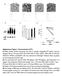
Khaled_Fig. S MITF TYROSINASE R²= MITF PDE4D R²= Variance from mean mrna expression
|
|
|
- Evangeline Cox
- 6 years ago
- Views:
Transcription
1 Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²= MALME3M SKMEL28 UACC257 MITF PDE4D R²= MITF PDE4B R²= SKMEL2 SKMEL5 UACC62 M4 MITF PDE4C R²=-.44 MDAMB435 Figure S PDE4D and PDE4B mrna expression correlate with MITF expression. Quantitative mrna levels for each gene in eight human melanomas were acquired from Affymetrix microarrays as described by (Du et al. 24).
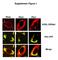
2 Khaled_Fig. S2 sictrl si MITF MITF Tub Figure S2 sirna mediated knock down of MITF. Human primary melanocytes were transfected 3 times with simitf. 72h after the first transfection, protein extracts were immunoblotted using antibodies against MITF (C5).
3 exons Khaled_Fig. S3 Nterm. Unique region PDE4D 5 UTR ucr ucr2 Catalytic domain C-term and 3 UTR D3 5 UTR D3 5 UTR Long isoforms D4 5 UTR D7 5 UTR D8 5 UTR Figure S4 schematic representation of PDE4D isoforms mrna structure. All PDE4D isoforms harbor a unique N- terminal region and are divided in three categories according to their length. The positions of the primers used in the study are displayed of the scheme. The scale of the length of each N-terminal region and of the length of each domain is not respected. D9 5 UTR D 5 UTR Short isoforms D2 5 UTR Super short isoform D6 5 UTR Pan isoforms PDE4D primers PCR primers qpcr primers
4 A Khaled_Fig. S4 53bp PDE4D 455bp PDE4D2 29bp PDE4D3 347bp PDE4D4 2bp PDE4D5 24bp PDE4D6 6bp PDE4D7 226bp PDE4D8 475bp PDE4D9 49bp NHM 5 heart Sk-n-sh B-Actin B Melanocytes Relative mrna expression 3 2 PDE4D PDE4D2 PDE4D3 PDE4D5 PDE4D6 PDE4D7 control fsk 2h fsk 6h fsk 2h PDE4D9 Figure S3 PDE4D isoform profiling. (A) cdna from sk-n-sh, cells, and 5 different primary human melanocytes (NHM) cultures was assessed by PCR. (B) Total mrna from normal human melanocytes exposed to 2 µm (fsk) was subjected to qpcr. The data are normalized to b-actin and each point is the mean ± SD of three experiments performed in duplicate. () P<.5.
5 Khaled_Fig. S5 A Relative mrna expression,4,2,8,6,4,2 Fibroblasts PDE4D PDE4D3 cont 2h 6h 2h B Fiborblasts PCREB tub 2h 6h 2h Figure S5 camp does not stimulate PDE4D or PDE4D3 mrna expression in normal human fibroblasts. (A) Total mrna of normal human fibroblasts exposed to 2 µm was subjected to qpcr. The data are normalized relative to b-actin and each point is the mean ± SD of three experiments performed in duplicate. (B) Extracts from primary human fibroblasts treated with as indicated were immunoblotted using antibodies against phospho-creb and a-tubulin.
6 Khaled_Fig. S6 A MITF si #2MITF sictrl simitf 2.5 cont 2h 6h 2h PDE4D cont 2h 6h 2h B MITF TUB cont 2h 6h 2h sictrl cont 2h 6h 2h simitf Figure S6 camp-induced upregulation of PDE4D3 is dependent upon MITF. (A) Primary human melanocytes were transfected with a non targeting sirna (sictrl) or sirnas targeting MITF and exposed to as indicated. Total mrna was subjected to qpcr. Results are expressed as fold stimulation and represent the mean ± SD of three independent experiments. () p<.5. (B) primary human melanocytes were transfected with simitf and treated as indicated. Protein extracts were immunoblotted using antibodies against MITF and a-tubulin.
