N-WASP Involvement in Dorsal Ruffle Formation in D V
|
|
|
- Briana Lamb
- 6 years ago
- Views:
Transcription
1 Molecular Biology of the Cell Vol. 18, , February 2007 N-WASP Involvement in Dorsal Ruffle Formation in D V Mouse Embryonic Fibroblasts John A. Legg,* Guillaume Bompard, John Dawson,* Hannah L. Morris,* Natalie Andrew,* Lisa Cooper, Simon A. Johnston,* Giorgos Tramountanis,* and Laura M. Machesky* Schools of *Biosciences and Medicine, The University of Birmingham, Birmingham, Edgbaston, West Midlands, B15 2TT, United Kingdom; and Centre de Recherches de Biochimie Macromoléculaire, Centre National de la Recherche Scientifique FRE2593, Montpellier, Cedex 5, France Submitted June 30, 2006; Revised November 22, 2006; Accepted November 30, 2006 Monitoring Editor: David Drubin The Wiskott Aldrich syndrome protein (WASP) family activates the Arp2/3 complex leading to the formation of new actin filaments. Here, we study the involvement of Scar1, Scar2, N-WASP, and Arp2/3 complex in dorsal ruffle formation in mouse embryonic fibroblasts (MEFs). Using platelet-derived growth factor to stimulate circular dorsal ruffle assembly in primary E13 and immortalized E9 Scar1 / and Scar1 null MEFs, we establish that Scar1 loss does not impair the formation of dorsal ruffles. Reduction of Scar2 protein levels via small interfering RNA (sirna) also did not affect dorsal ruffle production. In contrast, wiskostatin, a chemical inhibitor of N-WASP, potently suppressed dorsal ruffle formation in a dose-dependent manner. Furthermore, N-WASP and Arp2 sirna treatment significantly decreased the formation of dorsal ruffles in MEFs. In addition, the expression of an N-WASP truncation mutant that cannot bind Arp2/3 complex blocked the formation of these structures. Finally, N-WASP / fibroblast-like cells generated aberrant dorsal ruffles. These ruffles were highly unstable, severely depleted of Arp2/3 complex, and diminished in size. We hypothesize that N-WASP and Arp2/3 complex are part of a multiprotein assembly important for the generation of dorsal ruffles and that Scar1 and Scar2 are dispensable for this process. INTRODUCTION The actin cytoskeleton is crucial for numerous cellular processes, including cell motility, vesicle trafficking, and cell division. The generation of specialized F-actin structures, such as lamellipodia, dorsal ruffles, and filopodia enable actin to function in these diverse cellular events. One pathway that regulates the formation of these structures involves the Wiskott Aldrich Syndrome protein (WASP) family proteins and the Arp2/3 complex (Machesky et al., 1999; Suetsugu et al., 2003). Dorsal ruffles are short-lived, dynamic, F-actin enriched waves that occur across the top surface of the cell (for review, see Buccione et al., 2004). Also known as actin ribbons, these structures are generated by fibroblasts and differentiated epithelial cell types in response to agonists such as platelet-derived growth factor (PDGF) and epidermal growth factor. Dorsal ruffles are transient in nature, typically occurring after 2 min of agonist stimulation and disappearing within 30 min. Numerous cellular roles have been attributed to dorsal ruffles. These roles include preparing a static cell for subsequent movement (Krueger et al., 2003), macropinocytosis (Dowrick et al., 1993; Araki et al., 2000), This article was published online ahead of print in MBC in Press ( ) on December 20, D V The online version of this article contains supplemental material at MBC Online ( Address correspondence to: Laura M. Machesky (l.m.machesky@ bham.ac.uk) or John A. Legg (jal245@bham.ac.uk). and the internalization of cell surface receptors (Orth et al., 2006). In mammalian cells, there are five members of the WASP family, called WASP, N-WASP, and Scar/WAVE1-3 (for review, see Millard et al., 2004). These proteins are important activators of Arp2/3 complex-induced actin nucleation (Machesky and Insall, 1998; Miki et al., 1998; Machesky et al., 1999). At their C termini, all the WASP proteins contain G-actin and Arp2/3 complex binding motifs that enable them to form a trimer nucleus with the Arp2/3 complex and to induce de novo actin polymerization. At the extreme N termini, the Scar proteins have a unique Scar homology domain (SHD) (Bear et al., 1998). Instead of an SHD, WASP and N-WASP have Enabled/VASP homology 1/WASP homology 1 and Cdc42 and Rac interactive binding (CRIB) domains. The differences in domain structures at the N termini are important in defining the regulation of these proteins. Both WASP and N-WASP interact via their CRIB motifs with GTP-Cdc42, relieving an autoinhibited state and allowing them to bind and activate the Arp2/3 complex (Rohatgi et al., 1999; Higgs and Pollard, 2000). However, the Scar proteins do not possess a CRIB domain and their activity is regulated by the Scar complex proteins (reviewed in Bompard and Caron, 2004). Scar1 and Scar2 were proposed to have differential roles in lamellipodia and dorsal ruffle formation. Loss of Scar1 resulted in loss of dorsal ruffles but not lamellipodia, whereas Scar2 deficiency led to decreased lamellipodia but not dorsal ruffles (Suetsugu et al., 2003). In addition to Scar1 and Scar2, the WASP family member N-WASP has been localized to dorsal ruffles along with WIP, dynamin 2, and cortactin after PDGF stimulation (Anton et al., 2003; Krueger et al., 2003) by The American Society for Cell Biology
2 N-WASP in Dorsal Ruffle Formation Inhibition of dynamin-cortactin binding by the injection of anti-dynamin 2 antibodies or overexpression of truncation mutants abrogated dorsal ruffle formation, indicating that an interaction between these proteins is required for wave formation (Krueger et al., 2003). WIP presence is also important for dorsal ruffle formation (Anton et al., 2003). Microinjection of anti-wip antibodies or the absence of WIP in fibroblasts resulted in decreased and delayed PDGF-induced dorsal ruffle production. Anton et al. (2003) proposed that WIP, in conjunction with its binding partner cortactin, could both function in dorsal ruffle formation through activation of the Arp2/3 complex and stabilization of actin filaments. After PDGF stimulation, N-WASP deficient fibroblasts still formed lamellipodia; however, the effects on dorsal ruffle formation have not been studied in detail (Snapper et al., 2001). In this study, we investigated the roles of the WASP family proteins in dorsal ruffle formation after PDGF treatment. Using two separate sources of Scar1 null mouse embryonic fibroblasts (MEFs), Scar1 deficiency did not impair the generation or kinetics of dorsal ruffles. In addition, sirna knockdown of Scar1 and Scar2 protein levels did not result in the decreased formation of dorsal ruffles, indicating either functional redundancy between Scar1 and Scar2 or a lack of requirement in dorsal ruffles. In contrast, treatment of MEFs with wiskostatin, a chemical inhibitor of N-WASP, potently inhibited dorsal ruffle formation in a dose-dependent manner. Depletion of N-WASP and Arp2 levels with sirna oligonucleotides (oligos) resulted in a statistically significant reduction in the production of dorsal ruffles. In addition, ectopic expression of an N-WASP truncation mutant deficient in Arp2/3 complex binding inhibited dorsal ruffle formation. Therefore, a complex of N-WASP and Arp2/3 complex is required for dorsal ruffle formation in MEFs. Finally, N-WASP null fibroblast-like cells (FLCs) generated actin protrusions in response to PDGF, but these structures were highly unstable, reduced in frequency and size, and depleted of Arp2/3 complex. In conclusion, it seems that N-WASP and not Scar1 or Scar2, is the WASP family member required for robust dorsal ruffle formation in MEFs. MATERIALS AND METHODS Antibodies and Reagents All chemicals were purchased from Sigma Chemical (Poole, Dorset, United Kingdom) unless stated otherwise. Antibodies were from the following sources: anti-scar1 (Launay et al., 2003), anti-scar2, anti-scar3, and anti-p34arc (Upstate Biotechnology, Lake Placid, NY), anti-n-wasp (kind gift of Pontus Aspenstrom, Uppsala, Sweden), horseradish peroxidase-conjugated secondary antibody anti-rabbit (Jackson ImmunoResearch Laboratories, West Grove, PA), goat anti-rabbit fluorescein isothiocyanate (FITC) and Alexa-488 conjugates (Invitrogen, Paisley, United Kingdom). DNA Constructs N-WASP constructs were a kind gift of Adrian Thrasher (Hospital for Sick Children, London, United Kingdom). Scar A constructs were cloned into prk5-myc and have been described previously (Machesky and Insall, 1998; Machesky et al., 1999). Human WASP full-length and bovine N-WASP fulllength and N-WASP WWCA (residues 1-404) constructs were cloned into pegfpn1 (Clontech, Mountain View, CA). Cell Culture and Lysis The immortalized wild-type and Scar1 knockout cells were generated from E9 embryos and were a kind gift from Yoshifumi Itoh (Imperial College London, London, United Kingdom). The E9.5 N-WASP FLCs were a kind gift from Scott Snapper (Massachusetts General Hospital, Boston, MA). These cells were cultured in DMEM containing 10% fetal bovine serum (FBS). Cell lysates were