an assay that accurately reflects increases in the amount of SnoN protein in transfected neurons. We expressed a gene encoding SnoN fused to renilla luciferase (Ren-SnoN) in neurons, thus allowing us to use renilla luciferase activity as a surrogate for the amount of SnoN in transfected neurons (Figure 6H). Luciferase fusion proteins have been successfully used to quantify protein turnover of APC substrates (Lukas et al., 1999). In our experiments, a renilla fusion with the SnoN DBM mutant protein (Ren-SnoN DBM) had a significantly higher level of renilla luciferase activity than renilla-snon, suggesting that mutation of the Cdh1 recognition D box motif within SnoN leads to increased levels of SnoN protein in neurons (Figure 6H). Importantly, using the renilla fusion assay, we found a significant increase in the level of renilla-snon upon Cdh1 knockdown in granule neurons. In contrast, Cdh1 knockdown had little effect on the level of renilla-snon DBM activity (Figure 6I). These results suggest that Cdh1 promotes the degradation of SnoN protein. Taken together, our findings support the conclusion that Cdh1-APC promotes the ubiquitination and consequent proteasomal degradation of SnoN in neurons.
9 SnoN Regulation of Axonal Growth 397 Figure 7. SnoN Knockdown Impairs Granule Neuron Parallel Fiber Development In Vivo (A) The SnoN DBM expression plasmid or its control pcmv5 vector together with the GFP and Bcl-x L expression plasmids were injected into the cerebellum of P3 rat pups. Five days later at P8, cerebella were isolated from rat pups, and 10 mm coronal sections of the cerebella were subjected to immunohistochemistry using the GFP antibody. No apparent difference in parallel fiber patterning in cerebella was detected in SnoN DBMexpressing cerebella as compared to cerebella transfected with the control vector. Arrows point to parallel fibers in the molecular layer; asterisks indicate granule neurons in the internal granule layer. (B) The U6/snon-cmvGFP RNAi or U6/ cmvgfp control plasmid together with the Bcl-x L expression plasmid were injected into the cerebellum of P3 rat pups, and cerebella were analyzed as in Figure 7A. Representative images of newly generated neurons that extend axons in the EGL. Arrows point to transfected newly generated neurons. Scale bar, 50 mm. (C) Coronal sections of cerebella from pups were subjected to in vivo electroporation as described in (B). The external granule layer (EGL), molecular layer (ML), and internal granule layer (IGL) are indicated. Arrowheads indicate parallel fibers. Scale bars, 100 mm in large panels and 50 mm in small panels. (D) Quantification of parallel fibers. Transfected granule neurons in the IGL were counted in consecutive sections of the U6- cmvgfp- or U6/snon-cmvGFP-transfected cerebella. Axons were counted in the molecular layer (see Experimental Procedures). Graph indicates percentage of granule neurons that were associated with parallel fibers. Parallel fiber number in granule neurons is significantly reduced upon SnoN knockdown as compared to control U6-transfected neurons (p < 0.001, Student s t test; values indicate mean 6 SEM). A total of 476 neurons were measured. SnoN Is Required for the Development of Granule Neuron Parallel Fibers In Vivo The identification of SnoN as a key Cdh1-APC target protein that promotes axonal growth in primary granule neurons led us to characterize the in vivo function of SnoN in the cerebellar cortex. Since both gain-of-function and loss-of-function analyses implicated SnoN in axonal growth in primary neurons, we decided to employ both approaches in the assessment of SnoN function in vivo. To do this, we used an electroporation method to express SnoN DBM or to induce SnoN knockdown in the cerebellar cortex in postnatal rat pups (Figure 7A). In each case, we injected test plasmids into the cerebellar cortex of P3 rat pups and subjected these pups to electroporation. In both sets of experiments, a GFP expression plasmid or cassette was included. Five days after electroporation, the cerebellum was isolated and coronal sections of the cerebellum were subjected to immunohistochemistry with GFP antibodies to allow the assessment of granule neuron parallel fiber axons. GFP-positive granule neurons were present in the cerebellar cortex of electroporated animals. Most of the transfected GFP-positive granule neurons were in the IGL, with a smaller population in the EGL (Figures 7A 7C). In the gain-of-function analyses, the SnoN DBM granule neurons appeared to have normal parallel fibers of similar pattern as those in control pcmv5-transfected cerebella (Figure 7A). We were unable to assess whether SnoN DBM stimulates axonal growth in the cerebellar cortex, because it was not possible to measure the total length of individual parallel fibers in vivo (Figure 7A). In contrast to the gain-of-function analyses, the lossof-function analyses were informative. In these experiments, we injected the U6/snon-cmvGFP RNAi or control U6/cmvGFP plasmid into the cerebellar cortex of P3 rat pups and analyzed the cerebella 5 days later. Examination of the EGL neurons revealed that both control U6-transfected and SnoN hprnas-expressing granule neurons extended robust axons at this stage of development (Figure 7B). However, analysis of the IGL granule neurons revealed a striking SnoN knockdown-induced phenotype in parallel fiber development (Figure 7C). The control U6-transfected granule neurons residing in
