Supporting Information
|
|
|
- Sheryl Higgins
- 5 years ago
- Views:
Transcription
1 Supporting Information Lee et al /pnas SI Experimental Procedures Plasmid and Strain Construction. (p)odr-3::gfp::egl-4 was constructed by excising the coding region of GFP and the first few codons of EGL-4 from (p)egl-4::gfp::egl-4 (M. Fujiwara, Hakozaki, Fukuoka, Japan) (1) and placing this downstream of (p) odr-3 in the (p)odr-3::egl-4 vector (2) using MscI and BglII restriction sites. (p)odr-3::gfp::egl-4 was coinjected (50 ng/μl) with the AWC marker (p)odr-1::dsred (25 ng/μl) and coelomocyte marker (P. Sengupta, Waltham, MA) (p)ofm-1::gfp (25 ng/μl) into N2 animals, and the subsequent transgenic line was integrated by trimethylpsoralen (TMP) (3). The transgeneic line with 50% transmission of the transgene is exposed to TMP and UV, and the F2s are cloned out as described in Yandell et al. (3); however, the integrants are screened for 100% transmission of the coinjection marker. Integration events were mapped to chromosomes, and one on chromosome V was chosen. This was outcrossed to N2 through 10 generations to rid the genome of TMPinduced mutations. This strain is called pyis500[(p)odr-3::gfp:: EGL-4]. (p)odr-3::gfp::egl-4(δnls) was constructed by removing the original nuclear localization signal (NLS) with the flanking restriction sites BglII and AflII and replacing it with ΔNLS coding sequences from ref. 2. NLS::GFP::EGL-4 was constructed by site-directed mutagenesis (QuikChange; Stratagene) to place the Sv40 NLS (CCAAAGAAGAAGCGTAAGGTA) into (p)odr- 3::GFP::EGL-4 at the N terminus of the GFP-coding region. This was injected at 25 ng/μl. (p)hsp-16.2::nls:: EGL-4 was constructed by site-directed mutagenesis (QuikChange; Stratagene) to place the Sv40 NLS right after the start codon of EGL-4 in (p) hsp-16.2::egl-4 from ref. 2. The heat-shock promoter came from ppd49.78 (gift of A. Fire, Stanford, CA). (p)hsp-16.2::nls:: EGL- 4 was coinjected (5 ng/μl) with (p)str-2::dsred (100 ng/μl) and (p)ofm-1::gfp (100 ng/μl) GFP::EGL-4. GFP::EGL-4(T276A and T398A) and GFP::EGL-4(D611N) mutants were constructed by site-directed mutagenesis with the following primers: GCA- CAAGAGCCGCATCAGTTCAA, AATCAACTCAAGACAA GTGAAAGTC, and ACTGGATATCTGAAACTTGTGAATT TCGGATTCGCAAAGAAAC, respectively. GFP::EGL-4 T276A;T398A was constructed by site-directed mutagenesis of T276A followed by site-directed mutagenesis of T398A. Each construct was injected into egl-4(n479) at 25 ng/μl with (p)ofm-1:: GFP (25 ng/μl) as the coinjection marker. Lines were selected for equivalent expression levels of GFP::EGL-4. For analysis of behavior, we used a line that rescued a behavioral phenotype of egl-4 (n479). Typically, one in three lines expressing the EGL-4 cdna rescued adaptation or butanone chemotaxis of the egl-4 mutant. We obtained enough lines to find three rescuing lines, and we used the line with the average behavioral responses for further analysis. In cases where we failed to see rescue of an EGL-4 phenotype or when we were examining a transgene in the wild-type genetic background, we generally had at least six lines; we chose one with an average phenotype and one that did not show dominant effects. Behavioral Assays. Odors (Sigma) were diluted in EtOH as follows: 1:200 benzaldehyde (bz), 1:1,000 butanone, 1:100 isoamyl alcohol, and 1:1,000 diacetyl. Odor adaptations were performed either in liquid or on plates as described (2, 4). Adaptation was performed by diluting odors in the following way: 9 μl bz, 14 μl butanone, and 4 μl of diacetyl were diluted into 100 ml S-basal buffer, and populations of animals were exposed to the diluted odor for 30 min for short-term exposure or 80 min for long-term exposure. These treated populations were then subjected to chemotaxis assays. For isoamyl alcohol, animals were exposed to 12 μl isoamyl alcohol on agar plugs placed on the lid of a chemotaxis assay plate. Recovery from adaptation was performed in S-basal buffer at room temperature, and the length of recovery time was chosen based on initial experiments, which showed that the shortest time in which we could observe reliable recovery was 60 min for the 30-min exposed animals. Because we failed to observe recovery