7 + Nuclear extract Mouse IgG + αmitf-ab Competitor Wt Mut Khaled_Fig. S7 MITF supershift Free probe Figure S7 MITF Binds the PDE4D3 Ebox in vitro. Probe containing the Ebox from the human PDE4D3 promoter was used in EMSA to test in vitro binding of MITF to this sequence. UACC 257 melanoma nuclear extracts were used as a source of MITF, and a monoclonal anti-mitf antibody was used for supershift analysis. Supershifted complexe was observed on addition of wild-type biotinilated Ebox Wt probe in the presence of anti-mitf antibody. The supershifted band was competed away with increasing amounts of wild-type unlabeled probe but not with identical amounts of point-mutated unlabeled probe.
8 Khaled_Fig. S8 A Absorbance, variance from mean.5.5 control rolipram +rolipram 24h 48h 72h 96h B Cont Figure S8 Rolipram does not affect human primary melanocyte proliferation and morphology. (A) Human primary melanocyte were seeded in 96 well plate and treated as indicated. Every 24 h for 96h, WST- reagent was added to three well per condition and read at 44 nm 8 h after addition of the reagent. (B) Normal melanoma cells were treated as indicated for 48h. The pictures were taken at 4x magnification. rolipram Forskolin +rolipram
9 Khaled_Fig. S9.25 Pde4d sictrl si#pde4d3 si#2pde4d3 Figure S9 Efficiency of PDE4D sirna. Primary human melanocytes were transfected with a non targeting sirna (si cont) or a sirna specific for PDE4D. Total mrna was subjected to qpcr. The results are normalized relative to b-actin. () P<.5. Results are normalized relative to b-actin. Each data point is the mean ± SD of three experiments.
10 A Pde4d sictrl sipde4d3 DMSO PMEL7 LEF Day DMSO Day 2 t, 8h 8h B sictrl si PDE4d3 si PDE4d5 sipde4d3 sipde4d5 Pde4d sictrl + - sipde4d5 DMSO + - sipde4d h 8h 8h 8h sipde4d3 Khaled_Fig. S Figure S Forskolin pretreatment induces resistance that is reversible by PDE4D3 knockdown but not by PDE4D5 knockdown. (A) Primary human melanocytes were transfected with a non-targeting sirna (sictrl), sirnas targeting PDE4D3 or sirnas targeting PDE4D5. 24h after transfection, the cells were placed in minimal media for 4h and subsequently treated with for 4h.before being returned to minimal media. The following day, the cells were exposed to for the indicated times. Total mrna was subjected to qpcr. The results are normalized relative to b-actin. () p<.5. (B). Total mrna from experiments presented on panel (A) were subjected to qpcr. The results are normalized relative to b- actin. Each data point is the mean ± SD of three experiments.
HCT116 SW48 Nutlin: p53
 Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
CRE/CREB Reporter Assay Kit camp/pka Cell Signaling Pathway Catalog #: 60611
 Data Sheet CRE/CREB Reporter Assay Kit camp/pka Cell Signaling Pathway Catalog #: 60611 Background The main role of the camp response element, or CRE, is mediating the effects of Protein Kinase A (PKA)
Data Sheet CRE/CREB Reporter Assay Kit camp/pka Cell Signaling Pathway Catalog #: 60611 Background The main role of the camp response element, or CRE, is mediating the effects of Protein Kinase A (PKA)
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
/04/$15.00/0 Molecular Endocrinology 18(3): Copyright 2004 by The Endocrine Society doi: /me
 0888-8809/04/$15.00/0 Molecular Endocrinology 18(3):588 605 Printed in U.S.A. Copyright 2004 by The Endocrine Society doi: 10.1210/me.2003-0090 Increased Cytochrome P450 17 -Hydroxylase Promoter Function
0888-8809/04/$15.00/0 Molecular Endocrinology 18(3):588 605 Printed in U.S.A. Copyright 2004 by The Endocrine Society doi: 10.1210/me.2003-0090 Increased Cytochrome P450 17 -Hydroxylase Promoter Function
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Supporting Information
 Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Supplemental Online Material. The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/-
 #1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
#1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1.
 Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p53 activity
 IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p5 activity H6 5 5 H58 A59 H6 H58 A59 anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p5 activity H6 5 5 H58 A59 H6 H58 A59 anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation
 1 2 3 4 5 SUPPLEMENTAL DATA Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation Magalí Nazar, Juan Pablo Nicola, María Laura
1 2 3 4 5 SUPPLEMENTAL DATA Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation Magalí Nazar, Juan Pablo Nicola, María Laura
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information. Mitophagy Controls the Activities. of Tumor Suppressor p53. to Regulate Hepatic Cancer Stem Cells
 Molecular Cell, Volume 68 Supplemental Information Mitophagy Controls the Activities of Tumor Suppressor to Regulate Hepatic Cancer Stem Cells Kai Liu, Jiyoung Lee, Ja Yeon Kim, Linya Wang, Yongjun Tian,
Molecular Cell, Volume 68 Supplemental Information Mitophagy Controls the Activities of Tumor Suppressor to Regulate Hepatic Cancer Stem Cells Kai Liu, Jiyoung Lee, Ja Yeon Kim, Linya Wang, Yongjun Tian,
* ** ** * IB: p-p90rsk. p90rsk (Ser380) (arbitrary units) (Ser380) p90rsk. IB: p90rsk. Tubulin. IB: Tubulin. Ang II (200 nm) Ang II (200 nm)
 I: p-p9rsk I: p9rsk I: C I: p-p9rsk I: p9rsk 5 (ka) 5 5 (min) Ang II ( nm) p-p9rsk (Ser8) p9rsk p-p9rsk (Ser8) p9rsk (h) Mannitol 5 mm -Glucose 5 mm p9rsk (Ser8) (arbitrary units) p-p9rsk (Ser8) (arbitrary
I: p-p9rsk I: p9rsk I: C I: p-p9rsk I: p9rsk 5 (ka) 5 5 (min) Ang II ( nm) p-p9rsk (Ser8) p9rsk p-p9rsk (Ser8) p9rsk (h) Mannitol 5 mm -Glucose 5 mm p9rsk (Ser8) (arbitrary units) p-p9rsk (Ser8) (arbitrary
mir-24-mediated down-regulation of H2AX suppresses DNA repair
 Supplemental Online Material mir-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells Ashish Lal 1,4, Yunfeng Pan 2,4, Francisco Navarro 1,4, Derek M. Dykxhoorn
Supplemental Online Material mir-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells Ashish Lal 1,4, Yunfeng Pan 2,4, Francisco Navarro 1,4, Derek M. Dykxhoorn
Table S1. Primer sequences
 Table S1. Primer sequences Primers for quantitative PCR Tgf 1 Forward Tgf 1 Reverse Tgf 2Forward Tgf 2Reverse Tgf 3 Forward Tgf 3 Reverse Tgf r1 Forward Tgf r1 Reverse Tgf r2 Forward Tgf r2 Reverse Thbs1
Table S1. Primer sequences Primers for quantitative PCR Tgf 1 Forward Tgf 1 Reverse Tgf 2Forward Tgf 2Reverse Tgf 3 Forward Tgf 3 Reverse Tgf r1 Forward Tgf r1 Reverse Tgf r2 Forward Tgf r2 Reverse Thbs1
Supplemental Information
 Molecular Cell, Volume 59 Supplemental Information Interactions of Melanoma Cells with Distal Keratinocytes Trigger Metastasis via Notch Signaling Inhibition of MITF Tamar Golan, Arielle R. Messer, Aya
Molecular Cell, Volume 59 Supplemental Information Interactions of Melanoma Cells with Distal Keratinocytes Trigger Metastasis via Notch Signaling Inhibition of MITF Tamar Golan, Arielle R. Messer, Aya
Int. J. Mol. Sci. 2016, 17, 1259; doi: /ijms
 S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
SureSilencing sirna Array Technology Overview
 SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Hossain_Supplemental Figure 1
 Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Nature Immunology: doi: /ni Supplementary Figure 1. Zranb1 gene targeting.
 Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplemental Information. HEXIM1 and NEAT1 Long Non-coding RNA Form. a Multi-subunit Complex that Regulates. DNA-Mediated Innate Immune Response
 Molecular Cell, Volume 67 Supplemental Information HEXIM1 and NEAT1 Long Non-coding RNA Form a Multi-subunit Complex that Regulates DNA-Mediated Innate Immune Response Mehdi Morchikh, Alexandra Cribier,
Molecular Cell, Volume 67 Supplemental Information HEXIM1 and NEAT1 Long Non-coding RNA Form a Multi-subunit Complex that Regulates DNA-Mediated Innate Immune Response Mehdi Morchikh, Alexandra Cribier,
PrimePCR Assay Validation Report
 Gene Information Gene Name transforming growth factor, beta 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID TGFB1 Human This gene encodes a member of the
Gene Information Gene Name transforming growth factor, beta 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID TGFB1 Human This gene encodes a member of the
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination.
 Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
TDP-43 affects splicing profiles and isoform production of genes involved in the apoptotic and mitotic cellular pathways
 Nucleic Acids Research Advance Access published August 14, 2015 Nucleic Acids Research, 2015 1 doi: 10.1093/nar/gkv814 TDP-43 affects splicing profiles and isoform production of genes involved in the apoptotic
Nucleic Acids Research Advance Access published August 14, 2015 Nucleic Acids Research, 2015 1 doi: 10.1093/nar/gkv814 TDP-43 affects splicing profiles and isoform production of genes involved in the apoptotic
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by. Regulating Occludin
 Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by Regulating Occludin Chaithanya Chelakkot 1,ǂ, Jaewang Ghim 2,3,ǂ, Nirmal Rajasekaran 4, Jong-Sun Choi 5, Jung-Hwan
Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by Regulating Occludin Chaithanya Chelakkot 1,ǂ, Jaewang Ghim 2,3,ǂ, Nirmal Rajasekaran 4, Jong-Sun Choi 5, Jung-Hwan
Supplementary Information. Title BRD3 and BRD4 BET Bromodomain Proteins Differentially Regulate Skeletal Myogenesis
 Supplementary Information Title BRD3 and BRD4 BET Bromodomain Proteins Differentially Regulate Skeletal Myogenesis Authors Thomas C. Roberts 1,2, Usue Etxaniz 1, Alessandra Dall Agnese 1, Shwu-Yuan Wu
Supplementary Information Title BRD3 and BRD4 BET Bromodomain Proteins Differentially Regulate Skeletal Myogenesis Authors Thomas C. Roberts 1,2, Usue Etxaniz 1, Alessandra Dall Agnese 1, Shwu-Yuan Wu
Supplemental Data. Lee et al. Plant Cell. (2010) /tpc Supplemental Figure 1. Protein and Gene Structures of DWA1 and DWA2.
 Supplemental Figure 1. Protein and Gene Structures of DWA1 and DWA2. (A) Protein structures of DWA1 and DWA2. WD40 region was determined based on the NCBI conserved domain databases (B, C) Schematic representation
Supplemental Figure 1. Protein and Gene Structures of DWA1 and DWA2. (A) Protein structures of DWA1 and DWA2. WD40 region was determined based on the NCBI conserved domain databases (B, C) Schematic representation
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat
 Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Materials
 Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
SUPPLEMENTARY INFORMATION
 a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Supplemental Materials. Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans
 Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
RayBio Human NF-κB p65 Transcription Factor Activity Assay Kit
 RayBio Human NF-κB p65 Transcription Factor Activity Assay Kit Catalog #: TFEH-p65 User Manual Mar 13, 2017 3607 Parkway Lane, Suite 200 Norcross, GA 30092 Tel: 1-888-494-8555 (Toll Free) or 770-729-2992,
RayBio Human NF-κB p65 Transcription Factor Activity Assay Kit Catalog #: TFEH-p65 User Manual Mar 13, 2017 3607 Parkway Lane, Suite 200 Norcross, GA 30092 Tel: 1-888-494-8555 (Toll Free) or 770-729-2992,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
TransAM Kits are DNA-binding ELISAs that facilitate the study of transcription factor activation in mammalian tissue and cell culture extracts.
