A Genome wide RNAi Screen. in Human Cells. Eric E. Zhang,1,2,5 Andrew C. Liu,1,3,5 Tsuyoshi Hirota,1,2,5 Loren J. Miraglia,1 Genevieve Welch,1
|
|
|
- Belinda Holland
- 6 years ago
- Views:
Transcription
1 A Genome wide RNAi Screen for Modifiers of the Circadian Clock in Human Cells Eric E. Zhang,1,2,5 Andrew C. Liu,1,3,5 Tsuyoshi Hirota,1,2,5 Loren J. Miraglia,1 Genevieve Welch,1 Pagkapol Y. Pongsawakul,2 Xianzhong Liu,1 Ann Atwood,2 Jon W. Huss III,1 Jeff Janes,1 Andrew I. Su,1 John B. Hogenesch,4,* and Steve A. Kay2,* Journal talk Marika Fava Blanca Iachia y Baca Circadian Clock Introduction suprachiasmatic nuclei (SCN) Figure 1. Control of circadian rhythms by specific brain regions.
2 Circadian Clock in Mammals RORE. NR1Ds and RORs either repress or activate gene transcription from ROR elements. D box binding elements. bzip, DBP, TEF either repress or activate gene transcription from D box. Figure 2. Schematic representation of the principal components of the Circadian Clock in Mammals Genes Function Knockout mice of Rev erbα, Rorα o Rorβ Lower amplitude rhythms Abnormal period lenghts interindividual variability in both phenotypes Knockout mice of Pgc1α Long period locomotor activity Dose dependent knockdown of ROR and REV ERB in vitro potent effect on the baseline and the amplitude of circadian gene expression
3 Complex Interaction Single cells Organotypic Whole SCN cultures organism Different results Intercellular coupling in generating response Materials U2SO cells Bmal1 dluc and Per2 dluc reporter cell lines sirna libraries for the primary and secondary screens were purchased from QIAGEN. Additional sirnas from Invitrogen and Dharmacon used as controls in target validation experiments. GL2 and GL3 sirnas from QIAGEN Screen controls purchased from Invitrogen included CRY1 HSS102308, CRY2 HSS102311, and BMAL1 or ARNTL HSS CRY1 and NR1D1 sirnas used in dose dependent experiment were previously described.
4 Overview of Web Based Data Resource in Circadian BioGPS Figure 3. Screening data displayed in BioGPS openaccess database Methods RNA interference Inverse transfection on microarray LumiCycle assay Q PCR
5 RNA interference Figure 4. Schematic representation of the pathway for the mechanism of RNA interference Results Figure 5. A cell based genome wide sirna screen for circadian clock modifiers (A) A schematic diagram of the genome wide sirna screen including the primary screen, data mining, hit selection, secondary screen, and validation of several selected targets. In the primary screen, reporter cells were transfected with sirna in 384 well plates followed by kinetic bioluminescence recording. (B) Distribution of circadian parameters of the entire primary screen. (C) Cellular clock phenotypes of sirna ( ) p yp knockdown of known clock genes. Plots of cellular oscillations upon knockdown of BMAL1, CLOCK, PER1, PER2, CRY1, or CRY2 by two independent pairs of sirnas in the primary screen are presented. The spikes of initial 10 hr bioluminescence readings resulted from media change and were removed from the plot.
6 Primary Hit Selection 1028 short period hits 76 double hits 4230 long period hits 274 double hits 493 high amplitude hits 18 double hits No low amplitude traces, poor curve fitting and inconsistent period length data. Pi Primary Hit Selection: 254 genes + 3 S.D. from the mean. 89 single sirna pair hits in duplicate well. Total 343 genes selected. Secondary Confirmation Assay Figure 6. BMAL1 assay. 222 of 238 genes identified in secondary screen 47 of 89 single sirna pair confirmed.
7 Dose Dependent Phenotypic Validation Figure 7. Period and amplitude dependence by sirna ammount. Network Effects Figure 8. Knockdown of most screen hits led to dose dependent reduction or up reguleted trascript levels
8 The Expanded Clock Gene Network Figure 9. List of interactions identified in primary sirna screen. Interconnectedness between the clock and other biological processes Figure 10A. Regulation of cellular circadian clock by components in the insulin signaling pathway.
9 Interconnectedness between the clock and other biological processes Figure 10B. Effect of sirnas against genes involved in insulin signaling. Conclusions 1000 genes whose knockdown resulted in low amplitude circadian oscillations. 100 genes whose knockdown led to long or short period length of oscillation. protein interaction network analysis showed that some factors directly or indirectly interact with core clock components. the clock is interconnected with many biological pathways.
10 Thanks for the attention!!!
Delay in Feedback Repression by Cryptochrome 1 Is Required for Circadian Clock Function
 Delay in Feedback Repression by Cryptochrome 1 Is Required for Circadian Clock Function Maki Ukai-Tadenuma, 1,8 Rikuhiro G. Yamada, 1,8 Haiyan Xu, 4 Jürgen A. Ripperger, 5 Andrew C. Liu, 4 and Hiroki R.
Delay in Feedback Repression by Cryptochrome 1 Is Required for Circadian Clock Function Maki Ukai-Tadenuma, 1,8 Rikuhiro G. Yamada, 1,8 Haiyan Xu, 4 Jürgen A. Ripperger, 5 Andrew C. Liu, 4 and Hiroki R.
