Dynamic visualization of nervous system in live Drosophila
|
|
|
- Loraine Underwood
- 6 years ago
- Views:
Transcription
1 Proc. Natl. Acad. Sci. USA Vol. 96, pp , August 1999 Neurobiology Dynamic visualization of nervous system in live Drosophila BANGHUA SUN*, PEIZHANG XU*, AND PAUL M. SALVATERRA* *Division of Neuroscience and Graduate School of Biological Sciences, Beckman Research Institute of the City of Hope, 1450 East Duarte Road, Duarte, CA Communicated by Eugene Roberts, Beckman Research Institute of the City of Hope, Duarte, CA, June 15, 1999 (received for review April 22, 1999) ABSTRACT We have constructed transgenic Drosophila melanogaster lines that express green fluorescent protein (GFP) exclusively in the nervous system. Expression is controlled with transcriptional regulatory elements present in the 5 flanking DNA of the Drosophila Na,K -ATPase -subunit gene Nervana2 (Nrv2). This regulatory DNA is fused to the yeast transcriptional activator GAL4, which binds specifically to a sequence motif termed the UAS (upstream activating sequence). Drosophila lines carrying Nrv2-GAL4 transgenes have been genetically recombined with UAS GFP (S65T) transgenes (Nrv2-GAL4 UAS GFP) inserted on the same chromosomes. We observe strong nervous system-specific fluorescence in embryos, larvae, pupae, and adults. The GFP fluorescence is sufficiently bright to allow dynamic imaging of the nervous system at all of these developmental stages directly through the cuticle of live Drosophila. These lines provide an unprecedented view of the nervous system in living animals and will be valuable tools for investigating a number of developmental, physiological, and genetic neurobiological problems. Dynamic visualization of the nervous system in a living organism would provide new insights into neural development, maintenance, and function. In principle, the use of the green fluorescent protein (GFP) reporter gene would be an ideal tool to accomplish this goal because it is easily observed in transgenic model organisms and cultured cells (1 3). In several studies, discrete neuronal subsets of Drosophila or nematodes have been observed in living animals with GFP (4 6). A number of practical problems, however, must be overcome before GFP visualization can serve as a useful marker for dynamic-imaging studies in live animals. First, it is necessary to have a promoter or DNA-regulatory fragment driving expression exclusively in the nervous system. Second, the strength of expression must be sufficient to allow easy observation of GFP fluorescence in intact organisms. Third, expression must be robust at all developmental stages. Recently, we isolated and characterized tissue-specific transcriptional regulatory elements for the two different Drosophila Na,K -ATPase (sodium pump) -subunit genes (7). These two genes, termed Nervana 1 and 2, originally were identified and cloned (8, 9) on the basis of reactivity with anti-horseradish peroxidase (anti-hrp) antibodies, which are commonly used to identify Drosophila neurons (10). The Nervana 1 protein does not carry the carbohydrate epitope(s) recognized by anti-hrp and is expressed primarily in muscle cells. In contrast, Nervana 2 is decorated with the anti-hrp epitope. and its expression is restricted to nervous system (11). The tissue-specific expression of each isoform gene is controlled by separate cis-dna regulatory elements located in the 5 flanking DNA (7). The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C solely to indicate this fact. PNAS is available online at The sodium pump is a major component of the plasma membranes of all excitable cell types and serves to generate and maintain the asymmetric gradient of Na and K ions necessary to establish the membrane resting potential and drive a variety of secondary ionic exchanger and metaboliteuptake systems (12, 13). Because extremely high levels of pump subunits are expressed in neurons at all developmental stages, it seemed likely that the regulatory elements of the Nrv 2 gene would be an excellent candidate for a pan-neuronal promoter with appropriate strength, specificity, and temporal expression to use GFP fluorescence in dynamic-imaging studies of the nervous system. In the present study, we fused the 5 flanking DNA of the Nrv 2 gene to the coding sequence of the yeast transcriptional activator GAL4 and constructed transgenic fly lines. When these lines are genetically recombined with lines carrying cognate GAL4 recognition elements (UAS, upstream activating sequence) controlling expression of a GFP responder gene, nervous system-specific GFP fluorescence was observed. The levels of GFP fluorescence are particularly strong, allowing the nervous system to be dynamically observed in live animals at all developmental stages. MATERIALS AND METHODS Nrv2-GAL4 Construct. We previously cloned the 7.7- kilobase Nrv2 5 flanking DNA into the Drosophila P element transformation vector, pcasper AUG -galactosidase (14), in a modified NotI cloning site (7). The GAL4 gene from the pgatn vector (15) was subcloned into that DNA construct to replace the lacz reporter gene of pcasper AUG galactosidase by using the NotI and XbaI cloning sites. P Element Transformation. Standard P element-mediated germ-line transformation of Drosophila melanogaster was performed (16). The Nrv2-GAL4 construct was injected into w embryos at a concentration of 0.8 g/ l along with puchs 2 3 helper plasmid (0.1 g/ l). A total of 14 independent lines (eye color selection) were generated with insertion of the transgene on the X, second, or third chromosomes. Linkage analysis of the inserted P elements was carried out by examining their segregation from second or third chromosomes marked with Cy or Sb, respectively. X-linked inserts were determined by standard crosses. Generation of Nrv2-GAL4 UAS GFP Lines. Fly lines carrying both a Nrv2-GAL4 driver and a UAS GFP responder on the second or third chromosomes were generated by recombination (17). Flies with Nrv2-GAL4 insertion on the second or third chromosome were crossed with flies carrying a UAS GFP (S65T) responder gene on the second or third chromosome (obtained from the Bloomington Drosophila Stock Center, B-1521, B-1522), respectively. The F 1 female progeny were crossed with w males, and the F 2 larval progeny were observed by using a fluorescence microscope. The larvae Abbreviations: GFP, green fluorescent protein; HRP, horseradish peroxidase; UAS, upstream activating sequence. To whom reprint requests should be addressed. psalv@ coh.org
2 Neurobiology: Sun et al. Proc. Natl. Acad. Sci. USA 96 (1999) with GFP expression in nervous system were selected and reared to make the Nrv2-GAL4 UAS GFP homozygous lines. The experiments described in this paper all were performed by using these Nrv2-GAL4 UAS GFP lines. Double Labeling of Whole-Mount Embryos with Anti-GFP and Anti-HRP Antibodies. Embryos were collected from Nrv2-GAL4 UAS GFP flies and staged at 25 C. They were dechorionated, fixed, and processed for immunocytochemistry as described by Patel (18). After blocking in PBT NGS (0.1% Triton X-100/0.1% BSA/5.0% normal goat serum in PBS), the embryos were incubated with a 1:5 dilution of rat anti-gfp mab JFP-J1 (kindly provided by S. Fujita, Mitsubishi Kasei, Tokyo) and rabbit anti-hrp antibodies (1 g/ml, Sigma). After overnight incubation at 4 C, the embryos were washed and probed with FITC-labeled goat anti-rat IgG (1:200 dilution; Pierce) and rhodamine-labeled goat anti-rabbit IgG (1:200 dilution; Pierce) for 2 h at room temperature (8). After washing, preparations were mounted in 70% (vol/vol) glycerol (in PBS) and viewed under a fluorescence microscope. Visualization of GFP with Confocal Microscopy. Dechorionated embryos or first-instar larvae were directly mounted in a 1:1 mixture of glycerol and PBS. Samples were observed directly by using a Zeiss LSM 410 confocal scanning microscope. Excitation was with the argon laser (488-nm excitation), and emission was at 527 nm. Visualization of GFP by Using Wide Field Fluorescence Microscopy. Mechanically dechorionated live embryos, larvae, or pupae were mounted in a 1:1 mixture of glycerol/pbs in a small viewing chamber constructed of a filter paper support for a standard coverslip and viewed directly. Adult flies were sealed in an imaging chamber (Molecular Probes) and observed directly. Fly parts (head, abdomen, leg, and wing) were collected from adults by manual dissection and mounted in 70% (vol/vol) glycerol (in PBS). Samples were observed with a Nikon Diavert or Olympus Vannox fluorescence microscope under Hg illumination by using standard FITC fluorescence filters. Digital images were recorded by using either a Cambridge Research (Cambridge, MA) Real-14 cooled CCD camera