generated from a dish of confluent cells by incubating for 15 min in ice-cold lysis buffer (1% Triton-X-100, 50 mm Tris-HCl, ph 7.5, 150 mm NaCl, 1 mm EDTA, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, and 1 g/ml each of chymostatin, aprotinin, leupeptin, and pepstatin). Lysates were clarified by centrifugation at 15,000 g for 10 min at 4 C and the protein concentration was determined by Bio-Rad protein assay. Preparation of Primary Wild-Type and Scar1 Null MEFs The Scar1 null mice were a kind gift from Seung Kwak (Wyeth-Ayerst, Princeton, NJ). Heterozygote Scar1 / mice were mated, and the embryos were extracted from embryo day 13 (E13) pregnant mice. The embryo heads were removed and kept for genotyping (see methods below). All cell lines used subsequently for this report were probed with an anti-scar1 antibody to confirm the absence of Scar1 in the knockout MEFs. Dissected embryos were trypsinized and resuspended in DMEM containing 10% FBS and plated out into 10-cm dishes in an incubator at 37 C, 5% CO 2. After 24 h, the medium was changed, and the cells then left to reach confluence. Scar1, Scar2, Arp2, and N-WASP sirna Treatment All sirna oligos were purchased as sigenome SMARTpool mix of four oligos targeted specifically against mouse Scar1, Scar2, N-WASP, and Arp2 (Dharmacon, RNA Technologies, Lafayette, CO). We transfected 75 nmol sigenome duplex in MEFs by using Lipofectamine 2000 transfection reagent (Invitrogen) for 48 h per manufacturer s instructions. Polymerase Chain Reaction (PCR) Protocol for Genotyping The DNA preparation from tail tips and embryos was done following the protocol in Laird et al. (1991). Primers were designed so that the wild-type mice will only produce a 400-bp band, the heterozygotes will have 400- and 150-bp bands, and the Scar1 knockouts will have a 150-bp band (see Supplemental Figure 1 for details). The primers were diluted to a final concentration working stock of 6.6 M and used in the following PCR reaction: WAVE1_1F: 5 -CCT- TAGCTCATCATCAGGAC-3 ; WAVE1_2R: 5 -AGTAATTAGCACAAACCA- TGG-3 ; LTR2-2F: 5 -TGGCGTTACTTAAGCTAGCT-3 ; and PCR program: 1) 94 C for 3 min, 2) 94 C for 30 s, 3) 60 C for 45 s, 4) 72 C for 1 min, 5) go to 2 and repeat for 35 cycles, and 6) 72 C for 8 min. Immunofluorescence Microscopy Cells were fixed in 4% formaldehyde for 10 min, neutralized in 50 mm NH 4 Cl in phosphate-buffered saline (PBS) for 10 min, and permeabilized for 4 min in 0.1% Triton X-100/PBS. Cells were stained with 1:25 anti-scar1, 1:25 anti- Scar2, 1:50 anti-n-wasp, and 1:500 anti-cortactin antibodies. Secondary antibodies used were goat anti-rabbit FITC and Alexa-488 conjugates (Invitrogen). To visualize F-actin, tetramethylrhodamine B isothiocyanate (TRITC) phalloidin or Alexa-350 phalloidin (Invitrogen) diluted 1:40 in PBS were used. Coverslips were incubated in primary and secondary antibodies for 30 min with three washes in PBS between every step. The coverslips were mounted in Mowiol containing p-phenyldiamine antifade. Slides were viewed using an MRC100 confocal laser scanning microscope (Bio-Rad, Hercules, CA) or a using a Zeiss Axioskop2 microscope equipped with a digital camera C (Hamamatsu, Bridgewater, NJ) and 63 oil immersion objective and processed using Openlab (Improvision, Coventry, United Kingdom) and Adobe Photoshop 7.0 software (Adobe Systems, Mountain View, CA) for Macintosh. PDGF and Wiskostatin Assays The MEFs growing on glass coverslips were washed twice in PBS and incubated overnight in serum-free media (DMEM alone). The media were removed and PDGF-BB (Calbiochem, San Diego, CA) (10 g/ml stock) was diluted in serum-free media to desired concentration (10 ng/ml in all experiments except Supplemental Figure 3 where 2, 1, and 0.5 ng/ml were used) and added to the coverslips. In the wiskostatin experiments, the MEFs were preincubated with wiskostatin (Calbiochem) or DMSO carrier in serum-free media for 5 min before incubation in media containing wiskostatin and 10 ng/ml PDGF. After fixed incubation lengths at 37 C, 5% CO 2, the coverslips were washed in PBS, fixed, and stained. Time-Lapse Microscopy The MEFs were allowed to adhere to WillCo-35-mm glass bottom dishes (WPI, Sarasota, FL) and were serum starved overnight. The media were removed and replaced with 2 ml of prewarmed DMEM containing 10 ng/ml PDGF. The cells were then observed with phase-contrast microscopy at 5 s/frame by using a Zeiss Axiovert 100 microscope equipped with a QICAM- FAST B bit camera and 32 objective. Measurements of Dorsal Ruffle and Cell Areas After 5 min PDGF stimulation, the N-WASP FLCs were washed twice in PBS, fixed, and stained for F-actin. To determine the average area of the dorsal ruffles, 30 images were taken from random fields of view on the coverslips by using a Zeiss Axioskop2 microscope equipped with a Hamamatsu digital camera C and 63 oil immersion objective. These images were opened Vol. 18, February
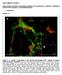
3 J. A. Legg et al. Figure 1. Dorsal ruffle production in E13 primary MEFs. (A) DNA isolated from embryos and mouse tail tips were used in a PCR genotyping reaction. Products were run on a 2% agarose gel. Lane 1, 100-bp ladder; lane 2, heterozygote; lane 3, wild-type; and lane 4, Scar1 null. (B) We analyzed 50 g of total protein from wildtype and Scar1 null MEFs by immunoblot with anti- Scar1, Scar2, and actin antibodies. (C) Wild-type and Scar1 null MEFs were stimulated with PDGF for 0, 2, 5, and 7 min. The cells were fixed and stained for F-actin and the percentage of cells generating at least one dorsal ruffle was determined. (D) The MEFs were PDGF stimulated for 2, 5, and 7 min, and the average number of dorsal ruffles (white arrows) per cell containing at least one dorsal ruffle was calculated. For C and D, 200 cells were counted per time point, and each point represents the average value from three independent experiments (error bars SD). Student s t test p values are shown on graphs. in ImageJ, and each dorsal ruffle circumference was outlined (McCarty et al., 2004). A Java plugin for ImageJ was then used to calculate the number of pixels contained within the outline of the dorsal ruffles. This was then repeated to measure the total areas of the cells. Imaging of a graticule then enabled the areas to be converted from pixels to micrometers. Each assay was repeated three times, measuring 30 dorsal ruffles per experiment. RESULTS Dorsal Ruffle Formation in Scar1 Null E13 Primary MEFs To investigate the role of Scar1 in dorsal ruffle formation, we generated wild-type and Scar1 null primary MEFs. The Scar1 knockout mice were generated using a retroviral genetrap insertion (Dahl et al., 2003). Scar1 null mice displayed numerous neuroanatomical abnormalities and postnatal lethality and so could not be used for breeding (Dahl et al., 2003). Therefore, heterozygote mice were bred to generate Scar1 null embryos. Embryonic day 13 (E13) embryos were used to generate primary MEFs. The embryo heads were used in genotyping PCR reactions (Figure 1A). For explanation of the genotyping protocol, refer to Supplemental Figure 1. Wild-type embryos produced a 400-bp band, and 680 Scar1 null embryos had a 150-bp PCR product. Protein lysates from wild-type and Scar1 null MEFs also were probed with anti-scar1, anti-scar2, and anti-actin antibodies (Figure 1B). The anti-scar1 antibody showed a strong band at approximately 66 kda in the wild-type lysate that was absent in the Scar1 null lane. This confirmed the loss of Scar1 protein expression in the knockout MEFs. Anti-Scar2 immunoblots showed the presence of a 70-kDa band in both wild-type and Scar1 null cells. Scar2 levels remained comparable between the two cell types, indicating there was no increase in Scar2 expression to compensate for Scar1 loss. Consistent with previous reports, an anti-scar3 blot showed no detectable expression of Scar3 in either wild-type or Scar1 knockout MEFs but did detect a strong band in mouse brain lysates and COS-7 cell lysates transfected with full-length Scar3-myc (data not shown; Suetsugu et al., 2003). Endogenous Scar1 and Scar2 localized to dorsal ruffles, and Scar2 (but not Scar1) consistently localized to peripheral lamellipodia (Supplemental Figure 2). To investigate the role of Scar1 in dorsal ruffle production, serum-starved MEFs were treated with 10 ng/ml PDGF for 0, 2, 5, and 7 min. Molecular Biology of the Cell