10 Neuron 398 the IGL displayed a normal anatomy including the typical T-shaped axons and association with parallel fibers in the molecular layer (ML). Parallel fibers extended in large numbers in and beyond the region of the transfected IGL granule neurons (Figure 7C). By contrast, although SnoN knockdown granule neurons were present in the IGL in similar numbers as control U6-transfected neurons, few of the SnoN knockdown neurons were associated with parallel fibers in the ML (Figure 7C). We quantified the effect of SnoN knockdown on parallel fibers in the cerebellar cortex. More than 90% of the control U6-transfected IGL granule neurons were associated with parallel fibers. In contrast, only 30% of the SnoN knockdown IGL granule neurons were associated with parallel fibers (Figure 7D). These remaining parallel fibers in the neurons appeared underdeveloped and failed to extend for long distances beyond the area of the transfected IGL neurons (Figure 7C). These results suggest that SnoN plays a key role in axonal growth and/or stabilization of IGL granule neurons. Consistent with this observation is the finding that SnoN is robustly expressed in granule neurons in the IGL but not in the EGL (Figure 2D). Taken together, our findings support the conclusion that SnoN specifically promotes the morphogenesis of granule neuron parallel fibers in the cerebellar cortex in vivo at a developmental stage following axonogenesis. Discussion In this study, we have characterized a mechanism by which the ubiquitin ligase Cdh1-APC controls axonal morphogenesis in the mammalian brain. Our findings indicate that Cdh1-APC operates in the nucleus to control axonal growth in granule neurons of the developing cerebellum. We have also identified the transcriptional corepressor SnoN as a key target of neuronal Cdh1- APC that promotes axonal growth. Cdh1 interacts with SnoN and stimulates the ubiquitin-dependent degradation of SnoN in primary neurons. Consistent with these results, epistasis analyses in neurons suggest that SnoN acts downstream of Cdh1 in the control of axonal growth. Knockdown of SnoN in the cerebellum by in vivo electroporation points to an essential function of SnoN in granule neuron parallel fiber development. Taken together, our findings suggest that Cdh1-APC and SnoN form a cell-intrinsic pathway that orchestrates axonal morphogenesis in the mammalian brain. Our study uncovers a function for SnoN in neurons as a potent promoter of axonal growth. In particular, our findings in primary neurons and in vivo support the view that SnoN promotes axonal elongation in the mammalian brain. The in vivo SnoN knockdown experiments raise the possibility that SnoN may also actively inhibit the destabilization of developing axons. In future studies, it will be interesting to determine how SnoN might impact on both axonal elongation and stabilization. Since SnoN is a member of the Ski/SnoN family of transcriptional corepressors (Akiyoshi et al., 1999; He et al., 2003; Luo et al., 1999; Nomura et al., 1999; Stroschein et al., 1999; Sun et al., 1999a, 1999b; Xu et al., 2000), it will be important to identify the transcription factors that act in concert with SnoN in neurons. Prior to our study, SnoN function was unknown in the nervous system. However, SnoN has been implicated as a protooncogene in a number of tissues outside the nervous system (Colmenares et al., 1991; Reed et al., 2001; Zhang et al., 2003), owing to its function as a key negative regulator of signaling induced by transforming growth factor-b (TGFb) (He et al., 2003; Luo et al., 1999; Stroschein et al., 1999). SnoN physically interacts with the TGFb-regulated transcription factors Smad2 and Smad3 and thereby represses Smad-dependent gene expression (Akiyoshi et al., 1999; He et al., 2003; Luo et al., 1999; Stroschein et al., 1999; Sun et al., 1999a; Xu et al., 2000). Conversely, TGFb signaling induces the Smad2- or Smad3-dependent recruitment of SnoN to Cdh1, leading to Cdh1-APC-mediated ubiquitination and consequent degradation of SnoN in proliferating cells (Stroschein et al., 2001; Wan et al., 2001). Thus, TGFb signaling triggers the release of a SnoNdependent repressor complex from Smad2 and Smad3, leading to the activation of TGFb-dependent transcription (Stroschein et al., 2001; Wan et al., 2001). In view of these observations, it will be important to determine if the Cdh1-APC-SnoN cell-intrinsic mechanism underlying axonal morphogenesis is subject to regulation by TGFb signaling in neurons, and to identify components of the TGFb-Smad signaling pathway that might control SnoN-dependent axonal growth in the brain. The finding that Cdh1-APC operates in the nucleus to control axonal growth supports the view that Cdh1-APC is a pivotal regulator of axonal development. This conclusion is bolstered by the identification of the transcriptional