T H E J O U R N A L O F C E L L B I O L O G Y
|
|
|
- Rose Webb
- 5 years ago
- Views:
Transcription
1 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Yamaguchi et al., S1
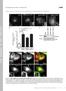
2 Figure S2. The expression levels of GFP-Akt1-PH and localization of GFP in MDA-MB-231 cells. (A) The expression levels of GFP-Akt1-PH were determined in transiently and stably transfected MDA-MB-231 cells. Relative fluorescence intensity of the GFP-Akt1-PH signals was calculated from microscopic images. Data are means + SEM of 38 transient cells and 35 stable cells analyzed. (B) MDA-MB-231 cells stably expressing a GFP construct were cultured on fluorescent gelatin-coated coverslips, stained for F-actin, and observed by confocal microscopy. Figure S1. PI3K activity is required for invadopodia formation in MDA-MB-231 cells. (A) MDA-MB-231 cells were cultured on fluorescent gelatin-coated coverslips for 7 h in the presence or absence of LY (LY) or wortmannin (WM) and stained for cortactin and F-actin to visualize invadopodia. Arrowheads denote degradation sites on the gelatin matrix. (B) Quantification of the gelatin degradation activity is shown. (C and D) Dose response curves of gelatin degradation obtained in the presence of increasing concentrations of LY (C) or wortmannin (D) are shown. (E and F) The percentage of cells with invadopodia (E) and the relative number of invadopodia per cell (F) were determined. Data are represented as means ± SEM of four (B and D F) and three (C) independent determinations. *, P < 0.02; and **, P < by Student s t tests. (G) Cells stably expressing GFP-actin were cultured on gelatin-coated glass-bottom dishes and observed using time-lapse fluorescent microscopy. Representative images of a cell before and 1 min after LY treatment are shown. Insets are magnified images of the boxed regions. Arrowheads denote invadopodia that were degraded after LY treatment. (H) Quantification of the fluorescence intensity at the invadopodia in GFP-actin expressing cells is shown. The arrowhead denotes the time point of DMSO or LY addition. Data are represented as mean ± SD of seven and six invadopodia for DMSO- and LY treated cells, respectively. *, P < by Student s t tests. S2
3 Figure S3. RT-PCR analysis of the expression of PI3K and Akt isoforms in MDA-MB-231 cells. (A) The expression of PI3K isoforms was determined by semiquantitative RT-PCR analysis. The expression of class I p110, p110, and p110 ; class II C2 and C2 ; and class III Vps34 was detected, whereas expression of class I p110 and class II C2 was not detected. This is consistent with the result obtained by real-time quantitative PCR analysis with a different set of primer pairs for the PI3K isoforms (shown in Fig. 2 A). (B) The expression of Akt isoforms was determined by real-time quantitative PCR analysis. All three Akt isoforms were expressed in MDA-MB-231 cells. Data are means + SEM of three independent experiments. S3
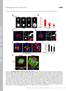
4 Figure S4. The function of p110 in invadopodia formation and localization of PDK1 and Akt at invadopodia in human breast cancer cells. (A) MDA-MB- 231 cells were transfected with three different sirnas that target distinct regions of the PIK3CA gene for 48 h, and the expression of p110 was examined by immunoblotting. (B) MDA-MB-231 cells transfected with the p110 sirnas were cultured on fluorescent gelatin-coated coverslips for 7 h, and the degradation area of gelatin was quantified. (C) BT-549 cells transfected with control or p110 sirna were cultured on fluorescent gelatin-coated coverslips for 7 h and stained for F-actin. (D) Gelatin degradation activity of BT549 cells transfected