in the 80-min exposed populations, we allowed them to recover as long as possible before the buffer-treated controls showed a decrease in chemotaxis that we attributed to starvation. Butanone repulsion was carried out as described in ref. 5. Each experiment was conducted on at least 3 separate days with >50 (usually 100) animals per assay. Transgenic extrachromosomal arrays carrying lines were scored on the fluorescence-dissecting scope using coelomocyte GFP as a marker for transgeneic individuals. The bars represent the means of all assays, and the error bars represent the SEM. Microscopic Assessment of GFP EGL-4 Subcellular Localization. To assess GFP EGL-4 localization, four to five L4 animals were placed onto a 9-cm OP50 seeded plate and incubated for 5 days at 22; their adult progeny were collected by washing plates with approximately 1 ml of S-basal buffer. Animals were washed 3 times in dh 2 0, exposed to odor or S-basal basal for 80, and then, placed onto a 1% agar pad containing 5 mm sodium azide. Twenty animals were observed at 63 times per experiment (Ziess Axioskop II), and each experiment was performed at least three times for each data point. The criteria for scoring GFP EGL-4 as nuclear were if the outline of the AWC nucleus was clearly visible and brighter than the cell body (similar to AWB); they were cytoplasmic if the nucleus was not visible. In ambiguous cases, the first ambiguous animal was scored as cytoplasmic and the next as nuclear etc. Double-blind experiments gave similar results. For butanone experiments, both AWC neurons were examined for each animal. For the AWC ON versus AWC OFF experiments, both AWC nuclei were examined, and the pstr-2:: dsred-expressing cell was assigned the AWC ON identity. The bars represent the mean values of the percentage of each population that exhibited nuclear GFP EGL-4 on each day, whereas the error bars represent the standard error of the mean. 1. Fujiwara M, Sengupta P, McIntire SL (2002) Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cgmp-dependent protein kinase. Neuron 36: L Etoile ND, et al. (2002) The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron 36: Yandell MD, Edgar LG, Wood WB (1994) Trimethylpsoralen induces small deletion mutations in Caenorhabditis elegans. Proc Natl Acad Sci USA 91: Colbert HA, Bargmann CI (1995) Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron 14: Tsunozaki M, Chalasani SH, Bargmann CI (2008) A behavioral switch: cgmp and PKC signaling in olfactory neurons reverses odor preference in C. elegans. Neuron 59: of5
2 Fig. S1. Immunologic characterization of endogenous EGL-4. (A) Western blot of 100 μl of either wild-type (N2, first two lanes) or egl-4(n479) mutant animals boiled for 30 min in Lamelli sample buffer and resolved on an 8% polyacrylamide gel with SDS. The first lane was incubated with preimmune serum, and the subsequent lanes were incubated with serum from a rabbit that had been exposed to a fragment of EGL-4. The left arrow indicates the mobility of full-length EGL-4, which is close to its predicted molecular weight of 87 kda. (B D) Fluorescence micrographs of the anterior ganglion of a wild-type animal that expressed GFP under the str-2 promoter to outline the AWC neuron are shown. The arrowhead points to the AWC neuron, the red line in C outlines the cell body, and the white indicates immunoreactivity with the same antiserum used in A above. Similarly treated egl-4(n479) animals showed no staining using this serum and these procedures (1). (E) Benzaldehyde exposure results in nuclear endogenous EGL-4. Animals were exposed to buffer (Upper) or benzaldehyde (Lower) for 80 min before fixing and staining (2), anti EGL-4 antibodies were followed by DAPI and rhodamine-conjugated secondary antibodies. The AWC cell body was visualized by pstr-2::gfp and outlined in green, whereas the nucleus, outlined in red, was visualized by DAPI. The staining seen in E is rhodamine. All eight benzaldehyde-exposed animals exhibited nuclear EGL-4 staining as seen in Upper, whereas 10 of 12 naïve animals showed the cytoplasmic EGL-4 seen in Lower. 1. Eastham-Anderson JR (2004) Nuclear translocation of EGL-4, a cgmp dependant (i.e. dependent) protein kinase, in response to odorant adaptation in C. elegans. Master s thesis (University of California, Davis, CA). 