corepressor SnoN as a key downstream target of Cdh1-APC in the control of axonal growth. These findings suggest that Cdh1-APC controls the development of axons by coordinating programs of gene expression leading to specific effects on axonal morphogenesis. Therefore, to gain insights into the mechanisms of Cdh1-APC control of axonal growth, it will be important in future studies to elucidate the programs of gene expression that are regulated by SnoN in neurons. Beyond SnoN, it will be also important to identify other substrates of Cdh1-APC in the control of axonal growth. Evidence for the existence of additional substrates of Cdh1-APC that might be relevant in axonal morphogenesis comes from our analyses of overexpression of the D box mutant SnoN (SnoN DBM) in the cerebellum. Although we found in these in vivo experiments that it was not possible to measure the anticipated gain-offunction effect of SnoN DBM of increased axonal length, we assessed whether overexpression of SnoN DBM might induce a defect in axonal patterning, similar to that with Cdh1 RNAi (Konishi et al., 2004). Interestingly, we found little difference in parallel fiber patterning in cerebella transfected with SnoN DBM as compared to control vector (Figure 7A). These data suggest the interesting possibility of additional substrates of neuronal Cdh1-APC that might mediate the ability of Cdh1-APC to control the patterning of axons. Studies in Drosophila and Xenopus model systems suggest that ubiquitin-dependent protein turnover locally within axon growth terminals regulates distinct behaviors of axon terminals, including their chemotropic responses and degeneration (Campbell and Holt, 2001; Watts et al., 2003). Although we have found that the nucleus is the prime site of action of Cdh1-APC in axonal
11 SnoN Regulation of Axonal Growth 399 growth in mammalian neurons, the possibility remains that Cdh1-APC might also contribute to the regulation of axonal growth locally in the axon growth cone. Cdh1-APC that resides outside the nucleus in mammalian neurons might also be relevant to other functions, including possibly synapse development and function, as demonstrated in nematodes and flies (Juo and Kaplan, 2004; van Roessel et al., 2004). Thus, the localization of Cdh1-APC in distinct subcellular compartments in neurons might endow this ubiquitin ligase with the ability to locally recruit its targets for degradation and regulate diverse functions in the mammalian brain. Experimental Procedures Cerebellar Granule Neuron Culture and Transfections Granule neurons were prepared from isolated cerebella of P6 Long- Evans rat pups as described (Shalizi et al., 2003). Neurons were transfected either 8 hr after plating or at 2 days in vitro (DIV) with a modified calcium phosphate method as described (Konishi et al., 2002). To test if the calcium phosphate transfection method ensures the efficient cotransfection of multiple plasmids, we cotransfected expression plasmids encoding GFP and DsRed and found that 92% of DsRed-positive neurons also expressed GFP. To rule out the possibility that the effects of RNAi or protein expression on axonal length were due to any effect of these manipulations on cell survival, the antiapoptotic protein Bcl-x L was coexpressed in all our experiments. The expression of Bcl-x L itself has little or no effect on axonal length (Konishi et al., 2004). Morphometry Analysis of the morphology of axons of the cerebellar granule neurons in vitro and in the slice overlay assay was carried out by capturing images of the neurons in a blinded manner using a Nikon eclipse TE2000 epifluorescence microscope. Measurements of axons were performed using SPOT imaging software as described (Gaudilliere et al., 2004). In Vivo Electroporation In vivo electroporation was performed as described (Konishi et al., 2004). In brief, the U6-cmvGFP or U6/snon-cmvGFP plasmid together with the Bcl-x L expression plasmid were diluted in PBS/ 0.3% fast green (3 4 ml with 4 mg/ml of U6 plasmids and 1 mg/ml of Bcl-x L plasmid) and injected into the cerebellum of P3 Sprague- Dawley rat pups. The pups were subjected to five electric pulses of 160 mv with 950 ms intervals. To analyze the parallel fibers in the in vivo electroporation experiments, the identity of transfected granule neurons in the EGL was confirmed based on the small size of the nuclei, as determined by staining with the DNA dye bisbenzimide (Hoechst 33258) and MEF2 immunoreactivity, a marker of granule neurons in the IGL. Transfected granule neurons were counted in consecutive sections of individual cerebella. Parallel fibers were counted in a restricted area of consecutive sections to prevent recounting. Slice Overlay Assay Slice overlay assay was performed as described (Konishi et al., 2004). In brief, cerebellar slices from P8 or P9 rat pups were prepared using a McIllwain Tissue Chopper. Slices (400 mm) were cultured on 0.4 mm membranes using the medium air interface method (MEM; 25 mm HEPES, 25% horse serum, 6.5 mg/ml D-glucose, 1 ml/ 100 ml PSG) for 24 hr at 36ºC/5% CO 2 before coculture with granule neurons. Granule neurons were isolated from P6 rats as described