or a Kodak DC120 color CCD camera. Image files were processed by using SCION IMAGE (Scion, Frederick, MD), IMAGE PRO (Media Cybernetics, Silver Spring, MD), or PHOTOSHOP (Adobe Systems, Mountain View, CA). RESULTS GFP Expression in Embryos. Exposure of embryos to the repeated periods of several minutes of UV light required for observation did not affect their viability. Most embryos ( 75%) completed development when they were observed repeatedly over a several-hour period and hatched into active larvae. GFP expression was easily monitored in the developing nervous system (both central and peripheral) at different embryonic stages (Fig. 1 A D). GFP fluorescence initially was detected in a group of cells at the anterior end of stage-12 embryos and a few cells in the ventral nerve cord, which are partially obscured by gut autofluorescence (Fig. 1A). As neural development proceeded, the fluorescence became visible in many cells in the brain and ventral nerve cord and was especially intense in some peripheral sensory neurons near the surface of the developing embryo (Fig. 1 B D). In stage-17 FIG. 1. GFP fluorescence and double-labeled antibody staining in embryos. A live stage-12 embryo (A) shows green fluorescent neurons in the anterior end (arrowhead) and in the ventral nerve cord (asterisk). The ventral nerve cord neurons are partially obscured by the strong yellow autofluorescence of the gut at this stage. In stage-15 embryos (B), some of the peripheral sensory neurons near the surface as well as many cells in the ventral nerve cord are visible. By stage 16 (C) and 17 (D) more cells in the ventral nerve cord and brain and a few peripheral sensory neurons show fluorescence. The same field of view is shown for a stage-17 embryo in E near the surface and F more internally. All embryos are oriented with anterior to the left and ventral to the bottom. A D are color charge-coupled device images, E and F are false-colored black and white charge-coupled device images. Fixed, stage-12 embryos were double stained with anti-gfp (G and H) and anti-hrp (I and J) antibodies. The hazy green area in G is yellow autofluorescence of the gut. The complete nervous system is stained with anti-gfp antibody (G) and is shown at higher power (H) or is stained with anti-hrp antibody (I) and is shown at higher power (J). Images in G and I as well as H and J were merged to produce K and L, respectively. There is a complete correspondence of the anti-gfp and anti-hrp staining. Note also the difference between GFP fluorescence in a live stage-12 embryo (A) and anti-gfp antibody staining of a similarly aged fixed embryo (G and H). The images in G L are false-colored. br, brain; gt, developing gut; vnc, ventral nerve cord. (Bar 100 m for whole embryos, 10 m for higher power views in H, J, and L.)
3 10440 Neurobiology: Sun et al. Proc. Natl. Acad. Sci. USA 96 (1999) embryos, the pattern of fluorescence is similar to early larval stages (Fig. 1 E and F). We also have observed embryos at different developmental stages by using confocal microscopy. In a stage-17 embryo, the GFP signal is easily observed in neurons and neural processes of both the central (brain, ventral nerve cord) and peripheral (Fig. 2 A and B) nervous system. A three-dimensional projection of confocal optical sections clearly reveals the extent and relationship of the GFP fluorescent cells and processes (Fig. 2B). An animated three-dimensional projection of confocal sections can be viewed as supplemental data on the PNAS web site ( A higher-power three-dimensional projection of optical sections through an early first-instar larval ventral nerve cord shows the cellular morphology and fluorescent nerve fibers (Fig. 2C). To establish the cellular specificity of GFP expression in Nrv2-GAL4 UAS GFP embryos, we compared the immunocytochemical staining of stage-12 embryos by using an FIG. 2. Confocal microscopy. (A) The nine small images are individual optical sections (spaced 6 m apart) taken from the anterior of a stage-17 embryo. (B) Twenty-eight optical sections from the same embryo (spaced 2 m apart) are projected to reconstruct the complete three-dimensional distribution of fluorescence. The single and double arrows indicate especially bright peripheral neurons. An animation of this projection series is available as supplemental data on the PNAS web site ( (C) A higher power view of fluorescence in the central part of the ventral nerve cord of an early first-instar larvae. Twenty optical sections (spaced 2.5 m apart) are projected to reconstruct the three-dimensional view. The fluorescence is clearly present in most central nervous system cells and processes. All images are oriented with anterior to the left and ventral down. [Bar 100 m (B) and 10 m (C).]
4 Neurobiology: Sun et al. Proc. Natl. Acad. Sci. USA 96 (1999) anti-gfp antibody to the staining pattern obtained with anti-hrp antibodies, a known pan-neuronal marker (10). Double labeling of whole-mount embryos with anti-gfp and anti-hrp antibodies showed an indistinguishable pattern of immunocytochemical staining, indicating that GFP expression is likely to be pan-neuronal at early embryonic stages (Fig. 1 G L). GFP expression is detected a few hours earlier by antibody staining than by fluorescence. This is most likely because of the time required for posttranslational modification (oxidation) required to produce GFP fluorescence (19, 20). One difference between the GFP expression pattern in laterstage embryos and anti-hrp staining is the absence of fluorescence in many of the v, v, l, and d peripheral sensory neurons (21). GFP Expression in Larvae. The exceptionally strong GFP signal in larvae allows direct visualization of the nervous system in fully active living animals. Especially striking when viewing fully active larvae is an appreciation of the dynamic mechanical forces that the brain and nerves are subjected to during movement. The brain and ventral ganglion as well as the nerve fibers are rapidly stretched and compressed as the animals move about. A QUICKTIME movie of a fluorescent larvae is available for viewing as supplemental data on the PNAS web site ( Extremely bright GFP fluorescence is specifically observed in the brain and ventral ganglion of all three larval instars. Because of the number of fluorescent cells, it is difficult to resolve individual cell types by using conventional fluorescence microscopy. The long longitudinal nerve fibers, segmental branches, and fine fibers at the distal ends of the segmental nerves are also clearly seen, as are the nerve fibers in the anterior part of the larvae (Fig. 3 A C). The bright fluorescent spots in the lateral regions of the larvae may be peripheral glial cells based on their position. Nervous system structures that do not fluoresce in larvae include most of the peripheral sensory neurons and the differentiating photoreceptor neurons of the third-instar larval eye disk. GFP Expression in Pupae. The intensity of GFP fluorescence is strong enough that we can monitor the morphological changes of the nervous system in developing pupae directly FIG. 3. GFP fluorescence in larvae, pupae, and adults. First- (A), second- (B), and third- (C) instar larvae show intense fluorescence in the brain and ventral ganglion as well as the longitudinal and segmental nerves and nerve branches. The large bright structures at the ends of segmental nerves may be peripheral glia (indicated by arrowheads). The specimens in A and C are viewed from the ventral surface whereas B is a lateral view. The image in C has been constructed from images of three smaller fields. In early pupae (D), the fluorescence is similar to late third-instar larvae. As metamorphosis proceeds, the nerves begin to appear wavy and frayed (E, and at higher power in F). Later stages of pupation show decreased fluorescence (G) and a shape more characteristic of adult central nervous system. Leg nerves are visible in late-stage pupae (H) or a few hours before hatching (I) and are indicated with arrowheads. Whole adults show bright fluorescence in the optic lobes, brain, thoracic ganglion, and many kinds of nerves. In J, the thoracic ganglion, leg, and probosal nerves are evident, while the optic lobe and brain staining is obscured by the red eye. An adult female abdomen shows major nerves with many finer branches at higher power (K). A dissected head viewed from the posterior shows the intense fluorescence in the brain, optic lobes, and proboscal nerves (L). The leg nerve (M) and wing nerve (N) are also clearly visible in dissected parts from adults. an, abdominal nerve; br, brain; ey, eye; ln, longitudinal nerve; ol, optic lobe; pb, proboscis; sn, segmental nerve; tg, thoracic ganglion; vnc, ventral nerve cord. (Bars 100 m.)