4 N-WASP in Dorsal Ruffle Formation At these time points, the cells were fixed and stained for F-actin and endogenous Arp2/3 complex (anti-p34). The percentage of cells with dorsal ruffles at each time point is shown in Figure 1C. At 0 min, the serum-starved cells had no dorsal ruffles, consistent with cells requiring agonist stimulation to generate dorsal ruffles. By 2 min, the cells started to generate dorsal ruffles, with 5% of the wild-type and the Scar1 null cells containing at least one dorsal ruffle. At 5 min, dorsal ruffle numbers have reached an optimum, with 30% of cells containing dorsal ruffles. The percentage of cells with dorsal ruffles then starts to decrease at 7 min as some ruffles close and the number tends to zero after 20 min (data not shown). Staining with anti-p34 showed a strong enrichment of endogenous Arp2/3 complex within the dorsal ruffles and at peripheral ruffles (data not shown). Importantly, at all time points, the Scar1 null cells produced the same percentage of dorsal ruffles as the wild-type cells, as shown by Student s t tests (see Figure 1C for p values). Figure 1D shows an example of a wild-type MEF producing three dorsal ruffles after 5 min of PDGF treatment. Therefore, because MEFs can produce multiple dorsal ruffles per cell, we calculated the average total number of dorsal ruffles per cell in a time course. Cells without dorsal ruffles were not included in this assay, therefore, no value was recorded at 0 min. Figure 1D shows the average number of dorsal ruffles per cell after 2, 5, and 7 min PDGF treatment. Interestingly, no optimal value was recorded at 5 min, with the response staying constant at two dorsal ruffles per cell at all time points. This is consistent with the dorsal ruffles forming, expanding, and contracting over a time scale of minutes and rarely splitting into multiple separate ruffles at later time points. Both the wild-type and Scar1 null cells showed the same constant response to the PDGF. Therefore, Scar1 is not essential for the generation of dorsal ruffles by the E13 primary MEFs in response to PDGF. Dorsal ruffles are highly dynamic structures, expanding and contracting over time. PDGF treatment of MEFs was therefore followed using real-time phase microscopy to determine if Scar1 loss affected dorsal ruffle dynamics. Representative movies are shown in Figure 2 (A1-A5 wild-type, B1-B5 Scar1 null MEFs; Supplemental Movies 1 and 2, respectively). After 1 min of PDGF treatment, dark punctate structures start occurring on the dorsal surface of the MEFs (A2 and B2 arrows). By 5 min, the dorsal ruffles have fully formed, consistent with the optimum peak seen in Figure 1C. These waves pulse, expand, and contract over time before finally closing (A5 and B5). Interestingly, phase bright macropinosomes formed upon closure of the dorsal ruffles in both wild-type and Scar1 null MEFs (A5 and B5 arrowheads). A magnified example of macropinosome formation is shown in images C1 C4 (Supplemental Movie 3). As the dorsal ruffle zips closed from right to left, significant numbers of phase-bright, 1- to 2- m-wide macropinosomes are formed (C2 C4, arrowheads). The visualization of these structures is consistent with reports that dorsal ruffles are involved in macropinocytosis (Dowrick et al., 1993; Araki et al., 2000). Imaging multiple dorsal ruffles from wild-type and Scar1 null cells did not reveal any qualitative differences in dorsal ruffle dynamics. Both cell types generated waves through dark punctate precursors, which then expanded into large dorsal ruffles. These structures constricted over time and closed normally to generate macropinosomes. Therefore, Scar1 deficiency does not detectably affect the dynamics of dorsal ruffle formation or closure. Figure 2. Time-lapse phase microscopy of the primary E13 wildtype and Scar1 null cells producing dorsal ruffles in response to 10 ng/ml PDGF treatment. A1 A6, wild-type cell. B1 B6, Scar1 null cell. C1 C4, enlarged movie of a dorsal ruffle in a Scar1 null cell. Arrows follow the formation of the dorsal ruffles. Arrowheads indicate phase bright macropinosome formation during dorsal ruffle closure. Time in seconds is indicated in top right corner. Bar, 20 m (see Movies 1, 2, and 3). Dorsal Ruffle Formation in E9 Scar1 Null Immortalized MEFs The results obtained in Figures 1 and 2 were particularly significant because they contradict previous findings that Scar1 is important for dorsal ruffle formation in MEFs (Suetsugu et al., 2003). This could be due to differences in the genetic background or preparation of the MEFs used for these studies, although the background C57BL/6 was reported to be the same in both cases. Because Suetsugu et al. (2003) used E9 MEFs, it is possible that the E13 primary MEFs have undergone further differentiation/gene expression changes to compensate for the loss of Scar1. Therefore, it was necessary to confirm the results with the same MEFs used in the previous study. We obtained immortalized E9 wild-type and Scar1 null MEFs (a gift from Y. Itoh and T. Takenawa, University of Tokyo, Tokyo, Japan) and repeated the PDGF assays to investigate dorsal ruffle production (Figure 3). Lysates from the E13 primary and E9 immortalized wild-type and Scar1 null MEFs were probed with anti-scar1, Scar2, and actin antibodies (Figure 3A). These immunoblots confirmed the loss of Scar1 in the immortalized knockout cells with Scar2 levels staying constant (Figure 3A). The overall intensity of the Scar1 and Scar2 staining between the wild-type and immortalized cells was also comparable, indicating that both MEF sources expressed similar levels of Vol. 18, February
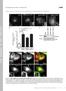
5 J. A. Legg et al. Scar1 and Scar2. None of the MEFs had any detectable Scar3 expression (data not shown). The immortalized MEFs were stimulated with 10 ng/ml PDGF, and the percentage of cells producing dorsal ruffles was determined (Figure 3, B and C). Phalloidin staining of the MEFs revealed that nearly 100% of the wild-type and Scar1 null cells produced dorsal ruffles after 5-min PDGF treatment (Figure 3C). This response was much stronger than with the primary MEFs, where only 30% of the cells produced dorsal ruffles after 5 min (Figure 1C). The dorsal ruffles in the immortalized MEFs were also much more persistent, with a significant percentage of cells retaining dorsal ruffles even after 15-min stimulation (Figure 3C). This difference between primary and immortalized MEFs in dorsal ruffle formation could result from additional genetic/ gene expression changes induced during cell transformation, which potentially increase the sensitivity to growth factor stimulation. Regardless of these differences, the wildtype and Scar1 null-immortalized MEFs responded identically to PDGF stimulation, with equal number of cells generating dorsal ruffles at all time points considered as shown by Student s t tests (Figure 3C). As mentioned, treatment of the immortalized MEFs with 10 ng/ml PDGF induced a robust, 100% response in dorsal ruffle formation. Taking into account a potential increase in sensitivity to PDGF in those cells, the lack of an obvious phenotype could therefore be due to saturating amount of growth factor. To test this hypothesis, decreasing amounts of PDGF were used to stimulate the immortalized MEFs, and the percentage of wild-type and Scar1 null cells producing dorsal ruffles was determined (Supplemental Figure 3). With 2 ng/ml PDGF (graph A), the response was skewed, with the response at 2 min dropping from 30 to 2% and the optimum moving from 5 to 7 min. At 1 and 0.5 ng/ml, the response was greatly reduced, with peak responses at 7 min of only 20 and 10%, respectively (graphs B and C). Independent of PDGF concentration or strength of response, the wild-type and Scar1 null immortalized MEFs produced a statistically similar response at all time points considered (Student s t test p values shown on graphs). Therefore, neither the E13 primary nor the E9 immortalized Scar null MEFs showed any defect in the production of dorsal ruffles, indicating that Scar1 is not essential for dorsal ruffle formation. Figure 3. Dorsal ruffle production in E9 immortalized MEFs. (A) We analyzed 50 g of total protein from E13 primary and E9 immortalized wild-type and Scar1 null MEFs by immunoblot with anti-scar1, Scar2, and actin antibodies. (B) Representative picture showing phalloidin staining of Scar1 null E9 MEFs after 5 min of PDGF treatment. Scale bar, 20 m. (C) Wild-type and Scar1 null E9 MEFs were stimulated with PDGF for 0, 2, 5, 7, 10, 12, and 15 min. For C, 200 cells were counted per time point, and each point represents the average value from three independent experiments. The cells were fixed and stained for F-actin, and the percentage of cells generating at least one dorsal ruffle was determined (error bars SD). Student s t test p values are shown on the graph. 