with control or p110 sirna was quantified. (E) Expression of p110 in Hs578T cells expressing control or p110 shrna was examined by immunoblotting. (F) Hs578T cells expressing control or p110 shrna were cultured on fluorescent gelatin-coated coverslips for 16 h and stained for F-actin. (G) Gelatin degradation activity of Hs578T cells expressing control or p110 shrna was quantified. (H) BT-549 and Hs578T cells were plated onto fluorescent gelatin-coated coverslips for 7 and 16 h, respectively, in the absence or presence of 100 nm PIK-75. The gelatin degradation areas were then quantified. (I) Representative images of BT549 and Hs578T cells treated with 100 nm PIK-75 are shown. (J) BT549 cells plated onto fluorescent gelatin-coated coverslips for 16 h were stained with the anti-akt and anti-pdk1 antibody. Arrowheads denote accumulation of Akt and PDK1 signals at the gelatin degradation sites. Insets in all images are magnified images of the boxed regions. Data are means + SEM of four (B, G, and H) and six (D) independent determinations. *, P < 0.01; and **, P < by Student s t tests. S4
5 Figure S5. The effects of expression of constitutively active Akt1 mutants on invadopodia formation. (A) MDA-MB-231 cells stably expressing HA-tagged E17K and E40K Akt1 were analyzed by immunoblotting. (B) Cells stably expressing the Akt constructs were cultured on fluorescent gelatin-coated coverslips for 7 h, and degraded areas on the gelatin matrix were quantified. Data are represented as means + SEM of four independent determinations. *, P < by Student s t tests. Video 1. LY treatment disassembles invadopodia in MDA-MB-231 cells expressing GFP-actin. MDA-MB-231 cells stably expressing GFP-actin were cultured on gelatin-coated glass-bottomed dishes and treated with 20 µm LY Cells were imaged by epifluorescence time-lapse microscopy using a microscope (IX81-ZDC-DSU). Frames were taken every minute for 30 min and are displayed at six frames per second. Corresponding still images are shown in Fig. S1 G. Video 2. LY treatment disassembles invadopodia in MDA-MB-231 cells expressing Venus-cortactin. MDA-MB-231 cells stably expressing Venus-cortactin were cultured on gelatin-coated glass-bottom dishes and treated with 20 µm LY Cells were imaged by epifluorescence time-lapse microscopy using a microscope (IX81-ZDC-DSU). Frames were taken every minute for 16 min and are displayed at six frames per second. Invadopodia enriched with cortactin disassembled immediately after addition of LY S5
6 Table S1. List of sirnas used in this study Gene symbol Gene ID Stealth RNAi PIK3CA 5290 #1 Stealth Select RNAi HSS #2 Stealth Select RNAi HSS #3 5 -GCGTTTCTGCTTTGGGACAACCATA-3 #4 5 -CAGGCAAAGACCGATTGCATAGGAA-3 #5 5 -GCAAACAGGGTTTGATAGCACTTAA-3 PIK3CB 5291 Stealth Select RNAi HSS PIK3CD 5293 Stealth Select RNAi HSS PIK3C2A 5286 Stealth Select RNAi HSS PIK3C2B 5287 Stealth Select RNAi HSS PIK3C Stealth Select RNAi HSS PDK #1 Stealth Select RNAi HSS #2 Stealth Select RNAi HSS Akt1 207 #1, 2 Validated Stealth DuoPak Akt2 208 #1, 2 Validated Stealth DuoPak Akt #1 Stealth Select RNAi HSS #2 Stealth Select RNAi HSS Control Negative Control Medium GC Duplex #2 S6