2. Nonet ML, Grundahl K, Meyer BJ, Rand JB (1993) Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell 73: Fig. S2. GFP EGL-4 and NLS-GFP EGL-4 are expressed at equivalent levels; 10 μl of packed worms were lysed and run on a 12% polyacrylamide gel. Proteins were transferred to nitrocellulaose and blotted with anti-gfp antibodies. GFP-tagged EGL-4 ran at the expected molecular weight of 114 kda, and soluble GFP was used as a loading control, because each population of transgeneic animals expressed GFP under the ofm-1 promoter as a coinjection marker. Thus, soluble GFP is used to normalize between populations. N2 animals expressing just the coinjection marker reveal the specificity of the anti-gfp antibody. 2of5
3 Fig. S3. Prolonged benzaldehyde exposure can cross-adapt the isoamyl-alcohol response in well-fed animals. (Upper) Exposure of fed wild-type animals expressing podr-3::gfp EGL-4 from an extrachromosomal array to buffer plus benzaldehyde for 80 min (dark bars) or buffer alone (light bars) are shown. The bars represent the mean percentage of animals with nuclear GFP EGL-4 in both AWCs over 3 separate days. On each day, >20 animals were scored. Error is SEM. (Lower) The same populations of animals were subjected to chemotaxis assays toward either benzaldehyde or isoamyl alcohol. The bars represent the mean chemotaxis index (CI) over 3 days, whereas the error is the standard error of the mean. 3of5
4 Fig. S4. Expression from the fos1a promoter was not appreciably affected by odor exposure. (Left) A micrograph of a naïve worm expressing cyan fluorescent protein (CFP) from the fos-1a promoter. (Right) The same strain of worm that had been exposed to benzaldehyde for 80 min. This was typical of >20 worms for each condition benzaldehyde chemotaxis index pstr-2 ::luciferase /unit DNA naive bz exposed naive bz exposed Fig. S5. Dynamic changes in str-2 expression cannot be seen during adaptation using a luciferase reporter. The destabilized luciferase reporter (1) was placed under the control of the str-2 promoter and the unc-54 3 UTR. Units of luciferase were assessed using standard luminometry and Promega luciferase assay kit. These values were normalized to transgene number through qpcr. Samples were obtained from naïve versus benzaldehyde exposed animals, and CIs are presented at the side. 1. Kaye JA, Rose NC, Goldsworthy B, Goga A, L Etoile ND (2009) A 3 UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron 61: of5
5 Table S1. str-2 expression in AWC neurons as a function of EGL-4 status Animals (percent) Genotype Heat 0 AWC pstr-2 ON 1 AWC pstr-2 ON 2 AWC pstr-2 ON n Wild type egl-4(loss of function)* egl-4(null); ex(podr-3::δnls EGL-4) egl-4(null); ex(phsp-16.2::nls EGL-4) Adult animals grown at 20 C were examined for pstr-2::rfp [in wild-type and egl-4(ks60) animals] or pstr-2:: DsRed [in wild-type and egl-4(n479) animals] in the AWC neurons. *The egl-4(loss of function) allele is ks60. The egl-4(null) allele was n479, and it was transgenic for the odr-3 promoter driving ΔNLS EGL-4. The egl-4(n479) was transgenic for the heat-inducible promoter driving NLS EGL-4. Animals were heat-treated for 2 h at 33 C and analyzed after min of recovery at room temperature. P < P = P = The P values for the significance of the difference between naïve and heat-treated animals were determined using a two-sample t test between proportions. 5of5
mod-1::mcherry unc-47::gfp RME ser-4::gfp vm2 vm2 vm2 vm2 VNC
 A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
Mos1 insertion. MosTIC protocol-11/2006. repair template - Mos1 transposase expression - Mos1 excision - DSB formation. homolog arm.
 MosTIC (Mos1 excision induced Transgene Instructed gene Conversion) Valérie Robert (vrobert@biologie.ens.fr) and Jean-Louis Bessereau (jlbesse@biologie.ens.fr) (November 2006) Introduction: MosTIC (Robert
MosTIC (Mos1 excision induced Transgene Instructed gene Conversion) Valérie Robert (vrobert@biologie.ens.fr) and Jean-Louis Bessereau (jlbesse@biologie.ens.fr) (November 2006) Introduction: MosTIC (Robert
Two classes of silencing RNAs move between Caenorhabditis elegans tissues.
 Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7866 1.8 Fraction Bordering Fraction Aggregating.6.4.2 1.8.6.4.2 Promoter Head neurons expressing npr-1 cdna Genotype WT - - URX AQR ALN AVM gcy-35 sax-7 gcy-32+ gpa-3 RMG ILs URX AQR
doi: 1.138/nature7866 1.8 Fraction Bordering Fraction Aggregating.6.4.2 1.8.6.4.2 Promoter Head neurons expressing npr-1 cdna Genotype WT - - URX AQR ALN AVM gcy-35 sax-7 gcy-32+ gpa-3 RMG ILs URX AQR
Nature Structural & Molecular Biology: doi: /nsmb.3308
 Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Figure S1. USP-46 is expressed in several tissues including the nervous system
 Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Nature Methods: doi: /nmeth Supplementary Figure 1. Neither driver nor effector alone displays expression of GFP
 Supplementary Figure 1 Neither driver nor effector alone displays expression of GFP Comparison of GFP fluorescence in a driver- only strain (a) or an effector-only strain (b) to their driver + effector
Supplementary Figure 1 Neither driver nor effector alone displays expression of GFP Comparison of GFP fluorescence in a driver- only strain (a) or an effector-only strain (b) to their driver + effector
Supplementary information
 Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1119481/dc1 Supporting Online Material for LIN-12/Notch Activation Leads to MicroRNA-Mediated Down-Regulation of Vav in C. elegans Andrew S. Yoo and Iva Greenwald* *To
www.sciencemag.org/cgi/content/full/1119481/dc1 Supporting Online Material for LIN-12/Notch Activation Leads to MicroRNA-Mediated Down-Regulation of Vav in C. elegans Andrew S. Yoo and Iva Greenwald* *To
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Methods
 Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Figure S1a. The modular dcas9 fusion system works efficiently to suppress and activate endogenous gene expression in C. elegans
 Figure S1a. The modular dcas9 fusion system works efficiently to suppress and activate endogenous gene expression in C. elegans A, a normal wild-type hermaphrodite N2 is shown. B, a dpy-5-supressed F1
Figure S1a. The modular dcas9 fusion system works efficiently to suppress and activate endogenous gene expression in C. elegans A, a normal wild-type hermaphrodite N2 is shown. B, a dpy-5-supressed F1
Construct Design and Cloning Guide for Cas9-triggered homologous recombination
 Construct Design and Cloning Guide for Cas9-triggered homologous recombination Written by Dan Dickinson (ddickins@live.unc.edu) and last updated December 2013. Reference: Dickinson DJ, Ward JD, Reiner
Construct Design and Cloning Guide for Cas9-triggered homologous recombination Written by Dan Dickinson (ddickins@live.unc.edu) and last updated December 2013. Reference: Dickinson DJ, Ward JD, Reiner
An antibiotic selection marker for nematode transgenesis
 nature methods An antibiotic selection marker for nematode transgenesis Rosina Giordano-Santini, Stuart Milstein, Nenad Svrzikapa, Domena Tu, Robert Johnsen, David Baillie, Marc Vidal & Denis Dupuy Supplementary
nature methods An antibiotic selection marker for nematode transgenesis Rosina Giordano-Santini, Stuart Milstein, Nenad Svrzikapa, Domena Tu, Robert Johnsen, David Baillie, Marc Vidal & Denis Dupuy Supplementary
unc-33/crmp and ankyrin organize microtubules and localize kinesin to polarize
 Supplementary Information unc-33/crmp and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting Tapan A. Maniar 1, Miriam Kaplan 1, George J. Wang 2, Kang Shen 2, Li Wei
Supplementary Information unc-33/crmp and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting Tapan A. Maniar 1, Miriam Kaplan 1, George J. Wang 2, Kang Shen 2, Li Wei
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
SUPPLEMENTARY INFORMATION
 Figure S1. lev-9 mutants are resistant to levamisole. The levamisole dose-response curves indicate that lev-9 mutants are partially resistant to levamisole similar to lev-10(kr26) mutants. unc-29(x29)
Figure S1. lev-9 mutants are resistant to levamisole. The levamisole dose-response curves indicate that lev-9 mutants are partially resistant to levamisole similar to lev-10(kr26) mutants. unc-29(x29)
Fig. S1. Phosphorylation of eif2 by PEK-1 is increased in ire-1 mutants. Fig. S2. ire-1
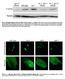 Fig. S1. Phosphorylation of eif2a by PEK-1 is increased in ire-1 mutants. Representative western blot of phosphorylated eif2a and tubulin of day-0 wild-type animals and ire-1 mutants treated with control,
Fig. S1. Phosphorylation of eif2a by PEK-1 is increased in ire-1 mutants. Representative western blot of phosphorylated eif2a and tubulin of day-0 wild-type animals and ire-1 mutants treated with control,
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Genome research in eukaryotes
 Functional Genomics Genome and EST sequencing can tell us how many POTENTIAL genes are present in the genome Proteomics can tell us about proteins and their interactions The goal of functional genomics
Functional Genomics Genome and EST sequencing can tell us how many POTENTIAL genes are present in the genome Proteomics can tell us about proteins and their interactions The goal of functional genomics
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA
 Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Caenorhabditis elegans: The Heavyweight Champ of Gene Knockout Technology
 Caenorhabditis elegans: The Heavyweight Champ of Gene Knockout Technology Rachel Pan The nematode (worm) Caenorhabditis elegans was the first multicellular organism to have its genome, or complete DNA
Caenorhabditis elegans: The Heavyweight Champ of Gene Knockout Technology Rachel Pan The nematode (worm) Caenorhabditis elegans was the first multicellular organism to have its genome, or complete DNA
Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions.
 Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions. 1 (a) Etoposide treatment gradually changes acetylation level and co-chaperone
Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions. 1 (a) Etoposide treatment gradually changes acetylation level and co-chaperone
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
SCREENING AND PRESERVATION OF DNA LIBRARIES
 MODULE 4 LECTURE 5 SCREENING AND PRESERVATION OF DNA LIBRARIES 4-5.1. Introduction Library screening is the process of identification of the clones carrying the gene of interest. Screening relies on a
MODULE 4 LECTURE 5 SCREENING AND PRESERVATION OF DNA LIBRARIES 4-5.1. Introduction Library screening is the process of identification of the clones carrying the gene of interest. Screening relies on a
Rapid selection of transgenic C. elegans using antibiotic resistance
 nature methods Rapid selection of transgenic C. elegans using antibiotic resistance Jennifer I Semple, Rosa Garcia-Verdugo & Ben Lehner Supplementary figures and text: Supplementary Figure 1 Supplementary
nature methods Rapid selection of transgenic C. elegans using antibiotic resistance Jennifer I Semple, Rosa Garcia-Verdugo & Ben Lehner Supplementary figures and text: Supplementary Figure 1 Supplementary
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supporting information
 Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Advanced Genetics. Why Study Genetics?
 Advanced Genetics Advanced Genetics Why Study Genetics? Why Study Genetics? 1. Historical and aesthetic appreciation Why Study Genetics? 1. Historical and aesthetic appreciation 2. Practical applications
Advanced Genetics Advanced Genetics Why Study Genetics? Why Study Genetics? 1. Historical and aesthetic appreciation Why Study Genetics? 1. Historical and aesthetic appreciation 2. Practical applications
GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts
 Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/323/5911/256/dc1 Supporting Online Material for HDAC4 Regulates Neuronal Survival in Normal and Diseased Retinas Bo Chen and Constance L. Cepko E-mail: bochen@genetics.med.harvard.edu,
www.sciencemag.org/cgi/content/full/323/5911/256/dc1 Supporting Online Material for HDAC4 Regulates Neuronal Survival in Normal and Diseased Retinas Bo Chen and Constance L. Cepko E-mail: bochen@genetics.med.harvard.edu,
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
b number of motif occurrences
 supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
Xu et al., Supplementary Figures 1-7
 Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
SUPPLEMENTARY INFORMATION. LIN-28 co-transcriptionally binds primary let-7 to regulate mirna maturation in C. elegans
 SUPPLEMENTARY INFORMATION LIN-28 co-transcriptionally binds primary let-7 to regulate mirna maturation in C. elegans Priscilla M. Van Wynsberghe 1, Zoya S. Kai 1, Katlin B. Massirer 2-4, Victoria H. Burton
SUPPLEMENTARY INFORMATION LIN-28 co-transcriptionally binds primary let-7 to regulate mirna maturation in C. elegans Priscilla M. Van Wynsberghe 1, Zoya S. Kai 1, Katlin B. Massirer 2-4, Victoria H. Burton
A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
Trasposable elements: Uses of P elements Problem set B at the end
 Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene. Andrew ElBardissi, The Pennsylvania State University
 Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
To investigate the heredity of the WFP gene, we selected plants that were homozygous
 Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplemental Materials and Methods. Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2,
 Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous
 Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Supplemental Information
 Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Partial list of differentially expressed genes from cdna microarray, comparing MUC18-
 Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplemental Data. Benstein et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Neuronal Activity and CaMKII Regulate Kinesin-Mediated Transport of Synaptic AMPARs