5 10442 Neurobiology: Sun et al. Proc. Natl. Acad. Sci. USA 96 (1999) through the pupal case during metamorphosis (Fig. 3 D I). The nervous system in early pupae is similar to that of third-instar larvae (Fig. 3D). As metamorphosis proceeds, the nerve-fiber bundles become wavy and begin to appear defasiculated (Fig. 3 E and F). Later, the fluorescence intensity of nerve fibers and cell bodies decreases, and eventually the fibers disappear in 1-day-old pupae (Fig. 3G). The central nervous system also undergoes morphological changes in overall shape, replacing the larval central nervous system shape with an adult shape. The GFP fluorescence is seen in brain, optic lobes, and thoracico-abdominal ganglion in the 1-day-old pupae (Fig. 3G). As metamorphosis continues, the intensity of GFP fluorescence increases along with the development of new nervous system structures (Fig. 3 H and I). GFP fluorescence can clearly be seen in peripheral nervous system structures, such as antennae, labial palps, maxillary palps, and even leg nerves, at later stages of pupation. The optic lobes are extremely bright in pupae close to eclosion (Fig. 3I). GFP Expression in Adults. Remarkably, we can even observe GFP fluorescence directly through the pigmented cuticle of intact living adults (Fig. 3J). Fluorescence is localized in brain, optic lobes, and thoracic ganglion, as well as in parts of the peripheral nervous system. For example, strong fluorescence is seen in antennae and the ocellus (photoreceptor organ located at the top of the head). In dissected adult fly parts, it is easier to recognize finer details of the various nerve-fiber tracts. In adult heads, in addition to the intense fluorescence in the brain and optic lobes, the maxillary and labial nerves in the proboscis are visible (Fig. 3L). GFP fluorescence is also clearly observed in the nerves of the abdomen, leg, and wing (Fig. 3 K, M, and N). DISCUSSION The Nrv2-GAL4 UAS GFP lines we describe here provide a view of the nervous system in live Drosophila at all developmental stages. The advantages of using Nrv2-specific transcriptional regulatory elements are the extremely robust expression and the apparently complete restriction of expression to the nervous system. Part of the robust expression is derived from the use of the strong yeast transcriptional activator GAL4, but part is also because of the high-level activity of the Nrv2 regulatory DNA. The sodium pump is an important homeostatic regulator with exceptionally high levels of expression in excitable cells, such as neurons. We also have observed even more robust muscle-specific GFP fluorescence in transgenic lines constructed by using the Nrv1 5 flanking DNA (unpublished observations). A number of laboratories have observed GAL4-mediated nervous system-specific expression of GFP in live Drosophila or in Caenorhabditis elegans; (1, 4, 22). Many of the Drosophila GAL4 lines that have been described show expression in subsets of glial and/or neurons (for recent review, see ref. 23). These subset-specific lines provide excellent tools for dynamic visualization of particular types of neurons or glia. In most cases, the GFP fluorescence patterns have been analyzed only during early embryogenesis and have not been described at later developmental stages. Most subset-specific expression of GFP relies on the use of rather specialized types of promoters or enhancer trap lines to direct GFP expression either directly or with the GAL4 activation system (23, 24). Many neuronal and glial genes, especially those with very specialized functions, may not be expressed at high enough levels to be useful for GFP fluorescence in living animals. Other homeostatic or housekeeping genes with potentially strong promoters, such as cellular actin or microtubule protein genes, also could be used in principle to drive high-level expression of GFP fluorescence. Unfortunately, most are expressed too broadly to target individual tissue types such as nervous system (25, 26). Fortunately, Drosophila has a single Nrv2 sodium pump -subunit gene whose expression is restricted to nervous system (9, 11). The expression of Nrv2 is independently regulated by separate transcriptional elements relative to the other Drosophila sodium pump -subunit gene (Nrv1). These separate regulatory elements are located in the 5 flanking DNA (7), and this independent regulation is essential to achieve the specificity of tissue-specific expression we report here. The Nrv2 gene is alternatively spliced to produce two different proteins (Nrv 2.1 and 2.2) with differential expression in head and thoracic ganglia (11), but apparently this further specificity occurs posttranscriptionally. In other species, individual - and -subunit genes of Na,K - ATPase are more numerous than in Drosophila. Several specific isoforms also are expressed at high levels in neurons in particular combinations (27), but the patterns are more complex than the case for Drosophila -subunit expression. Only a few genes have been described that are expressed specifically in most or all neurons throughout development. In addition to Nrv2 expression, the Drosophila elav gene product has been detected immunocytochemically in neurons at all developmental stages (28 30). We have compared the Nrv2- GAL4 UAS GFP lines described here with two different elav-gal4 lines (31). The patterns of elav-gal4- and Nrv2- GAL4-directed GFP fluorescence are similar, with a few notable differences. First, the GFP fluorescence is much more intense in our Nrv2-GAL4 UAS GFP lines. Typical integration times for the charge-coupled device images presented in this paper are less than 0.1 sec for Nrv2-GAL4 lines, whereas the equivalent stage elav-gal4 lines need integration times of 30 sec 1 min. Second, we do not see fluorescence in many peripheral sensory neurons in the Nrv2-GAL4 lines in late embryo and larval stages that are observed in elav-gal4 lines. Finally, we do not see fluorescence in developing photoreceptor cell neurons in third-instar larval eye disks in the Nrv2- GAL4 lines that are seen in elav-gal4 lines. The elav-gal4 lines thus seem to express GFP fluorescence in more types of neurons than the Nrv2-GAL4 lines. Our failure to detect fluorescence in the latter stage peripheral nervous system and photoreceptor neurons could indicate that the Nrv2 construct is missing some regulatory elements or that the signal in these types of neurons may be below the detection level of fluorescence microscopy. The ability to view the nervous system in living embryos makes our lines an excellent preparation to construct time lapse records of early nervous system development. One important aspect of Na,K -ATPase expression is that it is required to generate and maintain the ionic gradients essential for functionally active neurons and glia. Certain cells therefore may express the pump at different times in their developmental life histories, and such differences could indicate when the neurons become functionally active. It will be interesting to combine observations on our Nrv2-GAL4 UAS GFP lines with other markers of specific neuronal and glial cell types that may be recognized before the particular cells become functional. During larval stages, our lines can be used to observe the dynamics of nervous system movements accompanying the movement of larvae. They also can be useful for tracing the main axonal pathways. The ability to see GFP fluorescence through the pupal case also will make it possible to record time-lapse movies of the remodeling of the nervous system during metamorphosis. Our lines provide excellent visualization of particular neurons and nerve fibers that may be accessible for electrophysiological recording or lesioning studies. The strength of Drosophila as a model organism is in its genetic definition. We foresee many uses for our lines in genetic experiments. For example, the Nrv2-GAL4 UAS GFP could serve as a marker to view the effects of particular mutations on nervous system structure and development.