7 Table S2. List of primer pairs used for PCR analyses Gene symbol Gene ID Sequence 5-3 For real-time quantitative PCR PIK3CA 5290 TGGATGCTCTACAGGGCTTT GTCTGGGTTCTCCCAATTCA PIK3CB 5291 GCATTAAAAGGGAGCGAGTG CATGCCGTCGTAAAATCAGA PIK3CD 5293 CTGGCTGAAGTCCAAGAACC CTCGGATCATGATGTTGTCG PIK3CG 5294 ATACCATGATAGCGCCCTTG AATCACAGCGAACCTCTGCT PIK3C2A 5286 GAAAAACGAGGAATCCGACA CAGGGTTACTCCACCCAAGA PIK3C2B 5287 TTCCCTAGTCGCTTCGTGAT CAGTGGGTGGAAGAAGGTGT PIK3C2G 5288 TTCATCTCCCAGATGGCTCT AGTGGGGTCCGTACATTTTG PIK3C TCAGCCAAGCATTGTTGAAG TCCACTTTCGCGTTGTACTG Akt1 207 TCTATGGCGCTGAGATTGTG CTTAATGTGCCCGTCCTTGT Akt2 208 TATACCGCGACATCAAGCTG GGTCCCACAGAAGGTTTTCA Akt TGGACAAAGATGGCCACATA ACTGCTCGGCCATAGTCATT PPIB 5479 TTCTAGAGGGCATGGAGGTG (cyclophilin B) GCTTCTCCACCTCGATCTTG For semiquantitative RT-PCR PIK3CA 5290 GTCAATCGGTGACTGTGTGG TCCATCGTCTTTCACCATGA PIK3CB 5291 GGGAAAGCTCATCGTAGCTG CTTGATCTTGCAGCATTCCA PIK3CD 5293 CTGGTGCAGGTGCTCAAGTA TGAGAGCTCAGCTTGACGAA PIK3CG 5294 GGAACACCGACCTCACAGTT GAAGCTGAACTTTGCCCTTG PIK3C2A 5286 ACATCCGGAACCAACAAGAG TTCAGCTTGGGTGAGCTTTT PIK3C2B 5287 GGCCATGAGTTGTTTGAGGT CATCATGGGTGTAGCACAGG PIK3C2G 5288 CTCATTGGTGGCACCTACCT AGCCATCTGGGAGATGAATG PIK3C CTCACCAAAGCTCATCGACA GCTTGTGGTGTTTGCTCTCA PPIB 5479 GCACAGGAGGAAAGAGCATC (cyclophilin B) CTTCTCCACCTCGATCTTGC S7
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Paul et al.,
 Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Hong et al.,
 Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplementary Materials and Methods
 Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr
 Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Prashar et al., http://www.jcb.org/cgi/content/full/jcb.201304095/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. FBT phagocytosis in cells expressing PM-GFP. (A) T-PC
Supplemental material Prashar et al., http://www.jcb.org/cgi/content/full/jcb.201304095/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. FBT phagocytosis in cells expressing PM-GFP. (A) T-PC
supplementary information
 DOI: 10.1038/ncb2156 Figure S1 Depletion of p114rhogef with different sirnas. Caco-2 (a) and HCE (b) cells were transfected with individual sirnas, pools of the two sirnas or the On Target (OnT) sirna
DOI: 10.1038/ncb2156 Figure S1 Depletion of p114rhogef with different sirnas. Caco-2 (a) and HCE (b) cells were transfected with individual sirnas, pools of the two sirnas or the On Target (OnT) sirna
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
SUPPLEMENTAL FIGURES AND TABLES
 SUPPLEMENTAL FIGURES AND TABLES A B Flag-ALDH1A1 IP: α-ac HEK293T WT 91R 128R 252Q 367R 41/ 419R 435R 495R 412R C Flag-ALDH1A1 NAM IP: HEK293T + + - + D NAM - + + E Relative ALDH1A1 activity 1..8.6.4.2
SUPPLEMENTAL FIGURES AND TABLES A B Flag-ALDH1A1 IP: α-ac HEK293T WT 91R 128R 252Q 367R 41/ 419R 435R 495R 412R C Flag-ALDH1A1 NAM IP: HEK293T + + - + D NAM - + + E Relative ALDH1A1 activity 1..8.6.4.2
Supplementary Figure 1, Wiel et al
 Supplementary Figure 1, Wiel et al Supplementary Figure 1 ITPR2 increases in benign tumors and decreases in aggressive ones (a-b) According to the Oncomine database, expression of ITPR2 increases in renal
Supplementary Figure 1, Wiel et al Supplementary Figure 1 ITPR2 increases in benign tumors and decreases in aggressive ones (a-b) According to the Oncomine database, expression of ITPR2 increases in renal
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Prospéri et al.,
 Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Supplement Figure 1. Plin5 Plin2 Plin1. KDEL-DSRed. Plin-YFP. Merge
 Supplement Figure 1 Plin5 Plin2 Plin1 KDEL-DSRed Plin-YFP Merge Supplement Figure 2 A. Plin5-Ab MitoTracker Merge AML12 B. Plin5-YFP Cytochrome c-cfp merge Supplement Figure 3 Ad.GFP Ad.Plin5 Supplement
Supplement Figure 1 Plin5 Plin2 Plin1 KDEL-DSRed Plin-YFP Merge Supplement Figure 2 A. Plin5-Ab MitoTracker Merge AML12 B. Plin5-YFP Cytochrome c-cfp merge Supplement Figure 3 Ad.GFP Ad.Plin5 Supplement
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental Materials
 Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
B. C. Staurosporine BEZ235 DMSO. -actin. Suppl. Fig. 1 BEZ235 and BGT226 inhibit phosphorylation of Akt and downstream targets of PI3K and mtor.