Supporting Information
 Supporting Information Lande-Diner et al. 10.1073/pnas.1305980110 SI Text Because of the complex history of nomenclature regarding Mediator subunits and differences in protein names between mammals and
Supporting Information Lande-Diner et al. 10.1073/pnas.1305980110 SI Text Because of the complex history of nomenclature regarding Mediator subunits and differences in protein names between mammals and
A Genome-wide RNAi Screen for Modifiers of the Circadian Clock in Human Cells
 Resource A Genome-wide RNAi Screen for Modifiers of the Circadian Clock in Human Cells Eric E. Zhang, 1,2,5 Andrew C. Liu, 1,3,5 Tsuyoshi Hirota, 1,2,5 Loren J. Miraglia, 1 Genevieve Welch, 1 Pagkapol
Resource A Genome-wide RNAi Screen for Modifiers of the Circadian Clock in Human Cells Eric E. Zhang, 1,2,5 Andrew C. Liu, 1,3,5 Tsuyoshi Hirota, 1,2,5 Loren J. Miraglia, 1 Genevieve Welch, 1 Pagkapol
Les ARN de l Horloge et L Horloge des ARN. Franck Delaunay University of Nice and CNRS, France
 Les ARN de l Horloge et L Horloge des ARN Franck Delaunay University of Nice and CNRS, France Marseille 3 oct 27 Circadian rhythms Bioluminescence 11 1 9 8 7 12 6 1 2 3 4 5 Photosynthesis Hatching Locomotor
Les ARN de l Horloge et L Horloge des ARN Franck Delaunay University of Nice and CNRS, France Marseille 3 oct 27 Circadian rhythms Bioluminescence 11 1 9 8 7 12 6 1 2 3 4 5 Photosynthesis Hatching Locomotor
Symbol Accession Found in Human Red Blood Cell database?
 SUPPLEMENTARY INFORMATION doi:1.138/nature972 Symbol Accession Found in Human Red Blood Cell database? ARNTL (BMAL1) NP_125443 No CLOCK AAB83969 No CRY1 NP_66 No CRY2 AAH35161 No PER1 AAG1149 No PER2 NP_73728
SUPPLEMENTARY INFORMATION doi:1.138/nature972 Symbol Accession Found in Human Red Blood Cell database? ARNTL (BMAL1) NP_125443 No CLOCK AAB83969 No CRY1 NP_66 No CRY2 AAH35161 No PER1 AAG1149 No PER2 NP_73728
Impact of Retinoic acid induced-1 (Rai1) on Regulators of Metabolism and Adipogenesis
 Impact of Retinoic acid induced-1 (Rai1) on Regulators of Metabolism and Adipogenesis The mammalian system undergoes ~24 hour cycles known as circadian rhythms that temporally orchestrate metabolism, behavior,
Impact of Retinoic acid induced-1 (Rai1) on Regulators of Metabolism and Adipogenesis The mammalian system undergoes ~24 hour cycles known as circadian rhythms that temporally orchestrate metabolism, behavior,
SureSilencing sirna Array Technology Overview
 SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
WB: AB1786P. 75kDa. 63kDa. 48kDa
 in vitro electroporation The cell suspension (1X1 6 cells) was mixed with plasmid 1 μg pcdn3.1 intact vector or pcdn3.1 human drd3 in Opti-MEM Media (GICO). The expression plasmid of pcdn3.1 was obtained
in vitro electroporation The cell suspension (1X1 6 cells) was mixed with plasmid 1 μg pcdn3.1 intact vector or pcdn3.1 human drd3 in Opti-MEM Media (GICO). The expression plasmid of pcdn3.1 was obtained
Reference: Vol. 343, Issue 6166, pp Original title: Genome-scale CRISPR-Cas9 knockout screening in human cells
 Original title: Genome-scale CRISPR-Cas9 knockout screening in human cells Authors: Neville Sanjana, Ophir Shalem Original publication date: 08/28/2015 Annotator(s): Justin German, Ian Ahrens, Regg Strotheide,
Original title: Genome-scale CRISPR-Cas9 knockout screening in human cells Authors: Neville Sanjana, Ophir Shalem Original publication date: 08/28/2015 Annotator(s): Justin German, Ian Ahrens, Regg Strotheide,
Nature Structural & Molecular Biology: doi: /nsmb.3018
 Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
SUPPLEMENTARY DATA. Supplementary Table 1. Sequences of qpcr primers.
 Supplementary Table 1. Sequences of qpcr primers. Lpl Fatp1 (Slc27a1) Fatp4 (Slc27a4) Cd36 Acsl1 Gpam Agpat2 Dgat1 Dgat2 Plin1 Atgl Hsl Mgll Lpin1 Got2 Cav1 Cav2 Fitm2 Nr1h2 Nr1h3 Acsl4 Acsl5 Agpat6 Agpat9
Supplementary Table 1. Sequences of qpcr primers. Lpl Fatp1 (Slc27a1) Fatp4 (Slc27a4) Cd36 Acsl1 Gpam Agpat2 Dgat1 Dgat2 Plin1 Atgl Hsl Mgll Lpin1 Got2 Cav1 Cav2 Fitm2 Nr1h2 Nr1h3 Acsl4 Acsl5 Agpat6 Agpat9
Document S1. Supplemental Experimental Procedures and Three Figures (see next page)
 Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
2. Real-time reporter gene assay by photon counting system. 3. Real-time reporter gene assay by imaging system.
 Real-time Reporter Gene Assay Progress of real-time reporter assay using bioluminescence protein gene. 1. Typical reporter protein in Japan. 1. Typical reporter protein in Japan. 2. Real-time reporter
Real-time Reporter Gene Assay Progress of real-time reporter assay using bioluminescence protein gene. 1. Typical reporter protein in Japan. 1. Typical reporter protein in Japan. 2. Real-time reporter
Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila
 Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Supplementary Figure 1 (related to Figure 1, 2 and Table 1)
 Supplementary Figure 1 (related to Figure 1, 2 and Table 1) Role of E75 in regulating circadian rhythms. (A) E75 mrna expression in TG 27 controls, TG 27 >NE-30-49-10, TG 27 >UAS-E75 (II), TG 27 >UAS-E75
Supplementary Figure 1 (related to Figure 1, 2 and Table 1) Role of E75 in regulating circadian rhythms. (A) E75 mrna expression in TG 27 controls, TG 27 >NE-30-49-10, TG 27 >UAS-E75 (II), TG 27 >UAS-E75
Chapter 1. from genomics to proteomics Ⅱ
 Proteomics Chapter 1. from genomics to proteomics Ⅱ 1 Functional genomics Functional genomics: study of relations of genomics to biological functions at systems level However, it cannot explain any more
Proteomics Chapter 1. from genomics to proteomics Ⅱ 1 Functional genomics Functional genomics: study of relations of genomics to biological functions at systems level However, it cannot explain any more
PROTEOMICS AND FUNCTIONAL GENOMICS PRESENTATION KIMBERLY DONG
 PROTEOMICS AND FUNCTIONAL GENOMICS PRESENTATION KIMBERLY DONG A Lentiviral RNAi Library for Human and Mouse Genes Applied to an Arrayed Viral High-Content Screen Jason Moffat,1,2,4,10 Dorre A. Grueneberg,1,10
PROTEOMICS AND FUNCTIONAL GENOMICS PRESENTATION KIMBERLY DONG A Lentiviral RNAi Library for Human and Mouse Genes Applied to an Arrayed Viral High-Content Screen Jason Moffat,1,2,4,10 Dorre A. Grueneberg,1,10
Validation of Hits from an sirna Library Screen using Real-Time qpcr. Gregory L. Shipley The University of Texas Health Science Center- Houston
 Validation of Hits from an sirna Library Screen using Real-Time qpcr Gregory L. Shipley The University of Texas Health Science Center- Houston sirnas and Gene Silencing Small interfering RNAs or sirnas
Validation of Hits from an sirna Library Screen using Real-Time qpcr Gregory L. Shipley The University of Texas Health Science Center- Houston sirnas and Gene Silencing Small interfering RNAs or sirnas
Technical tips Session 4
 Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Optimization of RNAi Targets on the Human Transcriptome Ahmet Arslan Kurdoglu Computational Biosciences Program Arizona State University
 Optimization of RNAi Targets on the Human Transcriptome Ahmet Arslan Kurdoglu Computational Biosciences Program Arizona State University my background Undergraduate Degree computer systems engineer (ASU
Optimization of RNAi Targets on the Human Transcriptome Ahmet Arslan Kurdoglu Computational Biosciences Program Arizona State University my background Undergraduate Degree computer systems engineer (ASU
Animals. Balb/c mice (male) purchased 5 weeks postpartum, were adapted under
 Supplementary Methods Animals. Balb/c mice (male) purchased 5 weeks postpartum, were adapted under standard 12 h light: 12 h dark cycles (LD) for 2 weeks, and then transferred into the constant darkness
Supplementary Methods Animals. Balb/c mice (male) purchased 5 weeks postpartum, were adapted under standard 12 h light: 12 h dark cycles (LD) for 2 weeks, and then transferred into the constant darkness
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
C57BL/6N Female. C57BL/6N Male. Prl2 WT Allele. Prl2 Targeted Allele **** **** WT Het KO PRL bp. 230 bp CNX. C57BL/6N Male 30.
 A Prl Allele Prl Targeted Allele 631 bp 1 3 4 5 6 bp 1 SA β-geo pa 3 4 5 6 Het B Het 631 bp bp PRL CNX C 1 C57BL/6N Male (14) Het (7) (1) 1 C57BL/6N Female (19) Het (31) (1) % survival 5 % survival 5 ****
A Prl Allele Prl Targeted Allele 631 bp 1 3 4 5 6 bp 1 SA β-geo pa 3 4 5 6 Het B Het 631 bp bp PRL CNX C 1 C57BL/6N Male (14) Het (7) (1) 1 C57BL/6N Female (19) Het (31) (1) % survival 5 % survival 5 ****
long noncoding RNA Knockdown and detection of Thermo Scientific Lincode sirna and Solaris lncrna Expression Assays
 Thermo Scientific Lincode sirna and Solaris lncrna Expression Assays Knockdown and detection of long noncoding RNA C omprehensive pre-designed reagents for investigation of human lncrnas sirnas with enhanced
Thermo Scientific Lincode sirna and Solaris lncrna Expression Assays Knockdown and detection of long noncoding RNA C omprehensive pre-designed reagents for investigation of human lncrnas sirnas with enhanced
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets.