6 Neurobiology: Sun et al. Proc. Natl. Acad. Sci. USA 96 (1999) Existing axonal pathfinding or neurodegenerative mutants would be good candidate genes to examine in this regard (32 34). Our lines also could serve as a genetic background for generating new mutations that effect axonal pathfinding or neurodegeneration because they could easily be screened for phenotypes by simple observation with a fluorescence microscope. We have not attempted to increase the utility of our lines by modifying the GFP reporter gene. Others have produced GFP fusion proteins, for example, that incorporate protein motifs that direct the reporter to a particular intracellular compartment such as the plasma membrane (35), microtubules (36), neuronal processes (37), or nucleus (38). The flexible design of the GAL4 activation system will make it easy to incorporate these types of GFP-fusion reporters in future work. It also will be possible to direct a variety of other functional or mutant genes to the nervous system by using the Nrv2-GAL4 driver. One limitation of using GFP fluorescence is the time required for posttranslational oxidation to generate a fluorescent protein (19). We have used the S65T mutated form of GFP, which has been reported to speed up the acquisition of fluorescence (39), but generation of the fluorochrome still lags expression of GFP protein, as assessed by anti-gfp antibody staining. Future improvements in GFP design may lessen this time lag and thus make our lines more useful for studying early neurogenesis in living animals. We thank our colleagues Drs. T. Kitamoto, K. Ikeda, E. Roberts, L. Iverson, S. Novak, and R. Williamson for their helpful advice and criticism. This work was supported by a grant from the American Heart Association. 1. Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W. & Prasher, D. C. (1994) Science 263, Prasher, D. C. (1995) Trends Genet. 11, Gerdes, H. H. & Kaether, C. (1996) FEBS Lett. 389, Brand, A. (1995) Trends Genet. 11, Fleming, J. T., Squire, M. D., Barnes, T. M., Tornoe, C., Matsuda, K., Ahnn, J., Fire, A., Sulston, J. E., Barnard, E. A., Sattelle, D. B. & Lewis, J. A. (1997) J. Neurosci. 17, Labrousse, A. M., Shurland, D. L. & van der Bliek, A. M. (1998) Mol. Biol. Cell 9, Xu, P., Sun, B. & Salvaterra, P. M. (1999) Gene, in press. 8. Sun, B. & Salvaterra, P. M. (1995) J. Neurochem. 65, Sun, B. & Salvaterra, P. M. (1995) Proc. Natl. Acad. Sci. USA 92, Jan, L. Y. & Jan, Y. N. (1982) Proc. Natl. Acad. Sci. USA 79, Sun, B., Wang, W. & Salvaterra, P. M. (1998) J. Neurochem. 71, Sweadner, K. J. (1989) Biochim. Biophys. Acta 988, Lingrel, J. B., Orlowski, J., Shull, M. M. & Price, E. M. (1990) Prog. Nucleic Acid Res. Mol. Biol. 38, Thummel, C. S., Boulet, A. M. & Lipshitz, H. D. (1988) Gene 74, Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, Rubin, G. M. & Spradling, A. C. (1982) Science 218, Greenspan, R. J. (1997) Fly Pushing: The Theory and Practice of Drosophila Genetics (Cold Spring Harbor Lab. Press, Plainview, NY). 18. Patel, N. H. (1994) Methods Cell Biol. 44, Heim, R., Prasher, D. C. & Tsien, R. Y. (1994) Proc. Natl. Acad. Sci. USA 91, Inouye, S. & Tsuji, F. I. (1994) FEBS Lett. 341, Ghysen, A., Dambly-Chaudiere, C., Aceves, D., Jan, L. Y. & Jan, Y. N. (1986) Roux s Arch. Dev. Biol. 195, Yeh, E., Gustafson, K. & Boulianne, G. L. (1995) Proc. Natl. Acad. Sci. USA 92, Brand, A. (1999) Methods Cell Biol. 58, Itoh, K., Urban, J. & Technau, G. M. (1995) Roux s Arch. Dev. Biol. 204, Endow, S. A. (1999) Methods Cell Biol. 58, Edwards, K. A., Demsky, M., Montague, R. A., Weymouth, N. & Kiehart, D. P. (1997) Dev. Biol. 191, Blanco, G. & Mercer, R. W. (1998) Am. J. Physiol. 275, F633 F Robinow, S. & White, K. (1988) Dev. Biol. 126, Robinow, S. & White, K. (1991) J. Neurobiol. 22, Yao, K. M. & White, K. (1994) J. Neurochem. 63, Luo, L., Liao, Y. J., Jan, L. Y. & Jan, N. J. (1994) Genes Dev. 8, Baird, D. H., Koto, M. & Wyman, R. J. (1993) J. Neurobiol. 24, Hoang, B. & Chiba, A. (1998) J. Neurosci. 18, Holmes, A. L., Raper, R. N. & Heilig, J. S. (1998) Genetics 148, Lee, T. & Luo, L. (1999) Neuron 22, Theurkauf, W. E. & Hazelrigg, T. I. (1998) Development (Cambridge, U.K.) 125, Murray, M. J., Merritt, D. J., Brand, A. H. & Whitington, P. M. (1998) J. Neurobiol. 37, Davis, I., Girdham, C. H. & O Farrell, P. H. (1995) Dev. Biol. 170, Heim, R., Cubitt, A. B. & Tsien, R. Y. (1995) Nature (London) 373,
Trasposable elements: Uses of P elements Problem set B at the end
 Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Supplementary Methods
 Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
SUPPORTING ONLINE MATERIAL
 SUPPORTING ONLINE MATERIAL MATERIAL AND METHODS Fly stocks All Drosophila stocks were grown at 25 C on yeast-containing corn meal/molasses medium. The nanos (nos) alleles nos 18 and nos 53 have been previously
SUPPORTING ONLINE MATERIAL MATERIAL AND METHODS Fly stocks All Drosophila stocks were grown at 25 C on yeast-containing corn meal/molasses medium. The nanos (nos) alleles nos 18 and nos 53 have been previously
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
Supplementary Methods
 Caussinus and Gonzalez Supplementary Methods Mitotic clones Clones of NBs homozygous for numb 03235, mira ZZ176, pros 17 or apkc k06403 were generated by FLP/FRT mediated mitotic recombination (1). As
Caussinus and Gonzalez Supplementary Methods Mitotic clones Clones of NBs homozygous for numb 03235, mira ZZ176, pros 17 or apkc k06403 were generated by FLP/FRT mediated mitotic recombination (1). As
GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts
 Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
a) Stock 4: 252 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.
![a) Stock 4: 252 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males. a) Stock 4: 252 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.](/thumbs/91/106088469.jpg) Drosophila Problem Set 2018 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
Drosophila Problem Set 2018 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
a) Stock 4: 126 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.
![a) Stock 4: 126 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males. a) Stock 4: 126 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.](/thumbs/88/116538069.jpg) Drosophila Problem Set 2012 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
Drosophila Problem Set 2012 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
Visualizing Cells Molecular Biology of the Cell - Chapter 9
 Visualizing Cells Molecular Biology of the Cell - Chapter 9 Resolution, Detection Magnification Interaction of Light with matter: Absorbtion, Refraction, Reflection, Fluorescence Light Microscopy Absorbtion
Visualizing Cells Molecular Biology of the Cell - Chapter 9 Resolution, Detection Magnification Interaction of Light with matter: Absorbtion, Refraction, Reflection, Fluorescence Light Microscopy Absorbtion
fga phenotypes were also analyzed in the tracheal system, the organ responsible for
 SUPPLEMENTARY TEXT Tracheal defects fga phenotypes were also analyzed in the tracheal system, the organ responsible for oxygen delivery in the fly. In fga homozygous mutant alleles, we observed defects
SUPPLEMENTARY TEXT Tracheal defects fga phenotypes were also analyzed in the tracheal system, the organ responsible for oxygen delivery in the fly. In fga homozygous mutant alleles, we observed defects
Genome manipulation by homologous recombination in Drosophila Xiaolin Bi and Yikang S. Rong Date received (in revised form): 9th May 2003
 Xiaolin Bi is a post doctoral research fellow at the Laboratory of Molecular Cell Biology, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA. Yikang S. Rong is the principal
Xiaolin Bi is a post doctoral research fellow at the Laboratory of Molecular Cell Biology, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA. Yikang S. Rong is the principal
Lecture 20: Drosophila embryogenesis
 Lecture 20: Drosophila embryogenesis Mitotic recombination/clonal analyses Embrygenesis Four classes of genes: Maternal genes Gap genes Pair-rule genes Segment polarity genes Homeotic genes Read 140-141;
Lecture 20: Drosophila embryogenesis Mitotic recombination/clonal analyses Embrygenesis Four classes of genes: Maternal genes Gap genes Pair-rule genes Segment polarity genes Homeotic genes Read 140-141;
A Brief History of Light Microscopy And How It Transformed Biomedical Research
 A Brief History of Light Microscopy And How It Transformed Biomedical Research Suewei Lin Office: Interdisciplinary Research Building 8A08 Email: sueweilin@gate.sinica.edu.tw TEL: 2789-9315 Microscope
A Brief History of Light Microscopy And How It Transformed Biomedical Research Suewei Lin Office: Interdisciplinary Research Building 8A08 Email: sueweilin@gate.sinica.edu.tw TEL: 2789-9315 Microscope
Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling
 Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling Go-Woon Kim, Jong-Hoon Won, Ok-Kyung Lee, Sang-Soo Lee, Jeong-Hoon Han, Orkhon Tsogtbaatar, Sujin Nam, Yeon Kim, and Kyung-Ok Cho* Supplementary
Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling Go-Woon Kim, Jong-Hoon Won, Ok-Kyung Lee, Sang-Soo Lee, Jeong-Hoon Han, Orkhon Tsogtbaatar, Sujin Nam, Yeon Kim, and Kyung-Ok Cho* Supplementary
Lecture 8: Transgenic Model Systems and RNAi
 Lecture 8: Transgenic Model Systems and RNAi I. Model systems 1. Caenorhabditis elegans Caenorhabditis elegans is a microscopic (~1 mm) nematode (roundworm) that normally lives in soil. It has become one
Lecture 8: Transgenic Model Systems and RNAi I. Model systems 1. Caenorhabditis elegans Caenorhabditis elegans is a microscopic (~1 mm) nematode (roundworm) that normally lives in soil. It has become one
Illumatool ΤΜ Tunable Light System: A Non-Destructive Light Source For Molecular And Cellular Biology Applications. John Fox, Lightools Research.