 DMSO Staurosporine BGT226 A. B. C. BGT226 P-Akt 0 1 2 4 8 hrs. P-Akt 0 5 10 25 50 100 nm PS6 P-4E-BP1 -actin C-Caspase3 -actin PS6 P-4E-BP1 -actin Suppl. Fig. 1 and BGT226 inhibit phosphorylation of Akt
DMSO Staurosporine BGT226 A. B. C. BGT226 P-Akt 0 1 2 4 8 hrs. P-Akt 0 5 10 25 50 100 nm PS6 P-4E-BP1 -actin C-Caspase3 -actin PS6 P-4E-BP1 -actin Suppl. Fig. 1 and BGT226 inhibit phosphorylation of Akt
Nature Immunology: doi: /ni.3015
 Supplementary Figure 1 Role of RIP1-RIP3 and PGAM5 in RNA virus induced inflammasome activation. (a) LDH release from LPS-primed BMDMs from wild-type mice (WT), Rip3 -/- or Nlrp3 -/- mice infected with
Supplementary Figure 1 Role of RIP1-RIP3 and PGAM5 in RNA virus induced inflammasome activation. (a) LDH release from LPS-primed BMDMs from wild-type mice (WT), Rip3 -/- or Nlrp3 -/- mice infected with
Supplementary Information
 Supplementary Information promotes cancer cell invasion and proliferation by receptor-mediated endocytosis-dependent and -independent mechanisms, respectively Kensaku Shojima, Akira Sato, Hideaki Hanaki,
Supplementary Information promotes cancer cell invasion and proliferation by receptor-mediated endocytosis-dependent and -independent mechanisms, respectively Kensaku Shojima, Akira Sato, Hideaki Hanaki,
Antibodies used in this study Figure S1. Akt expression in neutrophils from WT and individual Akt isoform knockout mice
 ntibodies used in this study The anti β-actin monoclonal antibody (Sigma-ldrich, St. Louis, MO) was generated against a slightly modified human β-actin N-terminal peptide, c-sp-sp-sp-ile-la-la-leu-val-ile-
ntibodies used in this study The anti β-actin monoclonal antibody (Sigma-ldrich, St. Louis, MO) was generated against a slightly modified human β-actin N-terminal peptide, c-sp-sp-sp-ile-la-la-leu-val-ile-
Figure S1. GST-MDA-7 toxicity in GBM cells is dependent on cathepsin proteases, and weakly
 Supplemental Figure Legends. Figure S1. GST-MDA-7 toxicity in GBM cells is dependent on cathepsin proteases, and weakly dependent on caspase 8. GBM6 and GBM12 cells were treated 24 h after plating with
Supplemental Figure Legends. Figure S1. GST-MDA-7 toxicity in GBM cells is dependent on cathepsin proteases, and weakly dependent on caspase 8. GBM6 and GBM12 cells were treated 24 h after plating with
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1 sirna and shrna mediated depletion of ATP7A results in loss of melanosomal ATP7A staining. a-h, sirna mediated ATP7A depletion. Immunofluorescence microscopy (IFM) analysis of ATP7A
Supplementary Figure 1 sirna and shrna mediated depletion of ATP7A results in loss of melanosomal ATP7A staining. a-h, sirna mediated ATP7A depletion. Immunofluorescence microscopy (IFM) analysis of ATP7A
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplemental Information. DNp63 Inhibits Oxidative Stress-Induced Cell. Death, Including Ferroptosis, and Cooperates with
 Cell Reports, Volume 21 Supplemental Information DNp63 Inhibits Oxidative Stress-Induced Cell Death, Including Ferroptosis, and Cooperates with the BCL-2 Family to Promote Clonogenic Survival Gary X. Wang,
Cell Reports, Volume 21 Supplemental Information DNp63 Inhibits Oxidative Stress-Induced Cell Death, Including Ferroptosis, and Cooperates with the BCL-2 Family to Promote Clonogenic Survival Gary X. Wang,
THE JOURNAL OF CELL BIOLOGY
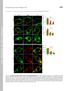 Supplemental Material THE JOURNAL OF CELL BIOLOGY Yeung et al., http://www.jcb.org/cgi/content/full/jcb.200903020/dc1 Figure S1. Assessment of the surface charge of maturing phagosomes. (A C and F H) RAW