682 Wiskostatin Inhibition of Dorsal Ruffle Formation in MEFs Studies on Scar1 did not reveal any obvious dorsal ruffle phenotype. Therefore, it was interesting to investigate the role of its related family member N-WASP. Endogenous N-WASP, along with its binding partners dynamin, cortactin, and WIP have all been localized to dorsal ruffles (Anton et al., 2003; Krueger et al., 2003). To investigate the endogenous localization of N-WASP relative to Arp2/3 complex and cortactin, PDGF-stimulated wild-type MEFs were stained using anti-arp2 or anti-cortactin and anti-n-wasp antibodies (Supplemental Figure 4). Staining of N-WASP and Arp2 showed a strong colocalization with F-actin within dorsal ruffles (Supplemental Figure 4, A D). N-WASP also colocalized with cortactin within dorsal ruffles (Supplemental Figure 4, E H), consistent with previous reports (Krueger et al., 2003). Therefore, N-WASP, Arp2/3 complex, and cortactin all colocalize in dorsal ruffles after PDGF stimulation. The activity of N-WASP is tightly regulated by the binding of numerous proteins and lipids (Higgs and Pollard, 2000). In its native form, N-WASP is autoinhibited due to intramolecular folding (Rohatgi et al., 1999). Wiskostatin is a chemical inhibitor of N-WASP. It functions by binding a cleft in the regulatory GTPase binding domain of N-WASP and stabilizing its native, autoinhibited conformation (Peterson et al., 2004). WASP expression is limited to hematopoietic cell lines, and it is not expressed in MEFs (Parolini et al., 1997). Therefore, it is possible to study N-WASP function in dorsal ruffle formation by inhibiting it with wiskostatin. When treated with wiskostatin alone, serum-starved immortalized wild-type and Scar1 null MEFs did not generate any dorsal ruffles (Figure 4A). Pretreatment of the MEFs with DMSO for 5 min, followed by 5 min 10 ng/ml PDGF treatment resulted in 100% of cells producing dorsal ruffles, independently of the Scar1 status. Finally, the MEFs were preincubated with 10 M wiskostatin to inhibit N- WASP and then treated for 5 min with PDGF and wiskostatin. Inhibition of N-WASP with wiskostatin abolished the formation of dorsal ruffles but not peripheral ruffles in both wild-type and Scar1 null cells. Figure 4C shows an example of a wild-type MEF treated with 10 M wiskostatin. No dorsal ruffles are present, but endogenous Arp2/3 complex can still localize into peripheral ruffles (arrows), consistent with reports that N-WASP null cells can still generate lamellipodia (Snapper et al., 2001). These data indicate that N- WASP, unlike Scar1 or Scar2, is important for dorsal ruffle formation. To further investigate the effects of wiskostatin, decreasing concentrations were used on wild-type MEFs to determine how potently it inhibited dorsal ruffle formation. Wiskostatin at 5 M still strongly inhibited dorsal ruffle formation (Figure 4B, 10%). As the concentration reached Molecular Biology of the Cell
6 N-WASP in Dorsal Ruffle Formation Figure 4. Wiskostatin treatment of E9 immortalized MEFs. (A) Serum-starved wild-type and Scar1 null MEFs were treated for 5 min with 10 M wiskostatin alone (Wisk), 10 ng/ml PDGF ( DMSO carrier) alone (PDGF), or PDGF and wiskostatin (PDGF Wisk). The cells were then fixed and stained with TRITC phalloidin, and the percentage of cells producing dorsal ruffles was determined. (B) Serum-starved wild-type E9 MEFs were pretreated with decreasing concentrations of wiskostatin for 5 min and then incubated with 10 ng/ml PDGF and wiskostatin. The same concentration of wiskostatin was used in the pretreatment as during the PDGF stimulation. After PDGF treatment, the average percentage of cells generating dorsal ruffles was determined. For A and B, 200 cells were counted per time point, and each point represents the average value from three independent experiments (error bars SD). (C) Representative image of a wild-type MEF treated with 10 M wiskostatin and 10 ng/ml PDGF. Cells were fixed and stained with TRITC phalloidin (A) and anti-p34 (B). Arrows indicate localization of Arp2/3 complex in peripheral ruffles. Bar, 20 m. 2.5, 1.25, and M, dorsal ruffle production returned rapidly to 100% levels. Therefore, 3 4 M wiskostatin is sufficient to inhibit 50% of dorsal ruffle production in these MEFs. This indicates that inhibition of N-WASP with wiskostatin has a potent inhibitory effect on dorsal ruffle formation. Scar1, Scar2, N-WASP, and Arp2 sirna Studies in MEFs To confirm the Scar1 data and examine the role of Scar2 in dorsal ruffle formation, sirna oligos specific to mouse Scar1 and Scar2 were used to specifically downregulate their protein levels. Wild-type immortalized MEFs transfected for 48 h with Scar1 and Scar2 sirna oligos showed a 70 and Figure 5. sirna studies on dorsal ruffle formation in E9 immortalized MEFs. (A) Wild-type MEFs were transfected for 48 h with sirna specific to mouse Scar1, Scar2, N-WASP, and Arp2. Knockdown of protein levels was shown by immunoblot with polyclonal antibodies specific to each protein. Anti-tubulin blots were included as protein loading controls. (B) The sirna-treated MEFs were serum starved and stimulated with 10 ng/ml PDGF for 5 min. The cells were fixed and stained with TRITC phalloidin, and the percentage of cells generating at least one dorsal ruffle was determined. N-WASP and Arp2 sirna treated cells were also cotransfected with human WASP-GFP and human Arp2-myc constructs. More than 200 were counted per time point, and each point represents the average value from three independent experiments (error bars SD). Student s t test p values are shown on graphs. 75% knockdown of protein levels, respectively, as determined by the intensity of bands produced after immunoblots with anti-scar1 and anti-scar2 antibodies (Figure 5A). Anti-tubulin loading controls confirmed roughly equal loadings between the control and sirna lanes. The sirnatreated cells were serum starved, treated for 5 min with PDGF, and fixed and stained for F-actin and endogenous Arp2/3 complex. The percentage of cells generating dorsal ruffles was then determined and plotted in Figure 5B. As a control, wild-type MEFs were treated with transfection reagent alone before PDGF treatment and staining. On average, 55% of wild-type cells treated with Lipofectamine 2000 alone generated dorsal ruffles. The 70 and 75% knockdown of Scar1 and Scar2, respectively, had no detectable effect on the percentage of cells producing dorsal ruffles (see Figure 5B for Student s t test p values). These experiments were repeated six times, and on each occasion neither Scar1 nor Scar2 sirna treatment resulted in a statistically significant decrease in dorsal ruffle production. These data confirm the previous Scar1 results and indicate that Scar2 is also not essential for dorsal ruffle formation. Vol. 18, February
7 J. A. Legg et al. To confirm the wiskostatin data, wild-type MEFs were treated with mouse N-WASP sirna oligos to knock down the endogenous protein levels of N-WASP. In addition, the Arp2/3 complex was targeted using Arp2 sirna (Figure 5, A and B). Knockdown of N-WASP and Arp2 protein levels was confirmed by immunoblot (Figure 5A). Depletion of N-WASP protein levels (85% reduction) resulted in a dramatic loss of dorsal ruffles. Control cells produced on average 55% dorsal ruffles; however, this number was reduced to 10% in N-WASP sirna-treated cells (p ). This N-WASP sirna inhibition was not as potent as wiskostatin treatment, which completely blocked dorsal ruffle production. This block is probably for two main reasons. First, the sirna did not completely deplete N-WASP protein levels, meaning the remaining 10% could be sufficient to retain dorsal ruffle production in a subset of cells. Second, it is possible that wiskostatin has other protein targets or cytotoxic effects, resulting in indirect inhibition of dorsal ruffle production (Guerriero and Weisz, 2006). However, we used 10 M or less wiskostatin in our assays, which gave only a weak off-target effect in the study of Guerriero and Weisz (2006). Interestingly, it was possible to partially retrieve the N-WASP sirna phenotype by cotransfection of human WASP-GFP (Figure 5B). In N-WASP sirna-treated cells retrieved by human WASP-GFP expression, WASP-GFP colocalizes with endogenous Arp2/3 complex and F-actin in dorsal ruffles (Supplemental Figure 5, A D, arrows). However, fewer than 10% of cells not transfected with WASP- GFP had dorsal ruffles. This is consistent with WASP-GFP being recruited to the dorsal ruffles and compensating for N-WASP loss. Therefore, inhibition of N-WASP function through wiskostatin or sirna treatment results in dorsal ruffle inhibition, and there is a degree of functional redundancy between the N-WASP and WASP in this structure. In addition to N-WASP, Arp2/3 complex was shown to be important in dorsal ruffle formation. Reduction of endogenous Arp2 levels (70% reduction; Figure 5A) resulted in a statistically significant inhibition in the percentage of cells generating dorsal ruffles (p 0.001; Figure 5B). The phenotype was not as severe as with N-WASP sirna treatment (20% compared with 10%). This was probably due to differences in the efficiency of the sirna treatments. Cotransfection of human Arp2-myc retrieved this phenotype back to control levels (Figure 5B). Similar to WASP-GFP, triple staining showed Arp2-myc colocalization with endogenous Arp2/3 complex and F-actin in dorsal ruffles (Supplemental Figure 5, E H, arrows). Therefore, like N-WASP, Arp2/3 complex function is necessary for dorsal ruffle formation in MEFs. Numerous roles have been attributed to dorsal ruffles, including the dissolution of stress fibers required to prepare a static cell for subsequent movement (Krueger et al., 2003). The consequence of N-WASP and Arp2 sirna treatment was evident in the stress fiber organization of the PDGF treated cells. Fifteen minutes into PDGF treatment, the wildtype, Scar1 null, and Scar2 sirna-treated immortalized MEFs contained large dorsal ruffles. Within the waves, there was a dramatic disassembly of the stress fibers (Supplemental Figure 6, arrows). However, PDGF treated N-WASP and Arp2 