 Neuron Supplemental Information Neuronal Activity and CaMKII Regulate Kinesin-Mediated Transport of Synaptic AMPARs Frédéric J. Hoerndli, Rui Wang, Jerry E. Mellem, Angy Kallarackal, Penelope J. Brockie,
Neuron Supplemental Information Neuronal Activity and CaMKII Regulate Kinesin-Mediated Transport of Synaptic AMPARs Frédéric J. Hoerndli, Rui Wang, Jerry E. Mellem, Angy Kallarackal, Penelope J. Brockie,
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
SUPPLEMENTARY INFORMATION
 SULEMENTARY INFORMATION doi:10.1038/nature12117 Supplementary Discussion Although loss of leads to initiation of postembryonic development even under starvation conditions, ectopic overexpression of under
SULEMENTARY INFORMATION doi:10.1038/nature12117 Supplementary Discussion Although loss of leads to initiation of postembryonic development even under starvation conditions, ectopic overexpression of under
Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway
 Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway Pelin Yaşar, Gamze Ayaz and Mesut Muyan SUPPLEMENTARY INFORMATION
Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway Pelin Yaşar, Gamze Ayaz and Mesut Muyan SUPPLEMENTARY INFORMATION
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10928 Materials and Methods 1. Plant material and growth conditions. All plant lines used were in Col-0 background unless otherwise specified. pif4-101 mutant
SUPPLEMENTARY INFORMATION doi:10.1038/nature10928 Materials and Methods 1. Plant material and growth conditions. All plant lines used were in Col-0 background unless otherwise specified. pif4-101 mutant
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION 1 Supplementary Figure S1. Proportion of HEK293T transfectants showing fluorescent foci after heat sho Blinded slides were counted for the appearance of fluorescent foci of tagrfp-t
SUPPLEMENTARY INFORMATION 1 Supplementary Figure S1. Proportion of HEK293T transfectants showing fluorescent foci after heat sho Blinded slides were counted for the appearance of fluorescent foci of tagrfp-t
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
File S1. Detailed protocol for CRISPR/Cas9 mediated genome editing with dual marker cassettes
 File S1 Detailed protocol for CRISPR/Cas9 mediated genome editing with dual marker cassettes A) Preparing sgrna vectors We ve modified the strategy to create new sgrna vectors starting with the klp 12
File S1 Detailed protocol for CRISPR/Cas9 mediated genome editing with dual marker cassettes A) Preparing sgrna vectors We ve modified the strategy to create new sgrna vectors starting with the klp 12
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/323/5920/1500/dc1 Supporting Online Material for RSY-1 Is a Local Inhibitor of Presynaptic Assembly in C. elegans Maulik R. Patel and Kang Shen To whom correspondence
www.sciencemag.org/cgi/content/full/323/5920/1500/dc1 Supporting Online Material for RSY-1 Is a Local Inhibitor of Presynaptic Assembly in C. elegans Maulik R. Patel and Kang Shen To whom correspondence
Supporting Information
 Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
WesternMAX Alkaline Phosphatase Chemiluminescent Detection Kits
 WesternMAX Alkaline Phosphatase Chemiluminescent Detection Kits Code N221-KIT N220-KIT Description WesternMAX Chemiluminescent AP Kit, Anti-Mouse Includes: Alkaline Phosphatase (AP) Conjugated Anti-Mouse
WesternMAX Alkaline Phosphatase Chemiluminescent Detection Kits Code N221-KIT N220-KIT Description WesternMAX Chemiluminescent AP Kit, Anti-Mouse Includes: Alkaline Phosphatase (AP) Conjugated Anti-Mouse
Aminoacid change in chromophore. PIN3::PIN3-GFP GFP S65 (no change) (5.9) (Kneen et al., 1998)
 Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Supplemental Online Material. The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/-
 #1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
#1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental Data. Hu et al. Plant Cell (2017) /tpc
 1 2 3 4 Supplemental Figure 1. DNA gel blot analysis of homozygous transgenic plants. (Supports Figure 1.) 5 6 7 8 Rice genomic DNA was digested with the restriction enzymes EcoRⅠ and BamHⅠ. Lanes in the