 Transcription Factor ELISAs TransAM sensitive quantitative transcription factor ELISAs TransAM Kits are DNA-binding ELISAs that facilitate the study of transcription factor activation in mammalian tissue
Transcription Factor ELISAs TransAM sensitive quantitative transcription factor ELISAs TransAM Kits are DNA-binding ELISAs that facilitate the study of transcription factor activation in mammalian tissue
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Silencer Select Pre-designed sirna Silencer Select Validated sirna Silencer Select Custom Designed sirna Custom Select sirna
 Catalog #: Various Silencer Select Pre-designed sirna Silencer Select Validated sirna Silencer Select Custom Designed sirna Custom Select sirna General Product Details and User Information Refer to page
Catalog #: Various Silencer Select Pre-designed sirna Silencer Select Validated sirna Silencer Select Custom Designed sirna Custom Select sirna General Product Details and User Information Refer to page
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Lecture #1. Introduction to microarray technology
 Lecture #1 Introduction to microarray technology Outline General purpose Microarray assay concept Basic microarray experimental process cdna/two channel arrays Oligonucleotide arrays Exon arrays Comparing
Lecture #1 Introduction to microarray technology Outline General purpose Microarray assay concept Basic microarray experimental process cdna/two channel arrays Oligonucleotide arrays Exon arrays Comparing
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Suat ERDOGAN* and Miles D. HOUSLAY INTRODUCTION
 Biochem. J. (1997) 321, 165 175 (Printed in Great Britain) 165 Challenge of human Jurkat T-cells with the adenylate cyclase activator forskolin elicits major changes in camp phosphodiesterase (PDE) expression
Biochem. J. (1997) 321, 165 175 (Printed in Great Britain) 165 Challenge of human Jurkat T-cells with the adenylate cyclase activator forskolin elicits major changes in camp phosphodiesterase (PDE) expression
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Supplementary Information
 Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Bronchial epithelium and its associated tissues act as a
 The Journal of Immunology A JNK-Independent Signaling Pathway Regulates TNF -Stimulated, c-jun-driven FRA-1 Protooncogene Transcription in Pulmonary Epithelial Cells 1 Pavan Adiseshaiah,* Dhananjaya V.
The Journal of Immunology A JNK-Independent Signaling Pathway Regulates TNF -Stimulated, c-jun-driven FRA-1 Protooncogene Transcription in Pulmonary Epithelial Cells 1 Pavan Adiseshaiah,* Dhananjaya V.
The microtubule-associated tau protein has intrinsic acetyltransferase activity. Todd J. Cohen, Dave Friedmann, Andrew W. Hwang, Ronen Marmorstein and
 SUPPLEMENTARY INFORMATION: The microtubule-associated tau protein has intrinsic acetyltransferase activity Todd J. Cohen, Dave Friedmann, Andrew W. Hwang, Ronen Marmorstein and Virginia M.Y. Lee Cohen
SUPPLEMENTARY INFORMATION: The microtubule-associated tau protein has intrinsic acetyltransferase activity Todd J. Cohen, Dave Friedmann, Andrew W. Hwang, Ronen Marmorstein and Virginia M.Y. Lee Cohen
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
PrimePCR Assay Validation Report
 Gene Information Gene Name SRY (sex determining region Y)-box 6 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID SOX6 Human This gene encodes a member of the
Gene Information Gene Name SRY (sex determining region Y)-box 6 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID SOX6 Human This gene encodes a member of the
ENCODE RBP Antibody Characterization Guidelines
 ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
SUPPLEMENTAL FIGURE LEGENDS. Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are
 SUPPLEMENTAL FIGURE LEGENDS Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are the amino acid sequences of human DDR2, mouse DDR2 and the closest homologs in zebrafish and C. Elegans.
SUPPLEMENTAL FIGURE LEGENDS Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are the amino acid sequences of human DDR2, mouse DDR2 and the closest homologs in zebrafish and C. Elegans.
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets.
 Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplement Figure 1. Plin5 Plin2 Plin1. KDEL-DSRed. Plin-YFP. Merge
 Supplement Figure 1 Plin5 Plin2 Plin1 KDEL-DSRed Plin-YFP Merge Supplement Figure 2 A. Plin5-Ab MitoTracker Merge AML12 B. Plin5-YFP Cytochrome c-cfp merge Supplement Figure 3 Ad.GFP Ad.Plin5 Supplement
Supplement Figure 1 Plin5 Plin2 Plin1 KDEL-DSRed Plin-YFP Merge Supplement Figure 2 A. Plin5-Ab MitoTracker Merge AML12 B. Plin5-YFP Cytochrome c-cfp merge Supplement Figure 3 Ad.GFP Ad.Plin5 Supplement
Quantitative Real Time PCR USING SYBR GREEN
 Quantitative Real Time PCR USING SYBR GREEN SYBR Green SYBR Green is a cyanine dye that binds to double stranded DNA. When it is bound to D.S. DNA it has a much greater fluorescence than when bound to
Quantitative Real Time PCR USING SYBR GREEN SYBR Green SYBR Green is a cyanine dye that binds to double stranded DNA. When it is bound to D.S. DNA it has a much greater fluorescence than when bound to
PrimePCR Assay Validation Report
 Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin)
Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin)
Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and
 Supplementary Tables Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and their side chain contacting residues in the second chain (B) a Interface Res. in Contacting
Supplementary Tables Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and their side chain contacting residues in the second chain (B) a Interface Res. in Contacting
Introduction to Microarray Analysis
 Introduction to Microarray Analysis Methods Course: Gene Expression Data Analysis -Day One Rainer Spang Microarrays Highly parallel measurement devices for gene expression levels 1. How does the microarray
Introduction to Microarray Analysis Methods Course: Gene Expression Data Analysis -Day One Rainer Spang Microarrays Highly parallel measurement devices for gene expression levels 1. How does the microarray
2/5/16. Honeypot Ants. DNA sequencing, Transcriptomics and Genomics. Gene sequence changes? And/or gene expression changes?
 2/5/16 DNA sequencing, Transcriptomics and Genomics Honeypot Ants "nequacatl" BY2208, Mani Lecture 3 Gene sequence changes? And/or gene expression changes? gene expression differences DNA sequencing, Transcriptomics
2/5/16 DNA sequencing, Transcriptomics and Genomics Honeypot Ants "nequacatl" BY2208, Mani Lecture 3 Gene sequence changes? And/or gene expression changes? gene expression differences DNA sequencing, Transcriptomics
Student Learning Outcomes (SLOS)
 Student Learning Outcomes (SLOS) KNOWLEDGE AND LEARNING SKILLS USE OF KNOWLEDGE AND LEARNING SKILLS - how to use Annhyb to save and manage sequences - how to use BLAST to compare sequences - how to get
Student Learning Outcomes (SLOS) KNOWLEDGE AND LEARNING SKILLS USE OF KNOWLEDGE AND LEARNING SKILLS - how to use Annhyb to save and manage sequences - how to use BLAST to compare sequences - how to get
466 Asn (N) to Ala (A) Generate beta dimer Interface
 Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
Supplemental Information. The TRAIL-Induced Cancer Secretome. Promotes a Tumor-Supportive Immune. Microenvironment via CCR2
 Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Real-time PCR. TaqMan Protein Assays. Unlock the power of real-time PCR for protein analysis
 Real-time PCR TaqMan Protein Assays Unlock the power of real-time PCR for protein analysis I can use my real-time PCR instrument to quantitate protein? Do protein levels correlate with related mrna levels
Real-time PCR TaqMan Protein Assays Unlock the power of real-time PCR for protein analysis I can use my real-time PCR instrument to quantitate protein? Do protein levels correlate with related mrna levels
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
EGFR (Phospho-Ser695)
 Assay Biotechnology Company www.assaybiotech.com Tel: 1-877-883-7988 Fax: 1-877-610-9758 EGFR (Phospho-Ser695) Colorimetric Cell-Based ELISA Kit Catalog #: OKAG02090 Please read the provided manual entirely
Assay Biotechnology Company www.assaybiotech.com Tel: 1-877-883-7988 Fax: 1-877-610-9758 EGFR (Phospho-Ser695) Colorimetric Cell-Based ELISA Kit Catalog #: OKAG02090 Please read the provided manual entirely
TECHNICAL BULLETIN. Color. Fig.1. Cell-Based protein phosphorylation procedure
 Cell-Based ELISA Kit for detecting phospho-stat3 (ptyr 705 ) in cultured cell lines adequate for 96 assays (1 96 well plate) Catalog Number RAB0444 Storage Temperature 20 C TECHNICAL BULLETIN Product Description