and transfected 3 hr later in suspension ( cells/2 ml DMEM) using a modified calcium phosphate method with the control U6 plasmid that also contained an expression cassette encoding GFP (U6-cmvGFP) or the U6/snon-cmvGFP together with an expression plasmid encoding Bcl-x L. Transfection reaction was terminated by adding a large volume of DMEM. Cells were pelleted and then plated on top of the cerebellar slices and cocultured for 3 days. Slices were then subjected to immunostaining using the GFP antibody. Slice integrity was assessed using the DNA dye bisbenzimide (Hoechst 33258). Supplemental Data The Supplemental Data include four supplemental figures and can be found with this article online at content/full/50/3/389/dc1/. Acknowledgments We thank Shirin Bonni for providing the SnoN RNAi and SnoN expression plasmids and for helpful discussions, Takahiko Matsuda and Connie Cepko for sharing equipment, Jan-Michael Peters for Cdh1 antibodies, Thijn Brummelkamp for the psuper Cdh1 RNAi plasmid, Zhigang Xie for help with time-lapse analyses, Xuecai Ge for help with experiments at an early stage of the study, and members of the Bonni laboratory for critical reading of the manuscript. Supported by an NIH grant (NS051255; A.B.), the Christopher Reeve Foundation (A.B.), the Deutsche Forschungsgemeinschaft (J.S.), and the Lefler Foundation (Z.Y.). A.B. is the recipient of a fellowship from the Alfred P. Sloan Foundation, a Robert H. Ebert Clinical Scholar Award from the Esther A. and Joseph Klingenstein Fund, an EJLB Foundation award, and a Sidney Kimmel Foundation Award. Received: August 10, 2005 Revised: September 12, 2005 Accepted: March 27, 2006 Published: May 3, 2006 References Akiyoshi, S., Inoue, H., Hanai, J., Kusanagi, K., Nemoto, N., Miyazono, K., and Kawabata, M. (1999). c-ski acts as a transcriptional co-repressor in transforming growth factor-b signaling through interaction with smads. J. Biol. Chem. 274, Altman, J., and Bayer, S.A. (1997). Development of the Cerebellar System: In Relation to Its Evolution, Structure, and Functions (Boca Raton, FL: CRC Press). Arlotta, P., Molyneaux, B.J., Chen, J., Inoue, J., Kominami, R., and Macklis, J.D. (2005). Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, Campbell, D.S., and Holt, C.E. (2001). Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32, Colmenares, C., Sutrave, P., Hughes, S.H., and Stavnezer, E. (1991). Activation of the c-ski oncogene by overexpression. J. Virol. 65, Dent, E.W., and Gertler, F.B. (2003). Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron 40, Dickson, B.J. (2002). Molecular mechanisms of axon guidance. Science 298, Gaudilliere, B., Konishi, Y., de la Iglesia, N., Yao, G., and Bonni, A. (2004). A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron 41, Goldberg, J.L. (2004). Intrinsic neuronal regulation of axon and dendrite growth. Curr. Opin. Neurobiol. 14, Harper, J.W., Burton, J.L., and Solomon, M.J. (2002). The anaphasepromoting complex: it s not just for mitosis any more. Genes Dev. 16, He, J., Tegen, S.B., Krawitz, A.R., Martin, G.S., and Luo, K. (2003). The transforming activity of Ski and SnoN is dependent on their ability to repress the activity of Smad proteins. J. Biol. Chem. 278, Henley, J., and Poo, M.M. (2004). Guiding neuronal growth cones using Ca2+ signals. Trends Cell Biol. 14, Huber, A.B., Kolodkin, A.L., Ginty, D.D., and Cloutier, J.F. (2003). Signaling at the growth cone: ligand-receptor complexes and the
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Cdh1-APC Controls Axonal Growth and Patterning in the Mammalian Brain
 Cdh1-APC Controls Axonal Growth and Patterning in the Mammalian Brain Yoshiyuki Konishi, Judith Stegmüller, Takahiko Matsuda, Shirin Bonni, Azad Bonni* *Corresponding author: azad_bonni@hms.harvard.edu
Cdh1-APC Controls Axonal Growth and Patterning in the Mammalian Brain Yoshiyuki Konishi, Judith Stegmüller, Takahiko Matsuda, Shirin Bonni, Azad Bonni* *Corresponding author: azad_bonni@hms.harvard.edu
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat
 Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/323/5911/256/dc1 Supporting Online Material for HDAC4 Regulates Neuronal Survival in Normal and Diseased Retinas Bo Chen and Constance L. Cepko E-mail: bochen@genetics.med.harvard.edu,
www.sciencemag.org/cgi/content/full/323/5911/256/dc1 Supporting Online Material for HDAC4 Regulates Neuronal Survival in Normal and Diseased Retinas Bo Chen and Constance L. Cepko E-mail: bochen@genetics.med.harvard.edu,
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of
 Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
SUPPLEMENTARY NOTE 3. Supplememtary Note 3, Wehr et al., Monitoring Regulated Protein-Protein Interactions Using Split-TEV 1
 SUPPLEMENTARY NOTE 3 Fluorescent Proteolysis-only TEV-Reporters We generated TEV reporter that allow visualizing TEV activity at the membrane and in the cytosol of living cells not relying on the addition
SUPPLEMENTARY NOTE 3 Fluorescent Proteolysis-only TEV-Reporters We generated TEV reporter that allow visualizing TEV activity at the membrane and in the cytosol of living cells not relying on the addition