sirna MEFs retained large numbers of stress fibers in the main body of the cells (Supplemental Figure 6A, arrowheads). Therefore, this result confirms previous reports that dorsal ruffle formation is associated with stress fiber disassembly and demonstrates that N-WASP and the Arp2/3 complex are required for this process (Krueger et al., 2003). The importance of Scar1, Scar2, Scar3, and N-WASP in dorsal ruffle formation was finally investigated using the 684 Figure 6. Effects of expressing truncation mutants of N-WASP and Scar proteins in E9 immortalized MEFs. Wild-type (white bars) and Scar1 null (gray bars) E9-immortalized MEFs were treated with transfection reagent alone (U) or transfected with Scar1 A (S1dA), Scar2 A (S2dA), Scar3 A (S3dA), wild-type N-WASP (Wt NW), or N-WASP WWCA (NWdWWCA) mutants as indicated. The cells were serum starved, stimulated with PDGF for 5 min, and the percentage of cells generating dorsal ruffles was determined. More than 200 cells were counted per time point, and each point represents the average value from three independent experiments (error bars SD). Student s t test p values are shown on graphs. ectopic expression of truncation mutants in wild-type and Scar1 null MEFs stimulated for 5 min with PDGF. To investigate Scar function, delta-a Scar constructs were used. These constructs lack the C-terminal amino acids essential for Arp2/3 complex binding (Machesky and Insall, 1998). Approximately 50% of control cells treated with transfection reagent alone generated dorsal ruffles (Figure 6). Transfection of Scar1 A, Scar2 A, or Scar3 A into wild-type MEFs (white bars) did not invoke a statistically significant decrease in dorsal ruffle production, consistent with none of the Scar proteins being essential for dorsal ruffle production. Wild-type N-WASP expression did not inhibit dorsal ruffle formation. However, transfection of an N-WASP truncation mutant deficient in Arp2/3 complex binding (N-WASP WWCA) resulted in a decrease in cells with dorsal ruffles (p 0.02; drop from 50 to 20%). This is probably due to the sequestration of endogenous SH3 domain proteins, such as dynamin and WIP, away from the Arp2/3 complex. Finally, Scar1 A and Scar2 A were expressed in Scar1 null MEFs (Figure 6, gray bars) to determine whether the presence of any Scar isoform is required for dorsal ruffle formation. Neither Scar1 A nor Scar2 A expression in the Scar1 null MEFs inhibited dorsal ruffle formation. Therefore, it seems that an N-WASP cortactin Arp2/3 ternary complex is important for generating dorsal ruffles, whereas Scar1, -2, and -3 are dispensable for this process. Dorsal Ruffle Formation in N-WASP Null FLCs Previous studies on N-WASP function have shown its activity is dispensable for cell migration, adhesion to fibronectin substrate, and lamellipodia extension (Snapper et al., 2001; Rogers et al., 2003). To further investigate N-WASP function in dorsal ruffle formation, E9.5 N-WASP / FLCs were obtained (Snapper et al., 2001). These cells differ from MEFs because they were transformed with simian virus 40 (SV40) large-t containing retrovirus to enable viability in cell cul- Molecular Biology of the Cell
8 N-WASP in Dorsal Ruffle Formation Figure 8. Analysis of dorsal ruffle formation in N-WASP FLCs. (A and B) The / RV-N-WASP and N-WASP / FLCs were PDGF stimulated for 0, 2, 5, and 7 min, and the percentage of cells generating any dorsal actin-rich protrusions (A) or only ring-shaped dorsal actin-rich protrusions (B) was calculated. More than 200 cells were counted per time point, and each point represents the average value from three independent experiments (error bars SD). Figure 7. PDGF treatment of N-WASP FLCs. (A) We analyzed 30 g of total protein from wild-type MEFs (positive control) and the N-WASP FLCs by immunoblot with anti-nwasp and tubulin antibodies. Loadings: lane 1, wild-type MEFs; lane 2, / RV-N- WASP; lane 3, N-WASP / ; and lane 4, N-WASP /. (B) The N-WASP / (A C), / RV-N-WASP (D F) and N-WASP / (G O) FLCs were PDGF stimulated for 5 min and fixed and stained with TRITC phalloidin and anti-p34. Arrows indicate collapsed dorsal ruffles and arrowheads show Arp2/3-enriched peripheral ruffles in the N-WASP / FLCs. Bar, 20 m. ture. Immunoblot analysis with anti-n-wasp antibody on protein lysates confirmed the absence of a 66-kDa band in the N-WASP null FLCs (N-WASP / ; Figure 7A). The wildtype SV40 FLCs (N-WASP / ) expressed similar levels of N-WASP as the MEF-positive control. Finally, SV40-FLCs infected with an N-WASP expressing retrovirus ( / RV- N-WASP) showed a marked increased in N-WASP protein levels. The N-WASP FLCs were stimulated for 5 min with PDGF and then fixed and stained for F-actin and endogenous Arp2/3 complex (Figure 7B). The N-WASP / (A C) and / RV-N-WASP (D F) FLCs generated large, circular dorsal ruffles that were strongly stained for endogenous Arp2/3 complex. To our surprise, the N-WASP / cells were able to generate dorsal actin protrusions in response to PDGF (G O). However, there were numerous obvious deficiencies associated with these structures. They were often collapsed, forming bright, punctate structures under F-actin staining (arrows in H and K). In cells that did generate full ringshaped dorsal ruffles, these were typically smaller than the equivalent structures in the N-WASP / and / RV-N- WASP FLCs and displayed greatly reduced staining for endogenous Arp2/3 complex (M O). However, the N-WASP / cells were still able to generate normal Arp2/3 complexenriched peripheral ruffles, consistent with N-WASP not functioning in lamellipodia formation (arrowheads in N; Snapper et al., 2001). Therefore, although not essential in these cells, N-WASP deficiency has resulted in highly abnormal dorsal ruffles that are unstable and depleted of Arp2/3 complex. To quantify these deficiencies, after 5 min of PDGF stimulation, the average areas of the dorsal ruffles in / RV- N-WASP and N-WASP / cells were quantified using a Java plugin for ImageJ (see Materials and Methods). The dorsal ruffles in / RV-N-WASP cells were on average 3 times larger than the N-WASP / cells (6500 m 2 compared with 2100 m 2 ). Measurement of the average areas of the cells showed that the difference in dorsal ruffle size was not due to N-WASP / FLCs being smaller than their / RV-N-WASP counterparts (20,000 m 2 compared with 19,000 m 2 ). Finally, the / RV-N-WASP and N-WASP / FLCs were stimulated for 0, 2, 5, and 7 min with PDGF to follow the generation of dorsal ruffles over time (Figure 8, A and B). At each time point, the cells were fixed, and the percentage of cells generating dorsal ruffles was determined. Consistent with Figures 1C and 3C, serum-starved FLCs did not gen- Vol. 18, February
9 J. A. Legg et al. erate dorsal ruffles. The / RV-N-WASP FLCs showed a strong and persistent response to PDGF, with 70% of cells generating dorsal ruffles at 2, 5, and 7 min (Figure 8A). The N-WASP / cells were severely deficient in producing dorsal ruffles, with only 20% generating dorsal ruffles at 2 min and a maximal response of 50% at 5 min. In this count, N-WASP / FLCs generating punctate, collapsed dorsal structures and full ring structures were both counted as dorsal ruffles. Therefore, the assay was repeated and the abnormal, collapsed structures generated by both / RV- N-WASP and N-WASP / FLCs were not included (Figure 8B). The graph for the / RV-N-WASP cells was largely unaffected, with 70% of cells generating large, ring dorsal ruffle structures at 2, 5, and 7 min. The N-WASP / response was greatly reduced, with 20% of cells generating dorsal ruffles at all time points. This demonstrates that a large proportion of N-WASP / FLCs generate small, abnormal dorsal ruffles. Movies of PDGF stimulated / RV-N-WASP and N-WASP / FLCs are included as Supplemental Movies 4 and 5. The / RV-N-WASP cells generated large, stable dorsal ruffles that expand and contract steadily over time, similar to the wild-type and Scar1 null MEFs in Figure 2. However, the N-WASP / cells displayed dramatic and persistent distortions at the dorsal surface, but these were unable to form into larger dorsal ruffle structures. In conclusion, N-WASP is required for the formation of large and stable dorsal ruffle waves as seen in primary and immortalized MEFs and FLCs. DISCUSSION We propose that N-WASP, but not Scar1- or Scar2-mediated recruitment of Arp2/3 complex is important for the formation of dorsal ruffles. Scar1 deficiency, in our experiments, did not affect the frequency, appearance or number of dorsal ruffles per cell or dorsal ruffle kinetics during live cell imaging; 70 and 75% reduction in Scar1 and Scar2 by sirna also did not alter the percentage of cells generating dorsal ruffles. Finally, Scar1 null MEFs obtained from John Scott s group (The Vollum Institute, Portland, OR) also did not show any qualitative differences in dorsal ruffle formation (data not shown; Soderling et al., 2003). Therefore, it is unlikely that Scar1 is essential for dorsal ruffle formation. These data are particularly interesting in light of a previous report where Scar1 and Scar2 were shown to have differential roles in dorsal ruffle