 Illumatool ΤΜ Tunable Light System: A Non-Destructive Light Source For Molecular And Cellular Biology Applications. John Fox, Lightools Research. Fluorescent dyes and proteins are basic analytical tools
Illumatool ΤΜ Tunable Light System: A Non-Destructive Light Source For Molecular And Cellular Biology Applications. John Fox, Lightools Research. Fluorescent dyes and proteins are basic analytical tools
Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex
 Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex 1.0. Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigen-specific antibodies coupled
Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex 1.0. Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigen-specific antibodies coupled
Mystery microscope images
 Mystery microscope images Equipment: 6X framed microscopy images 6X cards saying what each microscope image shows 6X cards with the different microscopy techniques written on them 6X cards with the different
Mystery microscope images Equipment: 6X framed microscopy images 6X cards saying what each microscope image shows 6X cards with the different microscopy techniques written on them 6X cards with the different
Evolutionary conservation of midline repulsion by Robo family receptors in flies and mice
 University of Arkansas, Fayetteville ScholarWorks@UARK Biological Sciences Undergraduate Honors Theses Biological Sciences 5-2018 Evolutionary conservation of midline repulsion by Robo family receptors
University of Arkansas, Fayetteville ScholarWorks@UARK Biological Sciences Undergraduate Honors Theses Biological Sciences 5-2018 Evolutionary conservation of midline repulsion by Robo family receptors
Lecture 20: Drosophila melanogaster
 Lecture 20: Drosophila melanogaster Model organisms Polytene chromosome Life cycle P elements and transformation Embryogenesis Read textbook: 732-744; Fig. 20.4; 20.10; 20.15-26 www.mhhe.com/hartwell3
Lecture 20: Drosophila melanogaster Model organisms Polytene chromosome Life cycle P elements and transformation Embryogenesis Read textbook: 732-744; Fig. 20.4; 20.10; 20.15-26 www.mhhe.com/hartwell3
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information
 Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Flybow genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster
 Nature Methods Flybow genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster Dafni Hadjieconomou, Shay Rotkopf, Cyrille Alexandre, Donald M Bell, Barry J Dickson & Iris
Nature Methods Flybow genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster Dafni Hadjieconomou, Shay Rotkopf, Cyrille Alexandre, Donald M Bell, Barry J Dickson & Iris
Creating mosaics in Drosophila
 Int. J. Dev. Biol. 42: 243-247 (1998) Creating mosaics in Drosophila NORBERT PERRIMON* Department of Genetics, Harvard Medical School, Boston,USA ABSTRACT The ability to create mosaic animals allows the
Int. J. Dev. Biol. 42: 243-247 (1998) Creating mosaics in Drosophila NORBERT PERRIMON* Department of Genetics, Harvard Medical School, Boston,USA ABSTRACT The ability to create mosaic animals allows the
Janos Szabad Department of Biology University of Szeged 6720 Szeged, Somogyi str
 Janos Szabad Department of Biology University of Szeged 6720 Szeged, Somogyi str. 4. E-mail: szabad.janos@med.u-szeged.hu - Through the use of antibodies - against the protein - against a fusion partner
Janos Szabad Department of Biology University of Szeged 6720 Szeged, Somogyi str. 4. E-mail: szabad.janos@med.u-szeged.hu - Through the use of antibodies - against the protein - against a fusion partner
Fluorescence Microscopy. Terms and concepts to know: 10/11/2011. Visible spectrum (of light) and energy
 Fluorescence Microscopy Louisiana Tech University Ruston, Louisiana Microscopy Workshop Dr. Mark DeCoster Associate Professor Biomedical Engineering 1 Terms and concepts to know: Signal to Noise Excitation
Fluorescence Microscopy Louisiana Tech University Ruston, Louisiana Microscopy Workshop Dr. Mark DeCoster Associate Professor Biomedical Engineering 1 Terms and concepts to know: Signal to Noise Excitation
Drosophila, Genetics, Behavior, and Optogenetics. David Deitcher BioNB 4910
 Drosophila, Genetics, Behavior, and Optogenetics David Deitcher BioNB 4910 Seymour Benzer Father of Drosophila Behavior Mutant Behaviors Identified by Benzer and Others Many of these mutants encode genes
Drosophila, Genetics, Behavior, and Optogenetics David Deitcher BioNB 4910 Seymour Benzer Father of Drosophila Behavior Mutant Behaviors Identified by Benzer and Others Many of these mutants encode genes
Name AP Biology Mrs. Laux Take home test #11 on Chapters 14, 15, and 17 DUE: MONDAY, DECEMBER 21, 2009
 MULTIPLE CHOICE QUESTIONS 1. Inducible genes are usually actively transcribed when: A. the molecule degraded by the enzyme(s) is present in the cell. B. repressor molecules bind to the promoter. C. lactose
MULTIPLE CHOICE QUESTIONS 1. Inducible genes are usually actively transcribed when: A. the molecule degraded by the enzyme(s) is present in the cell. B. repressor molecules bind to the promoter. C. lactose
mod-1::mcherry unc-47::gfp RME ser-4::gfp vm2 vm2 vm2 vm2 VNC
 A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
Cell Lineage in the Development of the Leech Nervous System
 Cell Lineage in the Development of the Leech Nervous System DAVID A. WEISBLAT Department of Molecular Biology, University of California, Berkeley, CA 94720 (U.S.A) INTRODUCTION Knowledge of the lines of
Cell Lineage in the Development of the Leech Nervous System DAVID A. WEISBLAT Department of Molecular Biology, University of California, Berkeley, CA 94720 (U.S.A) INTRODUCTION Knowledge of the lines of
λ N -GFP: an RNA reporter system for live-cell imaging
 λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
SUPPLEMENTARY INFORMATION
 Normalized dorsal trunk length (µm) Normalized dorsal trunk length (TC1-5) (µm) DOI: 1.138/ncb2467 a b c d btl>src64 Src42, btl>src64 e Src64 f g h DT Src64 DT Midgut Src64 Midgut i Src64 Atpα j k l DT
Normalized dorsal trunk length (µm) Normalized dorsal trunk length (TC1-5) (µm) DOI: 1.138/ncb2467 a b c d btl>src64 Src42, btl>src64 e Src64 f g h DT Src64 DT Midgut Src64 Midgut i Src64 Atpα j k l DT
Neural Stem Cell Characterization Kit
 Neural Stem Cell Characterization Kit Catalog No. SCR019 FOR RESEARCH USE ONLY Not for use in diagnostic procedures USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209 Europe +44 (0) 23 8026 2233
Neural Stem Cell Characterization Kit Catalog No. SCR019 FOR RESEARCH USE ONLY Not for use in diagnostic procedures USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209 Europe +44 (0) 23 8026 2233
A functional analysis of 5, intronic and promoter regions of the homeotic gene proboscipedia in Drosophila melanogaster
 Development 121, 2127-2141 (1995) Printed in Great Britain The Company of Biologists Limited 1995 2127 A functional analysis of 5, intronic and promoter regions of the homeotic gene proboscipedia in Drosophila
Development 121, 2127-2141 (1995) Printed in Great Britain The Company of Biologists Limited 1995 2127 A functional analysis of 5, intronic and promoter regions of the homeotic gene proboscipedia in Drosophila
The cell lineage of the muscles of the Drosophila head
 J. Embryol. exp. Morph. 85,285-294 (1985) 285 Printed in Great Britain The Company of Biologists Limited 1985 The cell lineage of the muscles of the Drosophila head K. VIJAY RAGHAVAN* A LUDWIN PINTO Molecular
J. Embryol. exp. Morph. 85,285-294 (1985) 285 Printed in Great Britain The Company of Biologists Limited 1985 The cell lineage of the muscles of the Drosophila head K. VIJAY RAGHAVAN* A LUDWIN PINTO Molecular
Supporting Information
 Supporting Information Willecke et al. 1.173/pnas.852115 A A' clones A'' A''' ' GFP clones ' fj- clones D D' Diap1 fj- clones Fig. S1. oundaries of and activity induce Hippo target genes. Third instar
Supporting Information Willecke et al. 1.173/pnas.852115 A A' clones A'' A''' ' GFP clones ' fj- clones D D' Diap1 fj- clones Fig. S1. oundaries of and activity induce Hippo target genes. Third instar
Supplementary Figure 1 TSG experimental strategy.