Flag-Rac Vector V12 V12 N17 C40. Vector C40 pakt (T308) Akt1. Myc-DN-PAK1 (N-SP)
 a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking
 Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Input. Pulldown GST. GST-Ahi1
 A kd 250 150 100 75 50 37 25 -GST-Ahi1 +GST-Ahi1 kd 148 98 64 50 37 22 GST GST-Ahi1 C kd 250 148 98 64 50 Control Input Hap1A Hap1 Pulldown GST GST-Ahi1 Hap1A Hap1 Hap1A Hap1 Figure S1. (A) Ahi1 western
A kd 250 150 100 75 50 37 25 -GST-Ahi1 +GST-Ahi1 kd 148 98 64 50 37 22 GST GST-Ahi1 C kd 250 148 98 64 50 Control Input Hap1A Hap1 Pulldown GST GST-Ahi1 Hap1A Hap1 Hap1A Hap1 Figure S1. (A) Ahi1 western
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression
 Supplemental figures Supplemental Figure. 1. Silencing expression of Celsr3 by shrna. To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression plasmids for the shrna
Supplemental figures Supplemental Figure. 1. Silencing expression of Celsr3 by shrna. To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression plasmids for the shrna
Supplementary Figure 1. Drawing of spinal cord open-book preparations and DiI tracing. Nature Neuroscience: doi: /nn.3893
 Supplementary Figure 1 Drawing of spinal cord open-book preparations and DiI tracing. Supplementary Figure 2 In ovo electroporation of dominant-negative PlexinA1 in commissural neurons induces midline
Supplementary Figure 1 Drawing of spinal cord open-book preparations and DiI tracing. Supplementary Figure 2 In ovo electroporation of dominant-negative PlexinA1 in commissural neurons induces midline
DCLK-immunopositive. Bars, 100 µm for B, 50 µm for C.
 Supplementary Figure S1. Characterization of rabbit polyclonal anti-dclk antibody. (A) Immunoblotting of COS7 cells transfected with DCLK1-GFP and DCLK2-GFP expression plasmids probed with anti-dclk antibody
Supplementary Figure S1. Characterization of rabbit polyclonal anti-dclk antibody. (A) Immunoblotting of COS7 cells transfected with DCLK1-GFP and DCLK2-GFP expression plasmids probed with anti-dclk antibody
PIASx Is a MEF2 SUMO E3 Ligase That Promotes Postsynaptic Dendritic Morphogenesis
 The Journal of Neuroscience, September 12, 2007 27(37):10037 10046 10037 Development/Plasticity/Repair PIASx Is a MEF2 SUMO E3 Ligase That Promotes Postsynaptic Dendritic Morphogenesis Aryaman Shalizi,
The Journal of Neuroscience, September 12, 2007 27(37):10037 10046 10037 Development/Plasticity/Repair PIASx Is a MEF2 SUMO E3 Ligase That Promotes Postsynaptic Dendritic Morphogenesis Aryaman Shalizi,
Supplementary Fig. 1 Kinetics of appearence of the faster migrating form of Bcl-10.
 α-cd3 + α-cd28: Time (min): + + + + + + + + + 0 5 15 30 60 120 180 240 300 360 360 n.s. Supplementary Fig. 1 Kinetics of appearence of the faster migrating form of. Immunoblot of lysates from Jurkat cells
α-cd3 + α-cd28: Time (min): + + + + + + + + + 0 5 15 30 60 120 180 240 300 360 360 n.s. Supplementary Fig. 1 Kinetics of appearence of the faster migrating form of. Immunoblot of lysates from Jurkat cells
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Methanol fixation allows better visualization of Kal7. To compare methods for
 Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
A Cdh1-APC/FMRP Ubiquitin Signaling Link Drives mglur-dependent Synaptic Plasticity in the Mammalian Brain
 Article A Cdh1-APC/FMRP Ubiquitin Signaling Link Drives mglur-dependent Synaptic Plasticity in the Mammalian Brain Highlights d Hippocampal mglur-ltd is markedly impaired in conditional Cdh1 knockout mice
Article A Cdh1-APC/FMRP Ubiquitin Signaling Link Drives mglur-dependent Synaptic Plasticity in the Mammalian Brain Highlights d Hippocampal mglur-ltd is markedly impaired in conditional Cdh1 knockout mice
A Cdh1-APC/FMRP Ubiquitin Signaling Link Drives mglur-dependent Synaptic Plasticity in the Mammalian Brain
 Article A Cdh1-APC/FMRP Ubiquitin Signaling Link Drives mglur-dependent Synaptic Plasticity in the Mammalian Brain Highlights d Hippocampal mglur-ltd is markedly impaired in conditional Cdh1 knockout mice
Article A Cdh1-APC/FMRP Ubiquitin Signaling Link Drives mglur-dependent Synaptic Plasticity in the Mammalian Brain Highlights d Hippocampal mglur-ltd is markedly impaired in conditional Cdh1 knockout mice
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
b alternative classical none
 Supplementary Figure. 1: Related to Figure.1 a d e b alternative classical none NIK P-IkBa Total IkBa Tubulin P52 (Lighter) P52 (Darker) RelB (Lighter) RelB (Darker) HDAC1 Control-Sh RelB-Sh NF-kB2-Sh
Supplementary Figure. 1: Related to Figure.1 a d e b alternative classical none NIK P-IkBa Total IkBa Tubulin P52 (Lighter) P52 (Darker) RelB (Lighter) RelB (Darker) HDAC1 Control-Sh RelB-Sh NF-kB2-Sh
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates
 Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates vascular calcification (VC). (a) Von Kossa staining shows that TSA potentiated the Pi-induced VC. Scale bar, 100
Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates vascular calcification (VC). (a) Von Kossa staining shows that TSA potentiated the Pi-induced VC. Scale bar, 100
Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions.
 Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions. 1 (a) Etoposide treatment gradually changes acetylation level and co-chaperone
Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions. 1 (a) Etoposide treatment gradually changes acetylation level and co-chaperone
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various
 Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated
 Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Dendritic targeting of mrna
 Dendritic targeting of mrna FMRP-dependent mglur-induced dendritic targeting of MAP1b and CaMKII mrna IMP-dependent NMDAR-induced dendritic targeting of β-action mrna Discussion (12/15) RNA and protein
Dendritic targeting of mrna FMRP-dependent mglur-induced dendritic targeting of MAP1b and CaMKII mrna IMP-dependent NMDAR-induced dendritic targeting of β-action mrna Discussion (12/15) RNA and protein
Supporting Information
 Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc RGLG Hormonal treatment H2O B RGLG µm ABA µm ACC µm GA Time (hours) µm µm MJ µm IA
 Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplemental Table 1: Sequences of real time PCR primers. Primers were intronspanning
 Symbol Accession Number Sense-primer (5-3 ) Antisense-primer (5-3 ) T a C ACTB NM_001101.3 CCAGAGGCGTACAGGGATAG CCAACCGCGAGAAGATGA 57 HSD3B2 NM_000198.3 CTTGGACAAGGCCTTCAGAC TCAAGTACAGTCAGCTTGGTCCT 60
Symbol Accession Number Sense-primer (5-3 ) Antisense-primer (5-3 ) T a C ACTB NM_001101.3 CCAGAGGCGTACAGGGATAG CCAACCGCGAGAAGATGA 57 HSD3B2 NM_000198.3 CTTGGACAAGGCCTTCAGAC TCAAGTACAGTCAGCTTGGTCCT 60
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Transcriptional regulation of BRCA1 expression by a metabolic switch: Di, Fernandez, De Siervi, Longo, and Gardner. H3K4Me3
 ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators.
 Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Name AP Biology Mrs. Laux Take home test #11 on Chapters 14, 15, and 17 DUE: MONDAY, DECEMBER 21, 2009
 MULTIPLE CHOICE QUESTIONS 1. Inducible genes are usually actively transcribed when: A. the molecule degraded by the enzyme(s) is present in the cell. B. repressor molecules bind to the promoter. C. lactose
MULTIPLE CHOICE QUESTIONS 1. Inducible genes are usually actively transcribed when: A. the molecule degraded by the enzyme(s) is present in the cell. B. repressor molecules bind to the promoter. C. lactose
7.06 Problem Set #3, 2006
 7.06 Problem Set #3, 2006 1. You are studying the EGF/Ras/MAPK pathway in cultured cells. When the pathway is activated, cells are signaled to proliferate. You generate various mutants described below.
7.06 Problem Set #3, 2006 1. You are studying the EGF/Ras/MAPK pathway in cultured cells. When the pathway is activated, cells are signaled to proliferate. You generate various mutants described below.