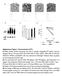 Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Concepts and Methods in Developmental Biology
 Biology 4361 Developmental Biology Concepts and Methods in Developmental Biology June 16, 2009 Conceptual and Methodological Tools Concepts Genomic equivalence Differential gene expression Differentiation/de-differentiation
Biology 4361 Developmental Biology Concepts and Methods in Developmental Biology June 16, 2009 Conceptual and Methodological Tools Concepts Genomic equivalence Differential gene expression Differentiation/de-differentiation
RNAi HTS and Data Analysis
 Part I RNAi HTS and Data Analysis 1 Introduction to Genome-Scale RNAi Research 1.1 RNAi: An Effective Tool for Elucidating Gene Functions and a New Class of Drugs RNAi is a mechanism in living cells that
Part I RNAi HTS and Data Analysis 1 Introduction to Genome-Scale RNAi Research 1.1 RNAi: An Effective Tool for Elucidating Gene Functions and a New Class of Drugs RNAi is a mechanism in living cells that
Gene expression analysis. Biosciences 741: Genomics Fall, 2013 Week 5. Gene expression analysis
 Gene expression analysis Biosciences 741: Genomics Fall, 2013 Week 5 Gene expression analysis From EST clusters to spotted cdna microarrays Long vs. short oligonucleotide microarrays vs. RT-PCR Methods
Gene expression analysis Biosciences 741: Genomics Fall, 2013 Week 5 Gene expression analysis From EST clusters to spotted cdna microarrays Long vs. short oligonucleotide microarrays vs. RT-PCR Methods
The Ups & Downs of Gene Expression:
 The Ups & Downs of Gene Expression: Using Lipid-Based Transfection and RT-qPCR to Deliver Perfect Knockdown and Achieve Optimal Expression Results Hilary Srere, Ph.D. Topics What is RNAi? Methods of Delivery
The Ups & Downs of Gene Expression: Using Lipid-Based Transfection and RT-qPCR to Deliver Perfect Knockdown and Achieve Optimal Expression Results Hilary Srere, Ph.D. Topics What is RNAi? Methods of Delivery
Role of REV-ERBα in the regulation of lung inflammation
 Role of REV-ERBα in the regulation of lung inflammation A thesis submitted to the University of Manchester for the degree of Doctor of Philosophy in the Faculty of Biology, Medicine and Health 2016 Marie
Role of REV-ERBα in the regulation of lung inflammation A thesis submitted to the University of Manchester for the degree of Doctor of Philosophy in the Faculty of Biology, Medicine and Health 2016 Marie
A probability-based approach for the analysis of large-scale RNAi screens
 A probability-based approach for the analysis of large-scale RNAi screens Renate König, Chih-yuan Chiang, Buu P Tu, S Frank Yan, Paul D DeJesus, Angelica Romero, Tobias Bergauer, Anthony Orth, Ute Krueger,
A probability-based approach for the analysis of large-scale RNAi screens Renate König, Chih-yuan Chiang, Buu P Tu, S Frank Yan, Paul D DeJesus, Angelica Romero, Tobias Bergauer, Anthony Orth, Ute Krueger,
System. Dynamic Monitoring of Receptor Tyrosine Kinase Activation in Living Cells. Application Note No. 4 / March
 System Application Note No. 4 / March 2008 Dynamic Monitoring of Receptor Tyrosine Kinase Activation in Living Cells www.roche-applied-science.com Dynamic Monitoring of Receptor Tyrosine Kinase Activation
System Application Note No. 4 / March 2008 Dynamic Monitoring of Receptor Tyrosine Kinase Activation in Living Cells www.roche-applied-science.com Dynamic Monitoring of Receptor Tyrosine Kinase Activation
sherwood - UltramiR shrna Collections
 sherwood - UltramiR shrna Collections Incorporating advances in shrna design and processing for superior potency and specificity sherwood - UltramiR shrna Collections Enabling Discovery Across the Genome
sherwood - UltramiR shrna Collections Incorporating advances in shrna design and processing for superior potency and specificity sherwood - UltramiR shrna Collections Enabling Discovery Across the Genome
Cellular Reprogramming and Controllability of Complex Systems
 Cellular Reprogramming and Controllability of Complex Systems University of Maryland and ISR April 25, 2016 Roger Brockett John A. Paulson School of Engineering and Applied Science Harvard University 1
Cellular Reprogramming and Controllability of Complex Systems University of Maryland and ISR April 25, 2016 Roger Brockett John A. Paulson School of Engineering and Applied Science Harvard University 1
Supplementary Figure 1. (A) Cell proliferative ability and (B) the invasiveness of
 LEGEND FOR SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Cell proliferative ability and (B) the invasiveness of LLC-1, LLC-3, and LLC-5 cell line series were quantified by BrdU assay after 72 h of
LEGEND FOR SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Cell proliferative ability and (B) the invasiveness of LLC-1, LLC-3, and LLC-5 cell line series were quantified by BrdU assay after 72 h of
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
Heparan Sulphate in Breast Cancer Low, Y.L.A 1 and Yip, C.W.G 2
 Heparan Sulphate in Breast Cancer Low, Y.L.A 1 and Yip, C.W.G 2 Department of Anatomy, Yong Loo Lin School of Medicine, National University of Singapore MD10, 4 Medical Drive, Singapore 117597 ABSTRACT
Heparan Sulphate in Breast Cancer Low, Y.L.A 1 and Yip, C.W.G 2 Department of Anatomy, Yong Loo Lin School of Medicine, National University of Singapore MD10, 4 Medical Drive, Singapore 117597 ABSTRACT
Experimental genetics - 2 Partha Roy
 Partha Roy Experimental genetics - 2 Making genetically altered animal 1) Gene knock-out k from: a) the entire animal b) selected cell-type/ tissue c) selected cell-type/tissue at certain time 2) Transgenic