 Supplementary Figure 1 TSG experimental strategy. (a-d) Protocol. (a) GR construct in entry vector pcr8-gw-topo. (b) AWM-2attB accepts open reading frames (ORFs) from entry vector through LR clonase reaction.
Supplementary Figure 1 TSG experimental strategy. (a-d) Protocol. (a) GR construct in entry vector pcr8-gw-topo. (b) AWM-2attB accepts open reading frames (ORFs) from entry vector through LR clonase reaction.
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/327/5969/1126/dc1 Supporting Online Material for A Nodule-Specific Protein Secretory Pathway Required for Nitrogen- Fixing Symbiosis Dong Wang, Joel Griffitts, Colby
www.sciencemag.org/cgi/content/full/327/5969/1126/dc1 Supporting Online Material for A Nodule-Specific Protein Secretory Pathway Required for Nitrogen- Fixing Symbiosis Dong Wang, Joel Griffitts, Colby
Methods and Materials
 SUMMARY OF PH.D. THESIS Molecular biological and genetic study of the sumoylation of Drosophila melanogaster p53 Pardi Norbert Supervisor: Dr. Boros Imre Doctoral School of Biology University of Szeged
SUMMARY OF PH.D. THESIS Molecular biological and genetic study of the sumoylation of Drosophila melanogaster p53 Pardi Norbert Supervisor: Dr. Boros Imre Doctoral School of Biology University of Szeged
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
New Tool for Late Stage Visualization of Oskar Protein in Drosophila melanogaster. Oocytes. Kathleen Harnois. Supervised by Paul Macdonald
 New Tool for Late Stage Visualization of Oskar Protein in Drosophila melanogaster Oocytes Kathleen Harnois Supervised by Paul Macdonald Institute of Cellular and Molecular Biology, The University of Texas
New Tool for Late Stage Visualization of Oskar Protein in Drosophila melanogaster Oocytes Kathleen Harnois Supervised by Paul Macdonald Institute of Cellular and Molecular Biology, The University of Texas
Map-Based Cloning of Qualitative Plant Genes
 Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
Gene specific primers. Left border primer (638) ala3-4 (Salk_082157) Gene specific primers. Left border primer (1343)
 Supplemental Data. Poulsen et al. (2008) The Arabidopsis P4-ATPase pump ALA3 localizes to the Golgi and requires a β subunit to function in lipid translocation and secretory vesicle formation. A ala3-1
Supplemental Data. Poulsen et al. (2008) The Arabidopsis P4-ATPase pump ALA3 localizes to the Golgi and requires a β subunit to function in lipid translocation and secretory vesicle formation. A ala3-1
X-gal Staining of Wholemount Drosophila Embryos. 3.7 % formaldehyde in 0.1 M phosphate buffer (ph 7.2) 0.1M Phosphate buffer (ph 7.2) and 0.
 X-gal Staining of Wholemount Drosophila Embryos Modified from The Little Blue Book (Gehring lab) Solutions 1. NaCl+TX: 3. PBT: 4. Fe/NaP Buffer: 0.7% NaCl and 0.3% Triton 3.7 % formaldehyde in 0.1 M phosphate
X-gal Staining of Wholemount Drosophila Embryos Modified from The Little Blue Book (Gehring lab) Solutions 1. NaCl+TX: 3. PBT: 4. Fe/NaP Buffer: 0.7% NaCl and 0.3% Triton 3.7 % formaldehyde in 0.1 M phosphate
Mos1 insertion. MosTIC protocol-11/2006. repair template - Mos1 transposase expression - Mos1 excision - DSB formation. homolog arm.
 MosTIC (Mos1 excision induced Transgene Instructed gene Conversion) Valérie Robert (vrobert@biologie.ens.fr) and Jean-Louis Bessereau (jlbesse@biologie.ens.fr) (November 2006) Introduction: MosTIC (Robert
MosTIC (Mos1 excision induced Transgene Instructed gene Conversion) Valérie Robert (vrobert@biologie.ens.fr) and Jean-Louis Bessereau (jlbesse@biologie.ens.fr) (November 2006) Introduction: MosTIC (Robert
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure S1. USP-46 is expressed in several tissues including the nervous system
 Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
CHAPTER 20 DNA TECHNOLOGY AND GENOMICS. Section A: DNA Cloning
 Section A: DNA Cloning 1. DNA technology makes it possible to clone genes for basic research and commercial applications: an overview 2. Restriction enzymes are used to make recombinant DNA 3. Genes can
Section A: DNA Cloning 1. DNA technology makes it possible to clone genes for basic research and commercial applications: an overview 2. Restriction enzymes are used to make recombinant DNA 3. Genes can
Nature Methods: doi: /nmeth.3250
 Supplementary Figure 1 In vitro and in vivo activities of additional QF variants. This figure characterises the additional QF variants generated to find the region responsible for QF toxicity. Some variants
Supplementary Figure 1 In vitro and in vivo activities of additional QF variants. This figure characterises the additional QF variants generated to find the region responsible for QF toxicity. Some variants
Materials and methods. by University of Washington Yeast Resource Center) from several promoters, including
 Supporting online material for Elowitz et al. report Materials and methods Strains and plasmids. Plasmids expressing CFP or YFP (wild-type codons, developed by University of Washington Yeast Resource Center)
Supporting online material for Elowitz et al. report Materials and methods Strains and plasmids. Plasmids expressing CFP or YFP (wild-type codons, developed by University of Washington Yeast Resource Center)
ApoTrack Cytochrome c Apoptosis ICC Antibody Kit: 2 color immunocytochemistry of cytochrome c and mitochondria.