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/4/167/ra20/dc1 Supplementary Materials for Poly(ADP-Ribose) (PAR) Binding to Apoptosis-Inducing Factor Is Critical for PAR Polymerase-1 Dependent Cell Death (Parthanatos)
www.sciencesignaling.org/cgi/content/full/4/167/ra20/dc1 Supplementary Materials for Poly(ADP-Ribose) (PAR) Binding to Apoptosis-Inducing Factor Is Critical for PAR Polymerase-1 Dependent Cell Death (Parthanatos)
Supplementary Information
 Supplementary Information Selective control of inhibitory synapse development by Slitrk3-PTPδ trans-synaptic interaction Hideto Takahashi 1, Kei-ichi Katayama 2, Kazuhiro Sohya 3,4, Hiroyuki Miyamoto 4,5,
Supplementary Information Selective control of inhibitory synapse development by Slitrk3-PTPδ trans-synaptic interaction Hideto Takahashi 1, Kei-ichi Katayama 2, Kazuhiro Sohya 3,4, Hiroyuki Miyamoto 4,5,
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
MeCP2. MeCP2/α-tubulin. GFP mir1-1 mir132
 Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in
 Supplementary information Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer s disease Zhentao Zhang, Mingke Song, Xia Liu, Seong Su Kang, Il-Sun Kwon, Duc
Supplementary information Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer s disease Zhentao Zhang, Mingke Song, Xia Liu, Seong Su Kang, Il-Sun Kwon, Duc
Nature Medicine doi: /nm.2558
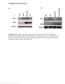 Supplementary. Fig. 1. (a) Sirt1 and mutant HTT (detected by HTT 81-90 antibody) protein levels were detected by Western blotting in cerebral cortex of N171-82Q mice. (b) Sirt1 and mutant HTT (detected
Supplementary. Fig. 1. (a) Sirt1 and mutant HTT (detected by HTT 81-90 antibody) protein levels were detected by Western blotting in cerebral cortex of N171-82Q mice. (b) Sirt1 and mutant HTT (detected
LiveReceptor AMPAR <Endogenous AMPAR Labeling Reagent>
 9-7 Hongo 2-Chome, Bunkyo-Ku Tokyo 113-0033, Japan LiveReceptor AMPAR Product Background Neurotransmitter receptors including glutamate receptors and GABA receptors
9-7 Hongo 2-Chome, Bunkyo-Ku Tokyo 113-0033, Japan LiveReceptor AMPAR Product Background Neurotransmitter receptors including glutamate receptors and GABA receptors
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Figure S1. USP-46 is expressed in several tissues including the nervous system
 Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
Supplementary Fig.1 Luton
 Supplementary Fig.1 Luton a 175 Brain Thymus Spleen Small Intestine Kidney Testis HeLa b 250 Lung Kidney MDCK c EFA6B si Control si Mismatch #637 #1564 #1770 83 62 47.5 175 IB: anti-efa6b #B1 130 66 Lysates
Supplementary Fig.1 Luton a 175 Brain Thymus Spleen Small Intestine Kidney Testis HeLa b 250 Lung Kidney MDCK c EFA6B si Control si Mismatch #637 #1564 #1770 83 62 47.5 175 IB: anti-efa6b #B1 130 66 Lysates
pdsipher and pdsipher -GFP shrna Vector User s Guide
 pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide,
 Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
A Survey of Genetic Methods
 IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
Supplemental Table S1. RT-PCR primers used in this study
 Supplemental Table S1. RT-PCR primers used in this study -----------------------------------------------------------------------------------------------------------------------------------------------
Supplemental Table S1. RT-PCR primers used in this study -----------------------------------------------------------------------------------------------------------------------------------------------
-RT RT RT -RT XBMPRII
 Base pairs 2000 1650 1000 850 600 500 400 Marker Primer set 1 Primer set 2 -RT RT RT -RT XBMPRII kda 100 75 50 37 25 20 XAC p-xac A 300 B Supplemental figure 1. PCR analysis of BMPRII expression and western
Base pairs 2000 1650 1000 850 600 500 400 Marker Primer set 1 Primer set 2 -RT RT RT -RT XBMPRII kda 100 75 50 37 25 20 XAC p-xac A 300 B Supplemental figure 1. PCR analysis of BMPRII expression and western
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
Supplementary Methods Plasmid constructs
 Supplementary Methods Plasmid constructs. Mouse cdna encoding SHP-1, amplified from mrna of RAW264.7 macrophages with primer 5'cgtgcctgcccagacaaactgt3' and 5'cggaattcagacgaatgcccagatcacttcc3', was cloned
Supplementary Methods Plasmid constructs. Mouse cdna encoding SHP-1, amplified from mrna of RAW264.7 macrophages with primer 5'cgtgcctgcccagacaaactgt3' and 5'cggaattcagacgaatgcccagatcacttcc3', was cloned
Xu et al., Supplementary Figures 1-7
 Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
isolated from ctr and pictreated mice. Activation of effector CD4 +
 Supplementary Figure 1 Bystander inflammation conditioned T reg cells have normal functional suppressive activity and ex vivo phenotype. WT Balb/c mice were treated with polyi:c (pic) or PBS (ctr) via
Supplementary Figure 1 Bystander inflammation conditioned T reg cells have normal functional suppressive activity and ex vivo phenotype. WT Balb/c mice were treated with polyi:c (pic) or PBS (ctr) via
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
Supplemental Figure 1. HepG2 cells were transfected with GLI luciferase reporter construct
 Supplemental Figure 1. HepG2 cells were transfected with GLI luciferase reporter construct (pgl38xgli), EWS-FLI1 luciferase reporter construct (NROB1-Luc) with or without GLI1, EWS- FLI1 and cdnas respectively.
Supplemental Figure 1. HepG2 cells were transfected with GLI luciferase reporter construct (pgl38xgli), EWS-FLI1 luciferase reporter construct (NROB1-Luc) with or without GLI1, EWS- FLI1 and cdnas respectively.
Data Sheet. Application Monitor glucocorticoid signaling pathway activity. Screen activators or inhibitors of the glucocorticoid signaling pathway.