1 2 3 4 Supplemental Figure 1. DNA gel blot analysis of homozygous transgenic plants. (Supports Figure 1.) 5 6 7 8 Rice genomic DNA was digested with the restriction enzymes EcoRⅠ and BamHⅠ. Lanes in the
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Firefly luciferase mutants as sensors of proteome stress
 Nature Methods Firefly luciferase mutants as sensors of proteome stress Rajat Gupta, Prasad Kasturi, Andreas Bracher, Christian Loew, Min Zheng, Adriana Villella, Dan Garza, F Ulrich Hartl & Swasti Raychaudhuri
Nature Methods Firefly luciferase mutants as sensors of proteome stress Rajat Gupta, Prasad Kasturi, Andreas Bracher, Christian Loew, Min Zheng, Adriana Villella, Dan Garza, F Ulrich Hartl & Swasti Raychaudhuri
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Nature Structural & Molecular Biology: doi: /nsmb.1583
 Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Color-Switch CRE recombinase stable cell line
 Color-Switch CRE recombinase stable cell line Catalog Number Product Name / Description Amount SC018-Bsd CRE reporter cell line (Bsd): HEK293-loxP-GFP- RFP (Bsd). RFP" cassette with blasticidin antibiotic
Color-Switch CRE recombinase stable cell line Catalog Number Product Name / Description Amount SC018-Bsd CRE reporter cell line (Bsd): HEK293-loxP-GFP- RFP (Bsd). RFP" cassette with blasticidin antibiotic
An N-terminal Region of Caenorhabditis elegans RGS Proteins EGL-10 and EAT-16 Directs Inhibition of G o Versus G q Signaling*
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 277, No. 49, Issue of December 6, pp. 47004 47013, 2002 2002 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. An N-terminal
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 277, No. 49, Issue of December 6, pp. 47004 47013, 2002 2002 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. An N-terminal
Protocols for cell lines using CRISPR/CAS
 Protocols for cell lines using CRISPR/CAS Procedure overview Map Preparation of CRISPR/CAS plasmids Expression vectors for guide RNA (U6-gRNA) and Cas9 gene (CMV-p-Cas9) are ampicillin-resist ant and stable
Protocols for cell lines using CRISPR/CAS Procedure overview Map Preparation of CRISPR/CAS plasmids Expression vectors for guide RNA (U6-gRNA) and Cas9 gene (CMV-p-Cas9) are ampicillin-resist ant and stable
Experimental Tools and Resources Available in Arabidopsis. Manish Raizada, University of Guelph, Canada
 Experimental Tools and Resources Available in Arabidopsis Manish Raizada, University of Guelph, Canada Community website: The Arabidopsis Information Resource (TAIR) at http://www.arabidopsis.org Can order
Experimental Tools and Resources Available in Arabidopsis Manish Raizada, University of Guelph, Canada Community website: The Arabidopsis Information Resource (TAIR) at http://www.arabidopsis.org Can order
Supporting Information
 Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Nature Medicine doi: /nm.2548
 Supplementary Table 1: Genotypes of offspring and embryos from matings of Pmm2 WT/F118L mice with Pmm2 WT/R137H mice total events Pmm2 WT/WT Pmm2 WT/R137H Pmm2 WT/F118L Pmm2 R137H/F118L offspring 117 (100%)
Supplementary Table 1: Genotypes of offspring and embryos from matings of Pmm2 WT/F118L mice with Pmm2 WT/R137H mice total events Pmm2 WT/WT Pmm2 WT/R137H Pmm2 WT/F118L Pmm2 R137H/F118L offspring 117 (100%)
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
H3K36me3 polyclonal antibody
 H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction
 Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Chapter 21. Acute Behavioral Responses to Pheromones in C. elegans (Adult Behaviors: Attraction, Repulsion) Heeun Jang and Cornelia I.
 Chapter 21 Acute Behavioral Responses to Pheromones in C. elegans (Adult Behaviors: Attraction, Repulsion) Abstract The pheromone drop test is a simple and robust behavioral assay to quantify acute avoidance
Chapter 21 Acute Behavioral Responses to Pheromones in C. elegans (Adult Behaviors: Attraction, Repulsion) Abstract The pheromone drop test is a simple and robust behavioral assay to quantify acute avoidance