 PROTOCOL ApoTrack Cytochrome c Apoptosis ICC Antibody Kit 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MSA07 Rev.1 DESCRIPTION ApoTrack Cytochrome c Apoptosis ICC Antibody Kit: 2 color immunocytochemistry
PROTOCOL ApoTrack Cytochrome c Apoptosis ICC Antibody Kit 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MSA07 Rev.1 DESCRIPTION ApoTrack Cytochrome c Apoptosis ICC Antibody Kit: 2 color immunocytochemistry
Lab 1 Microdissection 1. Microdissection
 Lab 1 Microdissection 1 Lab 1. Microdissection The goal of this techniques lab is to understand how to dissect and process tissues that have been incubated in bromodeoxyuridine (BrdU). BrdU is a synthetic
Lab 1 Microdissection 1 Lab 1. Microdissection The goal of this techniques lab is to understand how to dissect and process tissues that have been incubated in bromodeoxyuridine (BrdU). BrdU is a synthetic
Dino-Lite knowledge & education. Fluorescence Microscopes
 Dino-Lite knowledge & education Fluorescence Microscopes Dino-Lite Fluorescence models Smallest fluorescence microscope in the world Revolution to biomedical and educational applications Flexible Easy
Dino-Lite knowledge & education Fluorescence Microscopes Dino-Lite Fluorescence models Smallest fluorescence microscope in the world Revolution to biomedical and educational applications Flexible Easy
A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila
 A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila Joy S Wu & Liqun Luo Howard Hughes Medical Institute, Department of Biological Sciences, Neurosciences Program, Stanford
A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila Joy S Wu & Liqun Luo Howard Hughes Medical Institute, Department of Biological Sciences, Neurosciences Program, Stanford
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
Experimental Tools and Resources Available in Arabidopsis. Manish Raizada, University of Guelph, Canada
 Experimental Tools and Resources Available in Arabidopsis Manish Raizada, University of Guelph, Canada Community website: The Arabidopsis Information Resource (TAIR) at http://www.arabidopsis.org Can order
Experimental Tools and Resources Available in Arabidopsis Manish Raizada, University of Guelph, Canada Community website: The Arabidopsis Information Resource (TAIR) at http://www.arabidopsis.org Can order
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Confocal Microscopy Analyzes Cells
 Choosing Filters for Fluorescence A Laurin Publication Photonic Solutions for Biotechnology and Medicine November 2002 Confocal Microscopy Analyzes Cells Reprinted from the November 2002 issue of Biophotonics
Choosing Filters for Fluorescence A Laurin Publication Photonic Solutions for Biotechnology and Medicine November 2002 Confocal Microscopy Analyzes Cells Reprinted from the November 2002 issue of Biophotonics
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Prashar et al., http://www.jcb.org/cgi/content/full/jcb.201304095/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. FBT phagocytosis in cells expressing PM-GFP. (A) T-PC
Supplemental material Prashar et al., http://www.jcb.org/cgi/content/full/jcb.201304095/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. FBT phagocytosis in cells expressing PM-GFP. (A) T-PC
ApoTrack Cytochrome c Apoptosis ICC Antibody Kit
 ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Kit Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and non-apoptotic
ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Kit Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and non-apoptotic
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
ANAT2341 Embryology Introduction: Steve Palmer
 ANAT2341 Embryology Introduction: Steve Palmer Course overview Course lecturers specializations and roles Steve Palmer 9385 2957 Course overview Summary, aims and expected outcomes Course overview Graduate
ANAT2341 Embryology Introduction: Steve Palmer Course overview Course lecturers specializations and roles Steve Palmer 9385 2957 Course overview Summary, aims and expected outcomes Course overview Graduate
Sydney Brenner: Present
 Sydney Brenner: 1927 - Present Born in the small town of Germiston, South Africa Won a scholarship to the University of Witwatersrand at the age of 15 Received his PhD from Exeter College, Oxford Made
Sydney Brenner: 1927 - Present Born in the small town of Germiston, South Africa Won a scholarship to the University of Witwatersrand at the age of 15 Received his PhD from Exeter College, Oxford Made
2-step or indirect immunofluorescence 1. Substrate on which cells are plated: plastic vs. glass; coating vs. non
 Variables in standard immunostaining protocol 2-step or indirect immunofluorescence 1. Substrate on which cells are plated: plastic vs. glass; coating vs. non 2. Plating density: sparse vs. confluent 3.
Variables in standard immunostaining protocol 2-step or indirect immunofluorescence 1. Substrate on which cells are plated: plastic vs. glass; coating vs. non 2. Plating density: sparse vs. confluent 3.
SCREENING AND PRESERVATION OF DNA LIBRARIES
 MODULE 4 LECTURE 5 SCREENING AND PRESERVATION OF DNA LIBRARIES 4-5.1. Introduction Library screening is the process of identification of the clones carrying the gene of interest. Screening relies on a
MODULE 4 LECTURE 5 SCREENING AND PRESERVATION OF DNA LIBRARIES 4-5.1. Introduction Library screening is the process of identification of the clones carrying the gene of interest. Screening relies on a
-RT RT RT -RT XBMPRII
 Base pairs 2000 1650 1000 850 600 500 400 Marker Primer set 1 Primer set 2 -RT RT RT -RT XBMPRII kda 100 75 50 37 25 20 XAC p-xac A 300 B Supplemental figure 1. PCR analysis of BMPRII expression and western
Base pairs 2000 1650 1000 850 600 500 400 Marker Primer set 1 Primer set 2 -RT RT RT -RT XBMPRII kda 100 75 50 37 25 20 XAC p-xac A 300 B Supplemental figure 1. PCR analysis of BMPRII expression and western
Aminoacid change in chromophore. PIN3::PIN3-GFP GFP S65 (no change) (5.9) (Kneen et al., 1998)
 Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Wingless mutation in Drosophila melanogaster
 J. Biosci., Vol. 12, No. 1, March 1987, pp. 1-11. Printed in India. Wingless mutation in Drosophila melanogaster S. G. BHAT and P. BABU* Molecular Biology Unit, Tata Institute of Fundamental Research,
J. Biosci., Vol. 12, No. 1, March 1987, pp. 1-11. Printed in India. Wingless mutation in Drosophila melanogaster S. G. BHAT and P. BABU* Molecular Biology Unit, Tata Institute of Fundamental Research,
In vitro EGFP-CALI comprehensive analysis. Liang CAI. David Humphrey. Jessica Wilson. Ken Jacobson. Dept. of Cell and Developmental Biology
 In vitro EGFP-CALI comprehensive analysis Liang CAI David Humphrey Jessica Wilson Ken Jacobson Dept. of Cell and Developmental Biology 1/15 Liang s Rotation in Ken s Lab, Sep-Nov, 2003 1. CALI background
In vitro EGFP-CALI comprehensive analysis Liang CAI David Humphrey Jessica Wilson Ken Jacobson Dept. of Cell and Developmental Biology 1/15 Liang s Rotation in Ken s Lab, Sep-Nov, 2003 1. CALI background
SOX2 antibody - Embryonic Stem Cell Marker (ab15830) datasheet
 -Embryonic Stem Cell Marker (ab15830) 1/8 ページ d Products: Stem Cells >> Germline Stem Cells >> Embryonic Germ Cells SOX2 antibody - Embryonic Stem Cell Marker (ab15830) datasheet Product Name Product type
-Embryonic Stem Cell Marker (ab15830) 1/8 ページ d Products: Stem Cells >> Germline Stem Cells >> Embryonic Germ Cells SOX2 antibody - Embryonic Stem Cell Marker (ab15830) datasheet Product Name Product type
NGS Approaches to Epigenomics
 I519 Introduction to Bioinformatics, 2013 NGS Approaches to Epigenomics Yuzhen Ye (yye@indiana.edu) School of Informatics & Computing, IUB Contents Background: chromatin structure & DNA methylation Epigenomic
I519 Introduction to Bioinformatics, 2013 NGS Approaches to Epigenomics Yuzhen Ye (yye@indiana.edu) School of Informatics & Computing, IUB Contents Background: chromatin structure & DNA methylation Epigenomic
Description. Lipodin-Pro TM - Protein Transfection Reagent. 1. Kit Benefits
 Description Lipodin-Pro TM - Protein Transfection Reagent The delivery of proteins inside living cells represents an alternative to nucleic acids transfection and a powerful strategy for functional studies
Description Lipodin-Pro TM - Protein Transfection Reagent The delivery of proteins inside living cells represents an alternative to nucleic acids transfection and a powerful strategy for functional studies
Lecture 25 (11/15/17)
 Lecture 25 (11/15/17) Reading: Ch9; 328-332 Ch25; 990-995, 1005-1012 Problems: Ch9 (study-guide: applying); 1,2 Ch9 (study-guide: facts); 7,8 Ch25 (text); 1-3,5-7,9,10,13-15 Ch25 (study-guide: applying);
Lecture 25 (11/15/17) Reading: Ch9; 328-332 Ch25; 990-995, 1005-1012 Problems: Ch9 (study-guide: applying); 1,2 Ch9 (study-guide: facts); 7,8 Ch25 (text); 1-3,5-7,9,10,13-15 Ch25 (study-guide: applying);
Supporting Information
 Supporting Information Kaneuchi et al. 10.1073/pnas.1420589112 SI Materials and Methods Fly Strains. attp(zh-51d), P{y[+t7.7]=CaryIP}su(Hw)attP5, P{y[+t7.7]=CaryIP}su(Hw)attP2, matα4-gal-vp16, itpr ka901,