Partha Roy Experimental genetics - 2 Making genetically altered animal 1) Gene knock-out k from: a) the entire animal b) selected cell-type/ tissue c) selected cell-type/tissue at certain time 2) Transgenic
SI Materials and Methods Cell Culture. Transient transfection and endpoint luciferase reporter assays
 SI Materials and Methods Cell Culture. HEK 293T, U2OS and HepG2 cells were purchased from the American Type Culture Collection (ATCC). SW480 Per2-dluc reporter cells were a gift from Carrie Partch, Chemistry
SI Materials and Methods Cell Culture. HEK 293T, U2OS and HepG2 cells were purchased from the American Type Culture Collection (ATCC). SW480 Per2-dluc reporter cells were a gift from Carrie Partch, Chemistry
TOOLS sirna and mirna. User guide
 TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
New Technologies for RNAi Screening
 sirna is a hot topic! Craig C. Mello, PhD, Nobel Prize in Medicine 2006-1 - New Technologies for RNAi Screening Dr. Jörg Dennig Global Product Manager RNAi Joerg.Dennig@qiagen.com 1 New Technologies for
sirna is a hot topic! Craig C. Mello, PhD, Nobel Prize in Medicine 2006-1 - New Technologies for RNAi Screening Dr. Jörg Dennig Global Product Manager RNAi Joerg.Dennig@qiagen.com 1 New Technologies for
Supplementary Information
 Supplementary Information Negative regulation of initial steps in skeletal myogenesis by mtor and other kinases Raphael A. Wilson 1*, Jing Liu 1*, Lin Xu 1, James Annis 2, Sara Helmig 1, Gregory Moore
Supplementary Information Negative regulation of initial steps in skeletal myogenesis by mtor and other kinases Raphael A. Wilson 1*, Jing Liu 1*, Lin Xu 1, James Annis 2, Sara Helmig 1, Gregory Moore
HCT116 SW48 Nutlin: p53
 Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Galina Gabriely, Ph.D. BWH/HMS
 Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Trends in Medical Research -
 RNA Interference Tel : 02-2267-1740 / E-mail : kimys@inje.ac.kr, RNAi (RNA or RNA silencing) ( ) DNA small RNA (srnas) mrna,. DNA small RNA. DNA RNA small RNA. small RNA mirna (microrna), RNA RNAi. 1993
RNA Interference Tel : 02-2267-1740 / E-mail : kimys@inje.ac.kr, RNAi (RNA or RNA silencing) ( ) DNA small RNA (srnas) mrna,. DNA small RNA. DNA RNA small RNA. small RNA mirna (microrna), RNA RNAi. 1993
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Supplementary Information
 Supplementary Information Sam68 modulates the promoter specificity of NF-κB and mediates expression of CD25 in activated T cells Kai Fu 1, 6, Xin Sun 1, 6, Wenxin Zheng 1, 6, Eric M. Wier 1, Andrea Hodgson
Supplementary Information Sam68 modulates the promoter specificity of NF-κB and mediates expression of CD25 in activated T cells Kai Fu 1, 6, Xin Sun 1, 6, Wenxin Zheng 1, 6, Eric M. Wier 1, Andrea Hodgson
Supplementary Fig.1. Over-expression of RNase L in stable polyclonal cell line
 Supplemental Data mrna Protein A kda 75 40 NEO/vector NEO/RNase L RNASE L β-actin RNASE L β-actin B % of control (neo vector), normalized 240 ** 200 160 120 80 40 0 Neo Neo/RNase L RNase L Protein Supplementary
Supplemental Data mrna Protein A kda 75 40 NEO/vector NEO/RNase L RNASE L β-actin RNASE L β-actin B % of control (neo vector), normalized 240 ** 200 160 120 80 40 0 Neo Neo/RNase L RNase L Protein Supplementary
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
Computational Genomics. Reconstructing signaling and dynamic regulatory networks
 02-710 Computational Genomics Reconstructing signaling and dynamic regulatory networks Input Output Hidden Markov Model Input (Static transcription factorgene interactions) Bengio and Frasconi, NIPS 1995
02-710 Computational Genomics Reconstructing signaling and dynamic regulatory networks Input Output Hidden Markov Model Input (Static transcription factorgene interactions) Bengio and Frasconi, NIPS 1995
Online Supplementary Information
 Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Molecular Genetics Student Objectives
 Molecular Genetics Student Objectives Exam 1: Enduring understanding 3.A: Heritable information provides for continuity of life. Essential knowledge 3.A.1: DNA, and in some cases RNA, is the primary source
Molecular Genetics Student Objectives Exam 1: Enduring understanding 3.A: Heritable information provides for continuity of life. Essential knowledge 3.A.1: DNA, and in some cases RNA, is the primary source
colorimetric sandwich ELISA kit datasheet
 colorimetric sandwich ELISA kit datasheet For the quantitative detection of human ENO2 concentrations in serum, plasma and cell culture supernatants. general information Catalogue Number Product Name Species
colorimetric sandwich ELISA kit datasheet For the quantitative detection of human ENO2 concentrations in serum, plasma and cell culture supernatants. general information Catalogue Number Product Name Species
Dharmacon TM solutions for studying gene function
 GE Healthcare Capabilities Overview Dharmacon TM solutions for studying gene function RNAi Gene Expression Gene Editing RNA Interference Our RNAi products encompass the most complete portfolio of innovative
GE Healthcare Capabilities Overview Dharmacon TM solutions for studying gene function RNAi Gene Expression Gene Editing RNA Interference Our RNAi products encompass the most complete portfolio of innovative
Validation & Assay Performance Summary
 Validation & Assay Performance Summary CellSensor ESRE-bla HeLa Cell Line Cat. no. K87 CellSensor Cell-Based Assay Validation Packet This cell-based assay has been thoroughly tested and validated by Invitrogen
Validation & Assay Performance Summary CellSensor ESRE-bla HeLa Cell Line Cat. no. K87 CellSensor Cell-Based Assay Validation Packet This cell-based assay has been thoroughly tested and validated by Invitrogen
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
a Award Number: W81XWH TITLE:
 AD Award Number: W81XWH-04-1-0138 TITLE: Imaging Metastatic Prostate Cancer After Genetic Manipulation of Transcriptional Memory Regulators EZH2 and EED PRINCIPAL INVESTIGATOR: Lily Wu, M.D., Ph.D. CONTRACTING
AD Award Number: W81XWH-04-1-0138 TITLE: Imaging Metastatic Prostate Cancer After Genetic Manipulation of Transcriptional Memory Regulators EZH2 and EED PRINCIPAL INVESTIGATOR: Lily Wu, M.D., Ph.D. CONTRACTING
Supporting Information
 Supporting Information Patel et al. 10.1073/pnas. SI Materials and Methods Cells and Reagents. Hepatocyte-derived H35 cells were grown in high glucose DMEM (Gibco) media, supplemented with 10% FBS, 2%
Supporting Information Patel et al. 10.1073/pnas. SI Materials and Methods Cells and Reagents. Hepatocyte-derived H35 cells were grown in high glucose DMEM (Gibco) media, supplemented with 10% FBS, 2%
17.5 Eukaryotic Transcription Initiation Is Regulated by Transcription Factors That Bind to Cis-Acting Sites
 17.5 Eukaryotic Transcription Initiation Is Regulated by Transcription Factors That Bind to Cis-Acting Sites 1 Section 17.5 Transcription regulatory proteins, transcription factors, target cis-acting sites
17.5 Eukaryotic Transcription Initiation Is Regulated by Transcription Factors That Bind to Cis-Acting Sites 1 Section 17.5 Transcription regulatory proteins, transcription factors, target cis-acting sites
Lecture 22 Eukaryotic Genes and Genomes III
 Lecture 22 Eukaryotic Genes and Genomes III In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand full of genes.
Lecture 22 Eukaryotic Genes and Genomes III In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand full of genes.
University of Pennsylvania Perelman School of Medicine High Throughput Screening Core
 University of Pennsylvania Perelman School of Medicine High Throughput Screening Core Sara Cherry, Ph. D. David C. Schultz, Ph. D. dschultz@mail.med.upenn.edu (215) 573 9641 67 John Morgan Building Mission
University of Pennsylvania Perelman School of Medicine High Throughput Screening Core Sara Cherry, Ph. D. David C. Schultz, Ph. D. dschultz@mail.med.upenn.edu (215) 573 9641 67 John Morgan Building Mission
MeCP2. MeCP2/α-tubulin. GFP mir1-1 mir132
 Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
[Presented by: Andrew Howlett, Cruise Slater, Mahmud Hasan, Greg Dale]
![[Presented by: Andrew Howlett, Cruise Slater, Mahmud Hasan, Greg Dale] [Presented by: Andrew Howlett, Cruise Slater, Mahmud Hasan, Greg Dale]](/thumbs/96/128035026.jpg) Mutational Dissection [Presented by: Andrew Howlett, Cruise Slater, Mahmud Hasan, Greg Dale] Introduction What is the point of Mutational Dissection? It allows understanding of normal biological functions
Mutational Dissection [Presented by: Andrew Howlett, Cruise Slater, Mahmud Hasan, Greg Dale] Introduction What is the point of Mutational Dissection? It allows understanding of normal biological functions
Review of previous work: Overview of previous work on DNA methylation and chromatin dynamics
 CONTENTS Preface Acknowledgements Lists of Commonly Used Abbreviations Review of previous work: Overview of previous work on DNA methylation and chromatin dynamics Introduction Enzymes involved in DNA
CONTENTS Preface Acknowledgements Lists of Commonly Used Abbreviations Review of previous work: Overview of previous work on DNA methylation and chromatin dynamics Introduction Enzymes involved in DNA
Khaled_Fig. S MITF TYROSINASE R²= MITF PDE4D R²= Variance from mean mrna expression
 Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Investigating Compound Mechanism of Action
 Investigating Compound Mechanism of Action Using Genetic Interaction Screens Lisa Marshall, Priya Mitty, Michael Ollmann, and Paul Kassner Michael Ollmann, Ph.D. Principal Scientist, Therapeutic Discovery
Investigating Compound Mechanism of Action Using Genetic Interaction Screens Lisa Marshall, Priya Mitty, Michael Ollmann, and Paul Kassner Michael Ollmann, Ph.D. Principal Scientist, Therapeutic Discovery
detailed description. Cell proliferation and soft agar colony numbers were shown as relative values normalized to
 Requirement for BUB1B/BUBR1 in tumor progression of lung adenocarcinoma Supplementary Material Supplemental Figure S1. Schematic overview of mouse kinome sirna screen. See Materials and Methods for detailed
Requirement for BUB1B/BUBR1 in tumor progression of lung adenocarcinoma Supplementary Material Supplemental Figure S1. Schematic overview of mouse kinome sirna screen. See Materials and Methods for detailed
Talk with the editors of THE JOURNAL OF BIOLOGICAL CHEMISTRY
 Talk with the editors of THE JOURNAL OF BIOLOGICAL CHEMISTRY Part 1: What s new at the JBC? Mission Statement The Journal of Biological Chemistry encourages the submission of manuscripts based on original
Talk with the editors of THE JOURNAL OF BIOLOGICAL CHEMISTRY Part 1: What s new at the JBC? Mission Statement The Journal of Biological Chemistry encourages the submission of manuscripts based on original
Ectopic Gene Expression in Mammalian Cells
 Ectopic Gene Expression in Mammalian Cells Dr. Stefan Stein AG Grez Methodenseminar BCII Praktikum am Georg-Speyer-Haus 11.01. & 01.02.2012 1 Ectopic Gene Expression in Mammalian Cells Definition: ectopic
Ectopic Gene Expression in Mammalian Cells Dr. Stefan Stein AG Grez Methodenseminar BCII Praktikum am Georg-Speyer-Haus 11.01. & 01.02.2012 1 Ectopic Gene Expression in Mammalian Cells Definition: ectopic
Introduction into single-cell RNA-seq. Kersti Jääger 19/02/2014
 Introduction into single-cell RNA-seq Kersti Jääger 19/02/2014 Cell is the smallest functional unit of life Nucleus.ATGC.UACG. A Cell KLTSH. The complexity of biology How many cell types? How many cells?