Cell-Based ELISA Kit for detecting phospho-stat3 (ptyr 705 ) in cultured cell lines adequate for 96 assays (1 96 well plate) Catalog Number RAB0444 Storage Temperature 20 C TECHNICAL BULLETIN Product Description
Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death
 SUPPLEMENTAL DATA Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death Huajun Jin 1, Arthi Kanthasamy 1, Vellareddy Anantharam
SUPPLEMENTAL DATA Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death Huajun Jin 1, Arthi Kanthasamy 1, Vellareddy Anantharam
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Figure 1 Activated B cells are subdivided into three groups
 Supplementary Figure 1 Activated B cells are subdivided into three groups according to mitochondrial status (a) Flow cytometric analysis of mitochondrial status monitored by MitoTracker staining or differentiation
Supplementary Figure 1 Activated B cells are subdivided into three groups according to mitochondrial status (a) Flow cytometric analysis of mitochondrial status monitored by MitoTracker staining or differentiation
INOS. Colorimetric Cell-Based ELISA Kit. Catalog #: OKAG00807
 INOS Colorimetric Cell-Based ELISA Kit Catalog #: OKAG00807 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research Purposes Only. Not Intended
INOS Colorimetric Cell-Based ELISA Kit Catalog #: OKAG00807 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research Purposes Only. Not Intended
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl
 Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Supplementary Information
 Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
CytoGLOW. IKK-α/β. Colorimetric Cell-Based ELISA Kit. Catalog #: CB5358
 CytoGLOW IKK-α/β Colorimetric Cell-Based ELISA Kit Catalog #: CB5358 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research Purposes Only.
CytoGLOW IKK-α/β Colorimetric Cell-Based ELISA Kit Catalog #: CB5358 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research Purposes Only.
Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors
 Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors A Vector GR1 SacI BamHI CTCTGGCTAACTAGGC Insert 5/7nt - G TCGAGAGACCGATTGATCCG Insert 5/7nt - CCTAG 1 G CAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAAC
Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors A Vector GR1 SacI BamHI CTCTGGCTAACTAGGC Insert 5/7nt - G TCGAGAGACCGATTGATCCG Insert 5/7nt - CCTAG 1 G CAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAAC
SEM Immunocytochemistry for Cells & Materials
 SEM Immunocytochemistry for Cells & Materials R. Geoff Richards AO Research Institute, AO Foundation, Davos, Switzerland. Immunohistochemistry Immunocytochemistry can be performed on a biological specimen,
SEM Immunocytochemistry for Cells & Materials R. Geoff Richards AO Research Institute, AO Foundation, Davos, Switzerland. Immunohistochemistry Immunocytochemistry can be performed on a biological specimen,
TECHNICAL BULLETIN. Color. Fig.1. Cell-Based protein phosphorylation procedure
 Cell-Based ELISA Sampler Kit for detecting phospho-erk1/2 (pthr 202 /ptyr 204 ), phospho-jnk (pthr 183 /ptyr 185 ), and phospho-p38 MAPK (pthr 180 /ptyr 182 ) in cultured cell lines adequate for 192 assays
Cell-Based ELISA Sampler Kit for detecting phospho-erk1/2 (pthr 202 /ptyr 204 ), phospho-jnk (pthr 183 /ptyr 185 ), and phospho-p38 MAPK (pthr 180 /ptyr 182 ) in cultured cell lines adequate for 192 assays
Xfect Protein Transfection Reagent
 Xfect Protein Transfection Reagent Mammalian Expression Systems Rapid, high-efficiency, low-toxicity protein transfection Transfect a large amount of active protein Virtually no cytotoxicity, unlike lipofection
Xfect Protein Transfection Reagent Mammalian Expression Systems Rapid, high-efficiency, low-toxicity protein transfection Transfect a large amount of active protein Virtually no cytotoxicity, unlike lipofection
Soybean Microarrays. An Introduction. By Steve Clough. November Common Microarray platforms
 Soybean Microarrays Microarray construction An Introduction By Steve Clough November 2005 Common Microarray platforms cdna: spotted collection of PCR products from different cdna clones, each representing
Soybean Microarrays Microarray construction An Introduction By Steve Clough November 2005 Common Microarray platforms cdna: spotted collection of PCR products from different cdna clones, each representing
QUANTITATIVE RT-PCR PROTOCOL (SYBR Green I) (Last Revised: April, 2007)
 QUANTITATIVE RT-PCR PROTOCOL (SYBR Green I) (Last Revised: April, 007) Please contact Center for Plant Genomics (CPG) facility manager Hailing Jin (hljin@iastate.edu) regarding questions or corrections.