Supplemental Material THE JOURNAL OF CELL BIOLOGY Yeung et al., http://www.jcb.org/cgi/content/full/jcb.200903020/dc1 Figure S1. Assessment of the surface charge of maturing phagosomes. (A C and F H) RAW
Supplementary Information
 Supplementary Information Supplementary Figures Supplementary Figure 1. MLK1-4 phosphorylate MEK in the presence of RAF inhibitors. (a) H157 cells were transiently transfected with Flag- or HA-tagged MLK1-4
Supplementary Information Supplementary Figures Supplementary Figure 1. MLK1-4 phosphorylate MEK in the presence of RAF inhibitors. (a) H157 cells were transiently transfected with Flag- or HA-tagged MLK1-4
Supplementary Data. Supplementary Materials and Methods Quantification of delivered sirnas. Fluorescence microscopy analysis
 Supplementary Data Supplementary Materials and Methods Quantification of delivered sirnas After transfection, cells were washed in three times with phosphate buffered saline and total RNA was extracted
Supplementary Data Supplementary Materials and Methods Quantification of delivered sirnas After transfection, cells were washed in three times with phosphate buffered saline and total RNA was extracted
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Kimura et al.,
 Supplemental material JCB Kimura et al., http://www.jcb.org/cgi/content/full/jcb.201503023/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. TRIMs regulate IFN-γ induced autophagy. (A and B) HC image analysis
Supplemental material JCB Kimura et al., http://www.jcb.org/cgi/content/full/jcb.201503023/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. TRIMs regulate IFN-γ induced autophagy. (A and B) HC image analysis
Supplemental Methods Cell lines and culture
 Supplemental Methods Cell lines and culture AGS, CL5, BT549, and SKBR were propagated in RPMI 64 medium (Mediatech Inc., Manassas, VA) supplemented with % fetal bovine serum (FBS, Atlanta Biologicals,
Supplemental Methods Cell lines and culture AGS, CL5, BT549, and SKBR were propagated in RPMI 64 medium (Mediatech Inc., Manassas, VA) supplemented with % fetal bovine serum (FBS, Atlanta Biologicals,
Supporting Information
 Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Supplementary Information
 Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
Supplementary Information
 Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/282/ra53/dc1 Supplementary Materials for Estrogen Alters the Splicing of Type 1 Corticotropin-Releasing Hormone Receptor in Breast Cancer Cells Suchita Lal,
www.sciencesignaling.org/cgi/content/full/6/282/ra53/dc1 Supplementary Materials for Estrogen Alters the Splicing of Type 1 Corticotropin-Releasing Hormone Receptor in Breast Cancer Cells Suchita Lal,
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression
 Supplemental figures Supplemental Figure. 1. Silencing expression of Celsr3 by shrna. To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression plasmids for the shrna
Supplemental figures Supplemental Figure. 1. Silencing expression of Celsr3 by shrna. To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression plasmids for the shrna
Supplementary Information. A novel human endogenous retroviral protein inhibits cell-cell fusion. Supplementary Figures:
 Supplementary Information A novel human endogenous retroviral protein inhibits cell-cell fusion Jun Sugimoto, Makiko Sugimoto, Helene Bernstein, Yoshihiro Jinno and Danny J. Schust Supplementary Figures:
Supplementary Information A novel human endogenous retroviral protein inhibits cell-cell fusion Jun Sugimoto, Makiko Sugimoto, Helene Bernstein, Yoshihiro Jinno and Danny J. Schust Supplementary Figures:
J. Cell Sci. 128: doi: /jcs : Supplementary Material. Supplemental Figures. Journal of Cell Science Supplementary Material
 Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Aspergillus fumigatus CalA binds to integrin α 5 β 1 and mediates host cell invasion
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION ARTICLE NUMBER: 16211 DOI: 10.1038/NMICROBIOL.2016.211 Aspergillus fumigatus CalA binds to integrin α 5 β 1 and mediates host
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION ARTICLE NUMBER: 16211 DOI: 10.1038/NMICROBIOL.2016.211 Aspergillus fumigatus CalA binds to integrin α 5 β 1 and mediates host
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1.
 Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Supplemental Materials and Methods
 Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information
 Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana.
 SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix
 Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix Authors Kerim B. Kaylan, 1,2 Viktoriya Ermilova, 1,2 Ravi Chandra Yada,
Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix Authors Kerim B. Kaylan, 1,2 Viktoriya Ermilova, 1,2 Ravi Chandra Yada,
GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts
 Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2743 Figure S1 stabilizes cellular protein level, post-transcriptionally. (a, b) and DDR1 were RNAi-depleted from HEK.293.-CBG cells. Western blots with indicated antibodies (a). RT-PCRs
DOI: 10.1038/ncb2743 Figure S1 stabilizes cellular protein level, post-transcriptionally. (a, b) and DDR1 were RNAi-depleted from HEK.293.-CBG cells. Western blots with indicated antibodies (a). RT-PCRs
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell
 Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2.
 Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy
 Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
supplementary information
 Figure S1 ZEB1 full length mrna. (a) Analysis of the ZEB1 mrna using the UCSC genome browser (http://genome.ucsc.edu) revealed truncation of the annotated Refseq sequence (NM_030751). The probable terminus
Figure S1 ZEB1 full length mrna. (a) Analysis of the ZEB1 mrna using the UCSC genome browser (http://genome.ucsc.edu) revealed truncation of the annotated Refseq sequence (NM_030751). The probable terminus
Alteration of microrna expression correlates to fatty acidmediated. insulin resistance in mouse myoblasts
 Alteration of microrna expression correlates to fatty acidmediated insulin resistance in mouse myoblasts Supplemental Data Correlation coefficient matrix con PA PA 0.9425 1.0000 PA+OA 0.9407 0.9626 con
Alteration of microrna expression correlates to fatty acidmediated insulin resistance in mouse myoblasts Supplemental Data Correlation coefficient matrix con PA PA 0.9425 1.0000 PA+OA 0.9407 0.9626 con
CDK5 is essential for TGF-β1-induced epithelial-mesenchymal transition and breast cancer progression
 Supplementary information for: CDK5 is essential for TGF-β1-induced epithelial-mesenchymal transition and breast cancer progression Qian Liang, Lili Li, Jianchao Zhang, Yang Lei, Liping Wang, Dong-Xu Liu,
Supplementary information for: CDK5 is essential for TGF-β1-induced epithelial-mesenchymal transition and breast cancer progression Qian Liang, Lili Li, Jianchao Zhang, Yang Lei, Liping Wang, Dong-Xu Liu,
Xu et al., Supplementary Figures 1-7
 Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas.
 Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2880 Supplementary Figure 1 Sequence alignment of Deup1 and Cep63. The protein sequence alignment was generated by the Clustal X 2.0 multiple sequence alignment program using default parameters.
DOI: 10.1038/ncb2880 Supplementary Figure 1 Sequence alignment of Deup1 and Cep63. The protein sequence alignment was generated by the Clustal X 2.0 multiple sequence alignment program using default parameters.
Primers used for PCR of conductin, SGK1 and GAPDH have been described in (Dehner et al,
 Supplementary METHODS Flow Cytometry (FACS) For FACS analysis, trypsinized cells were fixed in ethanol, rehydrated in PBS and treated with 40μg/ml propidium iodide and 10μ/ml RNase for 30 min at room temperature.
Supplementary METHODS Flow Cytometry (FACS) For FACS analysis, trypsinized cells were fixed in ethanol, rehydrated in PBS and treated with 40μg/ml propidium iodide and 10μ/ml RNase for 30 min at room temperature.
HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid
 SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells
 Supplementary Information The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells Bing Zhao 1,3, Zhen Qi 1,3, Yehua Li 1,3, Chongkai Wang 2,
Supplementary Information The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells Bing Zhao 1,3, Zhen Qi 1,3, Yehua Li 1,3, Chongkai Wang 2,
SUPPLEMENTARY INFORMATION
 a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary methods Shoc2 In Vitro Ubiquitination Assay
 Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Flag-Rac Vector V12 V12 N17 C40. Vector C40 pakt (T308) Akt1. Myc-DN-PAK1 (N-SP)
 a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
M X 500 µl. M X 1000 µl
 GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
Figure S1. Specificity of polyclonal anti stabilin-1 and anti stabilin-2 antibodies Lysates of 293T cells transfected with empty vector, mouse
 Figure S1. Specificity of polyclonal anti stabilin-1 and anti stabilin-2 antibodies Lysates of 293T cells transfected with empty vector, mouse stabilin-1, or mouse stabilin-2 were immunoblotted using anti
Figure S1. Specificity of polyclonal anti stabilin-1 and anti stabilin-2 antibodies Lysates of 293T cells transfected with empty vector, mouse stabilin-1, or mouse stabilin-2 were immunoblotted using anti
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
cells (MLEC) that produce luciferase under the control of the PAI-1 promoter in response to
 Supplemental Materials and Methods TGF bioassay. To quantify the levels of active and total TGF, we used mink lung epithelial cells (MLEC) that produce luciferase under the control of the PAI-1 promoter
Supplemental Materials and Methods TGF bioassay. To quantify the levels of active and total TGF, we used mink lung epithelial cells (MLEC) that produce luciferase under the control of the PAI-1 promoter
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Inhibition of Twist1 mediated invasion by Chk2 promotes premature senescence in p53 defective cancer cells
 Inhibition of Twist1 mediated invasion by Chk2 promotes premature senescence in p53 defective cancer cells Debasis Nayak, a,1 Anmol Kumar, b Souneek Chakraborty, a,1 Reyaz ur Rasool, a,1 Hina Amin, a Archana
Inhibition of Twist1 mediated invasion by Chk2 promotes premature senescence in p53 defective cancer cells Debasis Nayak, a,1 Anmol Kumar, b Souneek Chakraborty, a,1 Reyaz ur Rasool, a,1 Hina Amin, a Archana
SUPPLEMENTARY INFORMATION. Small molecule activation of the TRAIL receptor DR5 in human cancer cells
 SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
Flow cytometry Stained cells were analyzed and sorted by SORP FACS Aria (BD Biosciences).
 Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