Introduction into single-cell RNA-seq Kersti Jääger 19/02/2014 Cell is the smallest functional unit of life Nucleus.ATGC.UACG. A Cell KLTSH. The complexity of biology How many cell types? How many cells?
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
SUPPORTING INFORMATION. A cleavage-responsive stem-loop hairpin for assaying guide RNA activity
 SUPPORTING INFORMATION A cleavage-responsive stem-loop hairpin for assaying guide RNA activity Tara R. deboer 1, Noreen Wauford 1, Jing-Yi Chung, Miguel Salvador Torres Perez, and Niren Murthy* University
SUPPORTING INFORMATION A cleavage-responsive stem-loop hairpin for assaying guide RNA activity Tara R. deboer 1, Noreen Wauford 1, Jing-Yi Chung, Miguel Salvador Torres Perez, and Niren Murthy* University
SUPPLEMENTAL MATERIALS IDENTIFICATION OF A FUNCTIONAL NETWORK OF HUMAN EPIGENETIC SILENCING FACTORS
 SUPPLEMENTAL MATERIALS IDENTIFICATION OF A FUNCTIONAL NETWORK OF HUMAN EPIGENETIC SILENCING FACTORS Andrey Poleshko, Margret B. Einarson, Natalia Shalginskikh, Rugang Zhang, Peter D. Adams 1, Anna Marie
SUPPLEMENTAL MATERIALS IDENTIFICATION OF A FUNCTIONAL NETWORK OF HUMAN EPIGENETIC SILENCING FACTORS Andrey Poleshko, Margret B. Einarson, Natalia Shalginskikh, Rugang Zhang, Peter D. Adams 1, Anna Marie
Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm
 Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm Anja Smith Director R&D Dharmacon, part of GE Healthcare Imagination at work crrna:tracrrna program Cas9 nuclease Active crrna is
Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm Anja Smith Director R&D Dharmacon, part of GE Healthcare Imagination at work crrna:tracrrna program Cas9 nuclease Active crrna is
Intracellular receptors specify complex patterns of gene expression that are cell and gene
 SUPPLEMENTAL RESULTS AND DISCUSSION Some HPr-1AR ARE-containing Genes Are Unresponsive to Androgen Intracellular receptors specify complex patterns of gene expression that are cell and gene specific. For
SUPPLEMENTAL RESULTS AND DISCUSSION Some HPr-1AR ARE-containing Genes Are Unresponsive to Androgen Intracellular receptors specify complex patterns of gene expression that are cell and gene specific. For
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Introduction to Bioinformatics
 Introduction to Bioinformatics If the 19 th century was the century of chemistry and 20 th century was the century of physic, the 21 st century promises to be the century of biology...professor Dr. Satoru
Introduction to Bioinformatics If the 19 th century was the century of chemistry and 20 th century was the century of physic, the 21 st century promises to be the century of biology...professor Dr. Satoru
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV
 1 Fall 2006 7.03 7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that
1 Fall 2006 7.03 7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that
7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV
 7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand
7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand
MOLECULAR BIOLOGY OF EUKARYOTES 2016 SYLLABUS
 03-442 Lectures: MWF 9:30-10:20 a.m. Doherty Hall 2105 03-742 Advanced Discussion Section: Time and place to be announced Probably Mon 4-6 p.m. or 6-8p.m.? Once we establish who is taking the advanced
03-442 Lectures: MWF 9:30-10:20 a.m. Doherty Hall 2105 03-742 Advanced Discussion Section: Time and place to be announced Probably Mon 4-6 p.m. or 6-8p.m.? Once we establish who is taking the advanced
Chapter 19 Genetic Regulation of the Eukaryotic Genome. A. Bergeron AP Biology PCHS
 Chapter 19 Genetic Regulation of the Eukaryotic Genome A. Bergeron AP Biology PCHS 2 Do Now - Eukaryotic Transcription Regulation The diagram below shows five genes (with their enhancers) from the genome
Chapter 19 Genetic Regulation of the Eukaryotic Genome A. Bergeron AP Biology PCHS 2 Do Now - Eukaryotic Transcription Regulation The diagram below shows five genes (with their enhancers) from the genome
Evaluation of RNA interference (RNAi) in Skin
 Evaluation of RNA interference (RNAi) in Skin Next Generation in RNAi Pamela A. Pavco VP Pharmaceutical Development 3rd International SCAR Club Meeting March 26, 2010 www.rxipharma.com Massachusetts, USA
Evaluation of RNA interference (RNAi) in Skin Next Generation in RNAi Pamela A. Pavco VP Pharmaceutical Development 3rd International SCAR Club Meeting March 26, 2010 www.rxipharma.com Massachusetts, USA