and peripheral ruffle formation, respectively (Suetsugu et al., 2003). Staining for endogenous Scar1 and Scar2 showed localization of both Scar1 and Scar2 to dorsal ruffles (Supplemental Figure 2). However, only Scar2 localized consistently to peripheral lamellipodia, and this is consistent with Scar2 functioning in lamellipodia formation (Suetsugu et al., 2003). The presence of Scar1 and Scar2 in similar localizations in dorsal ruffles presents the possibility that these proteins can functionally compensate for each other. Although we could achieve knockdown of either Scar1 or Scar2 individually, we were unable to observe MEFs with both proteins at undetectable levels. Complete removal of all Scar protein may be detrimental to MEF adhesion or integrity. However, expression of dominantnegative Scar2 A in the Scar1 null MEFs did not inhibit dorsal ruffle formation and indicates that the presence of Scar is not a prerequisite for dorsal ruffle formation. Numerous roles have been attributed to dorsal ruffles, including the dissolution of stress fibers in preparing a static cell for movement (Krueger et al., 2003), macropinocytosis (Dowrick et al., 1993; Araki et al., 2000), and the internalization of cell surface receptors (Orth et al., 2006). Evidence for 686 a role in macropinocytosis has come from observing the closure of dorsal ruffles by using real-time microscopy and uptake of fluorescent dextran (Dowrick et al., 1993; Araki et al., 2000). Peripheral ruffles have also been proposed to be the site of macropinocytosis (Suetsugu et al., 2003). Therefore, it seems that macropinocytosis can occur at both peripheral and dorsal surfaces. In addition, wild-type, Scar1 null, and Scar2 sirna-treated MEFs still made dorsal ruffles, which resulted in almost complete disruption of the stress fibers within the waves. In contrast, N-WASP- and Arp2 sirna-treated MEFs were unable to generate dorsal ruffles; therefore, the stress fibers remained unaffected. Our study confirms previous reports that dorsal ruffles are involved in the removal of stress fibers in static cells (Krueger et al., 2003). Invadopodia and podosomes are another class of membrane-deforming F-actin structures, which have striking similarities to dorsal ruffles (for review, see Buccione et al., 2004). Invadopodia and podosomes depend on a dynamin cortactin Arp2/3 complex-n-wasp multiprotein assembly. WASP/N WASP localize to all these structures, and targeted disruption of this complex using dominant-negative N-WASP constructs or depletion of N-WASP protein levels by using null cells or sirna inhibits invadopodia and podosome formation (Mizutani et al., 2002; Calle et al., 2004; Lorenz et al., 2004; Yamaguchi et al., 2005). Recent evidence supports a similar model for dorsal ruffle formation. Inhibition of dynamin cortactin binding by the injection of antidynamin 2 antibodies or overexpression of truncation mutants abrogated dorsal ruffle formation, indicating that an interaction between these proteins is required for wave formation (Krueger et al., 2003). Recently, overexpression of the dynamin-binding, multidomain scaffolding protein Tuba has been shown to induce dorsal ruffles, and this was inhibited by wiskostatin treatment (Kovacs et al., 2006), further supporting the idea of an N-WASP dynamin-containing actin assembly in dorsal ruffles. Analysis of N-WASP null fibroblast-like cells showed that N-WASP deficiency was not terminal for dorsal ruffle production in these cells. However, the dorsal ruffles of N- WASP null cells were severalfold smaller, depleted of Arp2/3 complex and highly unstable than the N-WASP overexpressing cells. We were unfortunately unable to quantify whether the wild-type fibroblast-like cell line produced the same number of dorsal ruffles as the overexpressing cells, because in our hands, the wild-type cell line did not grow well and formed clumps in tissue culture. Comparison between wild type and knockout would have obviously been the best control for the knockout cells, but we think that the significant differences observed between the knockout cells and overexpressing cells still strongly supports the role of N-WASP in dorsal ruffles. The ability of the fibroblast-like cells to generate dorsal ruffles is also complicated by their transformation with SV40 virus. Tyrosine phosphorylation and hyperactivation of Src targets, such as dynamin and cortactin, could potentially compensate for N-WASP loss and enable highly abnormal dorsal ruffles to form. The similarities between dorsal ruffles and podosomes support this hypothesis, because podosomes spontaneously form in Src-transformed cells (Shriver and Rohrschneider, 1981). Interestingly, the wiskostatin treated MEFs and N- WASP null FLCs were still able to recruit Arp2/3 complex and form normal peripheral ruffles. In contrast, Scar2 null MEFs reportedly cannot generate peripheral ruffles under some conditions (Suetsugu et al., 2003; Yan et al., 2003). Therefore, although morphologically similar structures, the Molecular Biology of the Cell
N-WASP INVOLVEMENT IN DORSAL RUFFLE FORMATION IN MOUSE EMBRYONIC FIBROBLASTS
 N-WASP INVOLVEMENT IN DORSAL RUFFLE FORMATION IN MOUSE EMBRYONIC FIBROBLASTS John A. Legg*, Guillaume Bompard, John Dawson*, Hannah L. Morris*, Natalie Andrew*, Lisa Cooper, Simon A. Johnston*, Giorgos
N-WASP INVOLVEMENT IN DORSAL RUFFLE FORMATION IN MOUSE EMBRYONIC FIBROBLASTS John A. Legg*, Guillaume Bompard, John Dawson*, Hannah L. Morris*, Natalie Andrew*, Lisa Cooper, Simon A. Johnston*, Giorgos
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Paul et al.,
 Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Flag-Rac Vector V12 V12 N17 C40. Vector C40 pakt (T308) Akt1. Myc-DN-PAK1 (N-SP)
 a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
Supplemental Information
 Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in
 Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
Supplemental data. Supplemental Materials and Methods
 Supplemental data Supplemental Materials and Methods Transfection of plasmid. Transfection of plasmids into FRTL5 cells was performed using Lipofectamine LTX with Plus reagent (Invitrogen) according to
Supplemental data Supplemental Materials and Methods Transfection of plasmid. Transfection of plasmids into FRTL5 cells was performed using Lipofectamine LTX with Plus reagent (Invitrogen) according to
Gα 13 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα 13 Activation Assay Kit Catalog Number: 80401 20 assays NewEast Biosciences 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα 13 Activation Assay Kit Catalog Number: 80401 20 assays NewEast Biosciences 1 Table of Content Product
Supplemental Data Supplementary Figure Legends and Scheme Figure S1.
 Supplemental Data Supplementary Figure Legends and Scheme Figure S1. UTK1 inhibits the second EGF-induced wave of lamellipodia formation in TT cells. A and B, EGF-induced lamellipodia formation in TT cells,
Supplemental Data Supplementary Figure Legends and Scheme Figure S1. UTK1 inhibits the second EGF-induced wave of lamellipodia formation in TT cells. A and B, EGF-induced lamellipodia formation in TT cells,
Antibodies used in this study Figure S1. Akt expression in neutrophils from WT and individual Akt isoform knockout mice
 ntibodies used in this study The anti β-actin monoclonal antibody (Sigma-ldrich, St. Louis, MO) was generated against a slightly modified human β-actin N-terminal peptide, c-sp-sp-sp-ile-la-la-leu-val-ile-
ntibodies used in this study The anti β-actin monoclonal antibody (Sigma-ldrich, St. Louis, MO) was generated against a slightly modified human β-actin N-terminal peptide, c-sp-sp-sp-ile-la-la-leu-val-ile-
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Supplementary Material
 Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration
 /, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
/, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
Cdc42 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Cdc42 Activation Assay Kit Catalog Number: 80701 20 assays 1 Table of Content Product Description 3 Assay
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Cdc42 Activation Assay Kit Catalog Number: 80701 20 assays 1 Table of Content Product Description 3 Assay
System. Dynamic Monitoring of Receptor Tyrosine Kinase Activation in Living Cells. Application Note No. 4 / March
 System Application Note No. 4 / March 2008 Dynamic Monitoring of Receptor Tyrosine Kinase Activation in Living Cells www.roche-applied-science.com Dynamic Monitoring of Receptor Tyrosine Kinase Activation
System Application Note No. 4 / March 2008 Dynamic Monitoring of Receptor Tyrosine Kinase Activation in Living Cells www.roche-applied-science.com Dynamic Monitoring of Receptor Tyrosine Kinase Activation
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
Apoptosis And Anti-tumor Effect Induced By Mtor Inhibitor And Autophagy Inhibitor In Human Osteosarcoma Cells
 Apoptosis And Anti-tumor Effect Induced By Mtor Inhibitor And Autophagy Inhibitor In Human Osteosarcoma Cells Ryosuke Horie. Kagawa University of medecine, Kita-gun, Japan. Disclosures: R. Horie: None.