QUANTITATIVE RT-PCR PROTOCOL (SYBR Green I) (Last Revised: April, 007) Please contact Center for Plant Genomics (CPG) facility manager Hailing Jin (hljin@iastate.edu) regarding questions or corrections.
GUGAUAAUGGAGCGAGAUUUUCUGUUGUGCUUGAUCUAACCAUGUGCUUGCGAGGUAUGA GAAAAACAUGGUUCCGUCAAGCACCAUGGAACGUCACGCAGCUUUCUACA
 Precursor mmu-mir-8- GUGAUAAUGGAGCGAGAUUUUCUGUUGUGCUUGAUCUAACCAUGUGCUUGCGAGGUAUGA GAAAAACAUGGUUCCGUCAAGCACCAUGGAACGUCACGCAGCUUUCUACA Precursor mmu-mir-8- GACCAGUUGCCGCGGGGCUUUCCUUUGUGCUUGAUCUAACCAUGUGGUGGAACGAUGGAA
Precursor mmu-mir-8- GUGAUAAUGGAGCGAGAUUUUCUGUUGUGCUUGAUCUAACCAUGUGCUUGCGAGGUAUGA GAAAAACAUGGUUCCGUCAAGCACCAUGGAACGUCACGCAGCUUUCUACA Precursor mmu-mir-8- GACCAGUUGCCGCGGGGCUUUCCUUUGUGCUUGAUCUAACCAUGUGGUGGAACGAUGGAA
measuring gene expression December 5, 2017
 measuring gene expression December 5, 2017 transcription a usually short-lived RNA copy of the DNA is created through transcription RNA is exported to the cytoplasm to encode proteins some types of RNA
measuring gene expression December 5, 2017 transcription a usually short-lived RNA copy of the DNA is created through transcription RNA is exported to the cytoplasm to encode proteins some types of RNA
DNA makes RNA makes Proteins. The Central Dogma
 DNA makes RNA makes Proteins The Central Dogma TRANSCRIPTION DNA RNA transcript RNA polymerase RNA PROCESSING Exon RNA transcript (pre-mrna) Intron Aminoacyl-tRNA synthetase NUCLEUS CYTOPLASM FORMATION
DNA makes RNA makes Proteins The Central Dogma TRANSCRIPTION DNA RNA transcript RNA polymerase RNA PROCESSING Exon RNA transcript (pre-mrna) Intron Aminoacyl-tRNA synthetase NUCLEUS CYTOPLASM FORMATION
Technical Review. Real time PCR
 Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
DNA Arrays Affymetrix GeneChip System
 DNA Arrays Affymetrix GeneChip System chip scanner Affymetrix Inc. hybridization Affymetrix Inc. data analysis Affymetrix Inc. mrna 5' 3' TGTGATGGTGGGAATTGGGTCAGAAGGACTGTGGGCGCTGCC... GGAATTGGGTCAGAAGGACTGTGGC
DNA Arrays Affymetrix GeneChip System chip scanner Affymetrix Inc. hybridization Affymetrix Inc. data analysis Affymetrix Inc. mrna 5' 3' TGTGATGGTGGGAATTGGGTCAGAAGGACTGTGGGCGCTGCC... GGAATTGGGTCAGAAGGACTGTGGC
(A) Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site.
 SUPPLEMENTRY INFORMTION SUPPLEMENTL FIGURES Figure S1. () Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site. () Western blot of ligated and unligated sciatic
SUPPLEMENTRY INFORMTION SUPPLEMENTL FIGURES Figure S1. () Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site. () Western blot of ligated and unligated sciatic
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX
 Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