Supplementary Information. ATM and MET kinases are synthetic lethal with. non-genotoxic activation of p53
 Supplementary Information ATM and MET kinases are synthetic lethal with non-genotoxic activation of p53 Kelly D. Sullivan 1, Nuria Padilla-Just 1, Ryan E. Henry 1, Christopher C. Porter 2, Jihye Kim 3,
Supplementary Information ATM and MET kinases are synthetic lethal with non-genotoxic activation of p53 Kelly D. Sullivan 1, Nuria Padilla-Just 1, Ryan E. Henry 1, Christopher C. Porter 2, Jihye Kim 3,
New Plant Breeding Technologies
 New Plant Breeding Technologies Ricarda A. Steinbrecher, PhD EcoNexus / ENSSER Berlin, 07 May 2015 r.steinbrecher@econexus.info distributed by EuropaBio What are the NPBTs? *RNAi *Epigenetic alterations
New Plant Breeding Technologies Ricarda A. Steinbrecher, PhD EcoNexus / ENSSER Berlin, 07 May 2015 r.steinbrecher@econexus.info distributed by EuropaBio What are the NPBTs? *RNAi *Epigenetic alterations
Stifel Nicolaus Healthcare Conference. September 2012
 Stifel Nicolaus Healthcare Conference September 212 Alnylam Forward Looking Statements This presentation contains forward-looking statements, within the meaning of Section 27A of the Securities Act of
Stifel Nicolaus Healthcare Conference September 212 Alnylam Forward Looking Statements This presentation contains forward-looking statements, within the meaning of Section 27A of the Securities Act of
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles
 Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles Long-term gene silencing shrna-specific design algorithm High titer, purified particles Thermo Scientific Dharmacon SMARTvector shrna
Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles Long-term gene silencing shrna-specific design algorithm High titer, purified particles Thermo Scientific Dharmacon SMARTvector shrna
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Arrayed shrna screening 11,415 hairpins representing 1383 unique genes. encompassing the murine protein kinome, GTPases, and components of the vesicle
 Supplemental Material for Strohecker et al. Identification of 6-phosphofructo-2- kinase/fructose-2,6-bisphosphatase (PFKFB4) as a Novel Autophagy Regulator by High Content shrna Screening Supplemental
Supplemental Material for Strohecker et al. Identification of 6-phosphofructo-2- kinase/fructose-2,6-bisphosphatase (PFKFB4) as a Novel Autophagy Regulator by High Content shrna Screening Supplemental
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
pdsipher and pdsipher -GFP shrna Vector User s Guide
 pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
How Targets Are Chosen. Chris Wayman 12 th April 2012
 How Targets Are Chosen Chris Wayman 12 th April 2012 A few questions How many ideas does it take to make a medicine? 10 20 20-50 50-100 A few questions How long does it take to bring a product from bench
How Targets Are Chosen Chris Wayman 12 th April 2012 A few questions How many ideas does it take to make a medicine? 10 20 20-50 50-100 A few questions How long does it take to bring a product from bench
Input. Pulldown GST. GST-Ahi1
 A kd 250 150 100 75 50 37 25 -GST-Ahi1 +GST-Ahi1 kd 148 98 64 50 37 22 GST GST-Ahi1 C kd 250 148 98 64 50 Control Input Hap1A Hap1 Pulldown GST GST-Ahi1 Hap1A Hap1 Hap1A Hap1 Figure S1. (A) Ahi1 western
A kd 250 150 100 75 50 37 25 -GST-Ahi1 +GST-Ahi1 kd 148 98 64 50 37 22 GST GST-Ahi1 C kd 250 148 98 64 50 Control Input Hap1A Hap1 Pulldown GST GST-Ahi1 Hap1A Hap1 Hap1A Hap1 Figure S1. (A) Ahi1 western
microrna Shifra Ben-Dor March 2010
 microrna Shifra Ben-Dor March 2010 Outline Biology of mirna Prediction of mirna mirna Databases Prediction of mirna Targets micrornas (mirna) Naturally expressed small RNAs Involved in regulation of target
microrna Shifra Ben-Dor March 2010 Outline Biology of mirna Prediction of mirna mirna Databases Prediction of mirna Targets micrornas (mirna) Naturally expressed small RNAs Involved in regulation of target
Transient silencing of the CDKN2A gene was achieved using a pool of four sirna duplexes
 Supplementary Methods Small interfering RNA (sirna) knockdown of CDKN2A (p16 INK4a /p14 ARF ) Transient silencing of the CDKN2A gene was achieved using a pool of four sirna duplexes that each targeted
Supplementary Methods Small interfering RNA (sirna) knockdown of CDKN2A (p16 INK4a /p14 ARF ) Transient silencing of the CDKN2A gene was achieved using a pool of four sirna duplexes that each targeted