Apoptosis And Anti-tumor Effect Induced By Mtor Inhibitor And Autophagy Inhibitor In Human Osteosarcoma Cells Ryosuke Horie. Kagawa University of medecine, Kita-gun, Japan. Disclosures: R. Horie: None.
This Document Contains:
 This Document Contains: 1. In-Cell Western Protocol II. Cell Seeding and Stimulation Supplemental Protocol III. Complete Assay Example: Detailing the Seeding, Stimulation and Detection of the A431 Cellular
This Document Contains: 1. In-Cell Western Protocol II. Cell Seeding and Stimulation Supplemental Protocol III. Complete Assay Example: Detailing the Seeding, Stimulation and Detection of the A431 Cellular
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the
 Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking
 Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
SUPPLEMENTARY INFORMATION
 The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
Nature Immunology: doi: /ni.1744
 Macrophage colony stimulating factor induces macrophage proliferation and survival through a pathway involving DAP12 and β-catenin Karel Otero, Isaiah R Turnbull, Pietro Luigi Poliani *, William Vermi
Macrophage colony stimulating factor induces macrophage proliferation and survival through a pathway involving DAP12 and β-catenin Karel Otero, Isaiah R Turnbull, Pietro Luigi Poliani *, William Vermi
Supplementary Materials and Methods
 Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
No wash 2 Washes 2 Days ** ** IgG-bead phagocytosis (%)
 Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
B. ADM: C. D. Apoptosis: 1.68% 2.99% 1.31% Figure.S1,Li et al. number. invaded cells. HuH7 BxPC-3 DLD-1.
 A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and
 SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Rab5 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Rab5 Activation Assay Kit Catalog Number: 83701 20 assays 24 Whitewoods Lane 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Rab5 Activation Assay Kit Catalog Number: 83701 20 assays 24 Whitewoods Lane 1 Table of Content Product
Single cell imaging of Bruton's Tyrosine Kinase using an irreversible inhibitor
 SUPPLEMENTARY INFORMATION Single cell imaging of Bruton's Tyrosine Kinase using an irreversible inhibitor Anna Turetsky 1,a, Eunha Kim 1,a, Rainer H. Kohler 1, Miles A. Miller 1, Ralph Weissleder 1,2,
SUPPLEMENTARY INFORMATION Single cell imaging of Bruton's Tyrosine Kinase using an irreversible inhibitor Anna Turetsky 1,a, Eunha Kim 1,a, Rainer H. Kohler 1, Miles A. Miller 1, Ralph Weissleder 1,2,
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
SUPPLEMENTAL MATERIAL. Supplemental Methods:
 SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
M X 500 µl. M X 1000 µl
 GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
Gα i Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα i Activation Assay Kit Catalog Number 80301 20 assays NewEast Biosciences, Inc 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα i Activation Assay Kit Catalog Number 80301 20 assays NewEast Biosciences, Inc 1 Table of Content Product
The following antibodies were used in this study: NuMA - rabbit-anti NuMA (gift from
 Materials and Methods Antibodies: The following antibodies were used in this study: NuMA - rabbit-anti NuMA (gift from D. A. Compton); Dynein Intermediate Chain - 74.1 (Chemicon, Temecula, CA); Dynein
Materials and Methods Antibodies: The following antibodies were used in this study: NuMA - rabbit-anti NuMA (gift from D. A. Compton); Dynein Intermediate Chain - 74.1 (Chemicon, Temecula, CA); Dynein
(B) Comparable expression of major integrin subunits and glycoproteins on the surface of resting WT and Lnk -/- platelets.
 Supplemental Figure S1. Characteristics of Lnk -/- platelets. (A) Electron micrographs of resting platelets showing the normal intracellular structure of Lnk -/ platelets. Samples were fixed with 4% PFA
Supplemental Figure S1. Characteristics of Lnk -/- platelets. (A) Electron micrographs of resting platelets showing the normal intracellular structure of Lnk -/ platelets. Samples were fixed with 4% PFA
Supplemental Materials and Methods
 Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
RheB Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based RheB Activation Assay Kit Catalog Number: 81201 20 assays NewEast Biosciences 1 FAX: 610-945-2008 Table
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based RheB Activation Assay Kit Catalog Number: 81201 20 assays NewEast Biosciences 1 FAX: 610-945-2008 Table
Human/Mouse/Rat Phospho-CREB (S133) Immunoassay. An ELISA-based assay using fluorogenic substrates to measure phosphorylated CREB in whole cells.
 Cell-Based ELISA Human/Mouse/Rat Phospho-CREB (S133) Immunoassay Catalog Number KCB2510 An ELISA-based assay using fluorogenic substrates to measure phosphorylated CREB in whole cells. This package insert
Cell-Based ELISA Human/Mouse/Rat Phospho-CREB (S133) Immunoassay Catalog Number KCB2510 An ELISA-based assay using fluorogenic substrates to measure phosphorylated CREB in whole cells. This package insert
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
J. Cell Sci. 128: doi: /jcs : Supplementary Material. Supplemental Figures. Journal of Cell Science Supplementary Material
 Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Arf6 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Arf6 Activation Assay Kit Catalog Number: 82401 20 assays NewEast Biosciences 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Arf6 Activation Assay Kit Catalog Number: 82401 20 assays NewEast Biosciences 1 Table of Content Product
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX
 Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand
 SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
Supplement Figure S1:
 Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
Supporting Online Material Y. Tang et al., published 1/24/03
 Y. Tang SOM, p. 1 Supporting Online Material Y. Tang et al., published 1/24/03 MATERIALS AND METHODS Construction of the Targeting Vector and Generation of Mice Carrying Mutations Targeting vector. Recombinant
Y. Tang SOM, p. 1 Supporting Online Material Y. Tang et al., published 1/24/03 MATERIALS AND METHODS Construction of the Targeting Vector and Generation of Mice Carrying Mutations Targeting vector. Recombinant
Description of supplementary material file
 Description of supplementary material file In the supplementary results we show that the VHL-fibronectin interaction is indirect, mediated by fibronectin binding to COL4A2. This provides additional information
Description of supplementary material file In the supplementary results we show that the VHL-fibronectin interaction is indirect, mediated by fibronectin binding to COL4A2. This provides additional information
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Page 1 of 5. Product Name Label Quantity Product No. Cy 3 10 µg (~0.75 nmol) MIR Cy µg (~7.5 nmol) MIR 7901
 Page 1 of 5 Label IT RNAi Delivery Control Product Name Label Quantity Product No. Cy 3 10 µg (~0.75 nmol) MIR 7900 Label IT RNAi Delivery Control Cy 3 100 µg (~7.5 nmol) MIR 7901 Fluorescein 10 µg (~0.75
Page 1 of 5 Label IT RNAi Delivery Control Product Name Label Quantity Product No. Cy 3 10 µg (~0.75 nmol) MIR 7900 Label IT RNAi Delivery Control Cy 3 100 µg (~7.5 nmol) MIR 7901 Fluorescein 10 µg (~0.75
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
Supplementary Fig.1 Luton
 Supplementary Fig.1 Luton a 175 Brain Thymus Spleen Small Intestine Kidney Testis HeLa b 250 Lung Kidney MDCK c EFA6B si Control si Mismatch #637 #1564 #1770 83 62 47.5 175 IB: anti-efa6b #B1 130 66 Lysates
Supplementary Fig.1 Luton a 175 Brain Thymus Spleen Small Intestine Kidney Testis HeLa b 250 Lung Kidney MDCK c EFA6B si Control si Mismatch #637 #1564 #1770 83 62 47.5 175 IB: anti-efa6b #B1 130 66 Lysates
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
T H E J O U R N A L O F C E L L B I O L O G Y
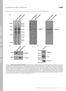 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Bays et al., http://www.jcb.org/cgi/content/full/jcb.201309092/dc1 Figure S1. Specificity of the phospho-y822 antibody. (A) Total cell
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Bays et al., http://www.jcb.org/cgi/content/full/jcb.201309092/dc1 Figure S1. Specificity of the phospho-y822 antibody. (A) Total cell
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Human/Mouse/Rat Phospho-Akt (S473) Immunoassay
 Cell-Based ELISA Human/Mouse/Rat Phospho-Akt (S473) Immunoassay Catalog Number KCB887 An ELISA-based assay using fluorogenic substrates to measure phosphorylated Akt in the context of a whole cell. This
Cell-Based ELISA Human/Mouse/Rat Phospho-Akt (S473) Immunoassay Catalog Number KCB887 An ELISA-based assay using fluorogenic substrates to measure phosphorylated Akt in the context of a whole cell. This
LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS.