Supporting Information Kaneuchi et al. 10.1073/pnas.1420589112 SI Materials and Methods Fly Strains. attp(zh-51d), P{y[+t7.7]=CaryIP}su(Hw)attP5, P{y[+t7.7]=CaryIP}su(Hw)attP2, matα4-gal-vp16, itpr ka901,
No wash 2 Washes 2 Days ** ** IgG-bead phagocytosis (%)
 Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Quantitative real-time RT-PCR analysis of the expression levels of E-cadherin
 Supplementary Information 1 Quantitative real-time RT-PCR analysis of the expression levels of E-cadherin and ribosomal protein L19 (RPL19) mrna in cleft and bud epithelial cells of embryonic salivary
Supplementary Information 1 Quantitative real-time RT-PCR analysis of the expression levels of E-cadherin and ribosomal protein L19 (RPL19) mrna in cleft and bud epithelial cells of embryonic salivary
BIO 315 Lab Exam I. Section #: Name:
 Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
QImaging Camera Application Notes Multicolor Immunofluorescence Imaging
 QImaging Camera Application Notes Multicolor Immunofluorescence Imaging In order to image localization of intracellular proteins with high specificity, it is frequently necessary to multiplex antibody
QImaging Camera Application Notes Multicolor Immunofluorescence Imaging In order to image localization of intracellular proteins with high specificity, it is frequently necessary to multiplex antibody
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Characterization of Histidine Decarboxylase in Drosophila Using an Internal FLAG Epitope
 Grand Valley State University ScholarWorks@GVSU Masters Theses Graduate Research and Creative Practice 12-2014 Characterization of Histidine Decarboxylase in Drosophila Using an Internal FLAG Epitope Maxwell
Grand Valley State University ScholarWorks@GVSU Masters Theses Graduate Research and Creative Practice 12-2014 Characterization of Histidine Decarboxylase in Drosophila Using an Internal FLAG Epitope Maxwell
FILE S1. Lab 4. One fly, two fly, red fly, white fly. (Okay, so I m no Dr. Seuss): Recombination mapping of Transposon Insertions in Drosophila
 2 SI FILE S1 Lab 4. One fly, two fly, red fly, white fly (Okay, so I m no Dr. Seuss): Recombination mapping of Transposon Insertions in Drosophila Introduction In a previous laboratory exercise, we studied
2 SI FILE S1 Lab 4. One fly, two fly, red fly, white fly (Okay, so I m no Dr. Seuss): Recombination mapping of Transposon Insertions in Drosophila Introduction In a previous laboratory exercise, we studied
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Chapter One. Construction of a Fluorescent α5 Subunit. Elucidation of the unique contribution of the α5 subunit is complicated by several factors
 4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
mcherry Monoclonal Antibody (16D7) Catalog Number M11217 Product data sheet
 Website: thermofisher.com Customer Service (US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 mcherry Monoclonal Antibody (16D7) Catalog Number M11217 Product data sheet Details
Website: thermofisher.com Customer Service (US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 mcherry Monoclonal Antibody (16D7) Catalog Number M11217 Product data sheet Details
Defense Technical Information Center Compilation Part Notice
 UNCLASSIFIED Defense Technical Information Center Compilation Part Notice ADP019695 TITLE: Fabricating Neural and Cardiomyogenic Stem Cell Structures by a Novel Rapid Prototyping--the Inkjet Printing Method
UNCLASSIFIED Defense Technical Information Center Compilation Part Notice ADP019695 TITLE: Fabricating Neural and Cardiomyogenic Stem Cell Structures by a Novel Rapid Prototyping--the Inkjet Printing Method
pbroad3-lacz An optimized vector for mouse and rat transgenesis Catalog # pbroad3-lacz
 pbroad3-lacz An optimized vector for mouse and rat transgenesis Catalog # pbroad3-lacz For research use only Version # 03B04-MT PRODUCT INFORMATION Content: - 20 µg of pbroad3-lacz provided as lyophilized
pbroad3-lacz An optimized vector for mouse and rat transgenesis Catalog # pbroad3-lacz For research use only Version # 03B04-MT PRODUCT INFORMATION Content: - 20 µg of pbroad3-lacz provided as lyophilized
Motivation From Protein to Gene
 MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
BIOSCI 0351: GENETICS LABORATORY COURSE INFORMATION- SPRING TERM 2016
 BioSci 0351, Spring, 2016 BIOSCI 0351: GENETICS LABORATORY COURSE INFORMATION- SPRING TERM 2016 GENERAL INFORMATION Time: Location: INSTRUCTORS: Office hours: Phone Numbers: Email: Mondays, 8:30AM 12:20PM
BioSci 0351, Spring, 2016 BIOSCI 0351: GENETICS LABORATORY COURSE INFORMATION- SPRING TERM 2016 GENERAL INFORMATION Time: Location: INSTRUCTORS: Office hours: Phone Numbers: Email: Mondays, 8:30AM 12:20PM
Fig. S1. Inhibition of Rh1 accumulation on the rhabdomere membrane in CG13089 PIG-U mutants. (A) Late pupae with dark wings were put into 0.
 Fig. S1. Inhibition of Rh1 accumulation on the rhabdomere membrane in CG13089 PIG-U mutants. (A) Late pupae with dark wings were put into 0.2% agar after the pupal case was partially removed. Two fluorescent
Fig. S1. Inhibition of Rh1 accumulation on the rhabdomere membrane in CG13089 PIG-U mutants. (A) Late pupae with dark wings were put into 0.2% agar after the pupal case was partially removed. Two fluorescent
Additional Practice Problems for Reading Period
 BS 50 Genetics and Genomics Reading Period Additional Practice Problems for Reading Period Question 1. In patients with a particular type of leukemia, their leukemic B lymphocytes display a translocation
BS 50 Genetics and Genomics Reading Period Additional Practice Problems for Reading Period Question 1. In patients with a particular type of leukemia, their leukemic B lymphocytes display a translocation
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Combined fluorescence and AFM imaging of cells
 Combined fluorescence and AFM imaging of cells Introduction Combining optical and AFM imaging of cells opens up many possibilities for correlating structural information about the cell surface with functional
Combined fluorescence and AFM imaging of cells Introduction Combining optical and AFM imaging of cells opens up many possibilities for correlating structural information about the cell surface with functional
OriGene GFC-Arrays for High-throughput Overexpression Screening of Human Gene Phenotypes
 OriGene GFC-Arrays for High-throughput Overexpression Screening of Human Gene Phenotypes High-throughput Gene Function Validation Tool Introduction sirna screening libraries enable scientists to identify
OriGene GFC-Arrays for High-throughput Overexpression Screening of Human Gene Phenotypes High-throughput Gene Function Validation Tool Introduction sirna screening libraries enable scientists to identify
(A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1
 SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
Supplemental Data. Hachez et al. Plant Cell (2014) /tpc Suppl. Figure 1A
 Suppl. Figure 1A Suppl. Figure 1B Supplemental Figure 1: Results of the commercial screening of interactants using split ubiquitin technique. (A) Isolated preys (192) using the bait construct pbt3-n- as
Suppl. Figure 1A Suppl. Figure 1B Supplemental Figure 1: Results of the commercial screening of interactants using split ubiquitin technique. (A) Isolated preys (192) using the bait construct pbt3-n- as
beta-galactosidase (A420) ctrl
 0.6 beta-galactosidase (A420) 0.4 0.2 ctrl 2A 22A 35B 36B 58A 64A 68E 75C 86Fa 86Fb 102F Fig. S1. Levels of induced expression at various attp sites. The induced β-galactosidase activity was measured from
0.6 beta-galactosidase (A420) 0.4 0.2 ctrl 2A 22A 35B 36B 58A 64A 68E 75C 86Fa 86Fb 102F Fig. S1. Levels of induced expression at various attp sites. The induced β-galactosidase activity was measured from
1a. What is the ratio of feathered to unfeathered shanks in the offspring of the above cross?
 1. Whether or not the shanks of chickens contains feathers is due to two independently assorting genes. Individuals have unfeathered shanks when they are homozygous for recessive genes at two loci; the
1. Whether or not the shanks of chickens contains feathers is due to two independently assorting genes. Individuals have unfeathered shanks when they are homozygous for recessive genes at two loci; the
A Survey of Genetic Methods
 IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15
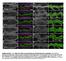 Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Blocking endocycle apoptosis page 1. Table S1: Relative abundance of mrna levels in the endocycling salivary gland versus mitotic
 Blocking endocycle apoptosis page 1 SUPPLEMENTAL DATA Table S1: Relative abundance of mrna levels in the endocycling salivary gland versus mitotic cycling brain-disc. Gene E/M 1 E rr /M rr E rr /E M rr
Blocking endocycle apoptosis page 1 SUPPLEMENTAL DATA Table S1: Relative abundance of mrna levels in the endocycling salivary gland versus mitotic cycling brain-disc. Gene E/M 1 E rr /M rr E rr /E M rr