 Data Sheet GAL4 Reporter Kit (Glucocorticoid Receptor Pathway) Catalog #: w70533 Background The glucocorticoid signaling pathway plays an important role in development, fluid homeostasis, cognition, immune
Data Sheet GAL4 Reporter Kit (Glucocorticoid Receptor Pathway) Catalog #: w70533 Background The glucocorticoid signaling pathway plays an important role in development, fluid homeostasis, cognition, immune
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/310/5763/1012/dc1 Supporting Online Material for A Calcium-Regulated MEF2 Sumoylation Switch Controls Postsynaptic Differentiation Aryaman Shalizi, Brice Gaudillière,
www.sciencemag.org/cgi/content/full/310/5763/1012/dc1 Supporting Online Material for A Calcium-Regulated MEF2 Sumoylation Switch Controls Postsynaptic Differentiation Aryaman Shalizi, Brice Gaudillière,
TOOLS sirna and mirna. User guide
 TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex
 POSTER PRESENTATION Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex Hai-Chon Lee *, Je-In Youn, Kyungwha Lee, Hwanyul Yong, Seung-Yong
POSTER PRESENTATION Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex Hai-Chon Lee *, Je-In Youn, Kyungwha Lee, Hwanyul Yong, Seung-Yong
Supplementary Figure 1. GST pull-down analysis of the interaction of GST-cIAP1 (A, B), GSTcIAP1
 Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
Supplement Figure S1:
 Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
Supplementary Information for. Regulation of Rev1 by the Fanconi Anemia Core Complex
 Supplementary Information for Regulation of Rev1 by the Fanconi Anemia Core Complex Hyungjin Kim, Kailin Yang, Donniphat Dejsuphong, Alan D. D Andrea* *Corresponding Author: Alan D. D Andrea, M.D. Alan_dandrea@dfci.harvard.edu
Supplementary Information for Regulation of Rev1 by the Fanconi Anemia Core Complex Hyungjin Kim, Kailin Yang, Donniphat Dejsuphong, Alan D. D Andrea* *Corresponding Author: Alan D. D Andrea, M.D. Alan_dandrea@dfci.harvard.edu
by Neurobasal medium (supplemented with B27, 0.5mM glutamine, and 100 U/mL
 Supplementary Materials and methods Neuronal cultures and transfection The hippocampus was dissected from E8 rat embryos, dissociated, and neurons plated onto glass coverslips coated with poly-ornithine
Supplementary Materials and methods Neuronal cultures and transfection The hippocampus was dissected from E8 rat embryos, dissociated, and neurons plated onto glass coverslips coated with poly-ornithine
Regulation of axonal and dendritic growth by the extracellular calcium-sensing
 Regulation of axonal and dendritic growth by the extracellular calcium-sensing receptor (CaSR). Thomas N. Vizard, Gerard W. O Keeffe, Humberto Gutierrez, Claudine H. Kos, Daniela Riccardi, Alun M. Davies
Regulation of axonal and dendritic growth by the extracellular calcium-sensing receptor (CaSR). Thomas N. Vizard, Gerard W. O Keeffe, Humberto Gutierrez, Claudine H. Kos, Daniela Riccardi, Alun M. Davies
T H E J O U R N A L O F C E L L B I O L O G Y
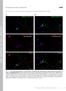 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kanaani et al., http://www.jcb.org/cgi/content/full/jcb.200912101/dc1 Figure S1. The K2 rabbit polyclonal antibody is specific for GAD67,
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kanaani et al., http://www.jcb.org/cgi/content/full/jcb.200912101/dc1 Figure S1. The K2 rabbit polyclonal antibody is specific for GAD67,
7.06 Cell Biology EXAM #2 March 20, 2003
 7.06 Cell Biology EXAM #2 March 20, 2003 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #2 March 20, 2003 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification.
 Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Nature Structural & Molecular Biology: doi: /nsmb.1583
 Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Fig Hypoxia causes growth retardation and developmental delay. (A) Morphology of wildtype zebrafish embryos at 48 hours post fertilization
 FIGURES AND TABLES Fig. 1-1. Hypoxia causes growth retardation and developmental delay. (A) Morphology of wildtype zebrafish embryos at 48 hours post fertilization (hpf) after 24 h of normoxia or hypoxia
FIGURES AND TABLES Fig. 1-1. Hypoxia causes growth retardation and developmental delay. (A) Morphology of wildtype zebrafish embryos at 48 hours post fertilization (hpf) after 24 h of normoxia or hypoxia
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Characterization of a novel gene involved in border cell migration
 Characterization of a novel gene involved in border cell migration Wei-Hao Li, He-Yen Chou and Li-Mei Pai Graduate Institute of Basic Medical Science Chang-Gung University, Tao-Yuan, Taiwan, R.O.C. Abstract
Characterization of a novel gene involved in border cell migration Wei-Hao Li, He-Yen Chou and Li-Mei Pai Graduate Institute of Basic Medical Science Chang-Gung University, Tao-Yuan, Taiwan, R.O.C. Abstract
7.06 Problem Set #3, Spring 2005
 7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Two classes of silencing RNAs move between Caenorhabditis elegans tissues.
 Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Learning Objectives. Define RNA interference. Define basic terminology. Describe molecular mechanism. Define VSP and relevance
 Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