 Supplemental Data: LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS. Scott Jepson, Bryan Vought, Christian H.
Supplemental Data: LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS. Scott Jepson, Bryan Vought, Christian H.
Methods Western blot analysis of plg Quantification of plasminogen accumulation by ELISA Immunohistochemical analysis
 Methods Western blot analysis of plg Wild-type mice first received a standardized burn wound and then were intravenously administered 2 mg of human plg (Omnio AB, Umeå, Sweden). 24 hours after wounding
Methods Western blot analysis of plg Wild-type mice first received a standardized burn wound and then were intravenously administered 2 mg of human plg (Omnio AB, Umeå, Sweden). 24 hours after wounding
2-step or indirect immunofluorescence 1. Substrate on which cells are plated: plastic vs. glass; coating vs. non
 Variables in standard immunostaining protocol 2-step or indirect immunofluorescence 1. Substrate on which cells are plated: plastic vs. glass; coating vs. non 2. Plating density: sparse vs. confluent 3.
Variables in standard immunostaining protocol 2-step or indirect immunofluorescence 1. Substrate on which cells are plated: plastic vs. glass; coating vs. non 2. Plating density: sparse vs. confluent 3.
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex
 Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex 1.0. Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigen-specific antibodies coupled
Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex 1.0. Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigen-specific antibodies coupled
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Online Supporting Material for. The Bisecting GlcNAc on N-Glycans Inhibits Growth. Factor Signaling and Retards Mammary Tumor
 Online Supporting Material for The Bisecting GlcNAc on N-Glycans Inhibits Growth Factor Signaling and Retards Mammary Tumor Progression Yinghui Song 1, Jason A. Aglipay 1, Joshua D. Bernstein 2, Sumanta
Online Supporting Material for The Bisecting GlcNAc on N-Glycans Inhibits Growth Factor Signaling and Retards Mammary Tumor Progression Yinghui Song 1, Jason A. Aglipay 1, Joshua D. Bernstein 2, Sumanta
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplementary Material - Methods
 Novel Protein-Protein Interactions in the Schizophrenia interactome Supplementary Material - Methods Experimental validations of predicted interactions Table S1-1: Protein pairs that were validated by
Novel Protein-Protein Interactions in the Schizophrenia interactome Supplementary Material - Methods Experimental validations of predicted interactions Table S1-1: Protein pairs that were validated by
Supplementary material and methods
 Inhibitory effect of caffeic acid on ADP-induced thrombus formation and platelet activation involves mitogen-activated protein kinases Yu Lu 1,2,3,#, Quan Li 3,4,#, Yu-Ying Liu 3,4, Kai Sun 3,4, Jing-Yu
Inhibitory effect of caffeic acid on ADP-induced thrombus formation and platelet activation involves mitogen-activated protein kinases Yu Lu 1,2,3,#, Quan Li 3,4,#, Yu-Ying Liu 3,4, Kai Sun 3,4, Jing-Yu
Supplementary Protocol. sirna transfection methodology and performance
 Supplementary Protocol sirna transfection methodology and performance sirna oligonucleotides, DNA construct and cell line. Chemically synthesized 21 nt RNA duplexes were obtained from Ambion Europe, Ltd.
Supplementary Protocol sirna transfection methodology and performance sirna oligonucleotides, DNA construct and cell line. Chemically synthesized 21 nt RNA duplexes were obtained from Ambion Europe, Ltd.
ApoTrack Cytochrome c Apoptosis ICC Antibody Kit
 ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Kit Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and non-apoptotic
ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Kit Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and non-apoptotic
The Transfection Collection TCF/LEF Transient Pack Wnt / -catenin Signaling Pathway Catalog #: 79273
 Data Sheet The Transfection Collection TCF/LEF Transient Pack Wnt / -catenin Signaling Pathway Catalog #: 79273 Background The Wnt / -catenin signaling pathway controls a large and diverse set of cell
Data Sheet The Transfection Collection TCF/LEF Transient Pack Wnt / -catenin Signaling Pathway Catalog #: 79273 Background The Wnt / -catenin signaling pathway controls a large and diverse set of cell
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Xu et al., Supplementary Figures 1-7
 Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
pdsipher and pdsipher -GFP shrna Vector User s Guide
 pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
We performed RT-PCR, cloning, sequencing and qrt-pcr in murine melanoma. cell lines and melanocytic tumors from RET-mice in accordance with the method
 Supplementary Material and Methods Quantitative RT-PCR (qrt-pcr) We performed RT-PCR, cloning, sequencing and qrt-pcr in murine melanoma cell lines and melanocytic tumors from RET-mice in accordance with
Supplementary Material and Methods Quantitative RT-PCR (qrt-pcr) We performed RT-PCR, cloning, sequencing and qrt-pcr in murine melanoma cell lines and melanocytic tumors from RET-mice in accordance with
Alpha or beta human chorionic gonadotropin knockdown decrease BeWo cell fusion by
 Alpha or beta human chorionic gonadotropin knockdown decrease BeWo cell fusion by down-regulating PKA and CREB activation Sudha Saryu Malhotra 1, Pankaj Suman 2 and Satish Kumar Gupta 1 * 1 Reproductive
Alpha or beta human chorionic gonadotropin knockdown decrease BeWo cell fusion by down-regulating PKA and CREB activation Sudha Saryu Malhotra 1, Pankaj Suman 2 and Satish Kumar Gupta 1 * 1 Reproductive
Flow cytometry Stained cells were analyzed and sorted by SORP FACS Aria (BD Biosciences).
 Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde
 Supplementary text Supplementary materials and methods Histopathological examination Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin wax with
Supplementary text Supplementary materials and methods Histopathological examination Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin wax with
Replication-independent chromatin loading of Dnmt1 during G2 and M phases
 Replication-independent chromatin loading of Dnmt1 during G2 and M phases Hariharan P. Easwaran 1, Lothar Schermelleh 2, Heinrich Leonhardt 1,2,* and M. Cristina Cardoso 1,* 1 Max Delbrück Center for Molecular
Replication-independent chromatin loading of Dnmt1 during G2 and M phases Hariharan P. Easwaran 1, Lothar Schermelleh 2, Heinrich Leonhardt 1,2,* and M. Cristina Cardoso 1,* 1 Max Delbrück Center for Molecular
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
-RT RT RT -RT XBMPRII
 Base pairs 2000 1650 1000 850 600 500 400 Marker Primer set 1 Primer set 2 -RT RT RT -RT XBMPRII kda 100 75 50 37 25 20 XAC p-xac A 300 B Supplemental figure 1. PCR analysis of BMPRII expression and western
Base pairs 2000 1650 1000 850 600 500 400 Marker Primer set 1 Primer set 2 -RT RT RT -RT XBMPRII kda 100 75 50 37 25 20 XAC p-xac A 300 B Supplemental figure 1. PCR analysis of BMPRII expression and western
The NESH/Abi-3-based WAVE2 complex is functionally distinct from the Abi-1-based WAVE2 complex
 Sekino et al. Cell Communication and Signaling (2015) 13:41 DOI 10.1186/s12964-015-0119-5 RESEARCH Open Access The NESH/Abi-3-based WAVE2 complex is functionally distinct from the Abi-1-based WAVE2 complex
Sekino et al. Cell Communication and Signaling (2015) 13:41 DOI 10.1186/s12964-015-0119-5 RESEARCH Open Access The NESH/Abi-3-based WAVE2 complex is functionally distinct from the Abi-1-based WAVE2 complex
T H E J O U R N A L O F C E L L B I O L O G Y
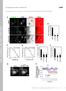 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Yamaguchi et al., http://www.jcb.org/cgi/content/full/jcb.201009126/dc1 S1 Figure S2. The expression levels of GFP-Akt1-PH and localization
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Yamaguchi et al., http://www.jcb.org/cgi/content/full/jcb.201009126/dc1 S1 Figure S2. The expression levels of GFP-Akt1-PH and localization
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary methods
 Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Electric Supplement Information
 Electric Supplement Information Preparation of DNA-AuNPs Thiol-modified oligonucleotide (5 -Cy5- ATCTCGGCTCTGCTAGCGAAAAAAAAAA-(C3H6)-SH-3, 5 15.5 OD) was added to 13 nm citrate-stabilized AuNPs (~3 nmol
Electric Supplement Information Preparation of DNA-AuNPs Thiol-modified oligonucleotide (5 -Cy5- ATCTCGGCTCTGCTAGCGAAAAAAAAAA-(C3H6)-SH-3, 5 15.5 OD) was added to 13 nm citrate-stabilized AuNPs (~3 nmol
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Methanol fixation allows better visualization of Kal7. To compare methods for
 Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Androgen Receptor (Phospho-Tyr363) Colorimetric Cell-Based ELISA Kit. Catalog #: OKAG02138
 Androgen Receptor (Phospho-Tyr363) Colorimetric Cell-Based ELISA Kit Catalog #: OKAG02138 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research
Androgen Receptor (Phospho-Tyr363) Colorimetric Cell-Based ELISA Kit Catalog #: OKAG02138 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research
