Immunostaining of ovaries, S2 cells and OSCs was performed as previously described
|
|
|
- Warren Holmes
- 5 years ago
- Views:
Transcription
1 Supplemental Material: Supplemental Materials and Methods Immunofluorescence Immunostaining of ovaries, S2 cells and OSCs was performed as previously described (Saito et al. 2006; Saito et al. 2009). The primary antibodies used were: culture supernatants of anti- hybridoma cells (1:1 dilution), culture supernatants of anti-piwi hybridoma cells (P4D2; 1:1 dilution) (Saito et al. 2006), a mouse monoclonal antibody for the myc-tag (1:1,000 dilution; Sigma), a guinea pig antibody against TJ (1:2,000 dilution) (Li et al. 2003) and a rabbit polyclonal antibody against Yb (1:250 dilution) (Szakmary et al. 2009). The secondary antibodies used were: Alexa 488-conjugated anti-mouse IgG (Molecular Probes), Alexa 546-conjugated anti-mouse IgG (Invitrogen), Alexa 488-conjugated anti-rabbit IgG (Invitrogen), Alexa 546-conjugated anti-guinea pig IgG (Invitrogen) and Alexa 546-conjugated anti-rabbit IgG (Invitrogen). DNA was stained with 4,6-diamidino-2-phenylindole (DAPI). Before fixation, the cells were incubated with MitoTracker Orange CMXRos (Invitrogen) targeted at the active mitochondria in living cells. All images were collected using a confocal microscope (Zeiss LSM5 EXCITER; Zeiss). Immunoprecipitation Immunoprecipitation from S2 lysates was performed essentially as previously described (Miyoshi et al. 2005). Immunoprecipitation from OSCs was also performed essentially 1
2 as previously described (Saito et al. 2009). Briefly, OSCs were homogenized in homogenization buffer [20 mm HEPES-KOH (ph7.3), 2 mm MgCl 2, 1 mm dithiothreitol (DTT), 150 mm NaCl, 2 µg/ml pepstatin, 2 µg/ml leupeptin, 0.5% aprotinin and 0.1% NP-40] to prepare OSC lysates. Anti-Piwi (P3G11) (Saito et al. 2006), anti- and anti-yb (Szakmary et al. 2009) antibodies were immobilized on Dynabeads protein G (Invitrogen). Reaction mixtures were rocked at 4 C for 2 h and the beads were washed four times with homogenization buffer. Total RNAs were isolated from the immunoprecipitates with phenol-chloroform and were precipitated with ethanol. RNAs were dephosphorylated with calf intestinal phosphatase (TaKaRa) and labeled with 32 P-gATP using T4 PNK (TaKaRa) for visualization. Analysis of pirna-intermediate chemical structures Total RNAs were isolated from the anti- immunoprecipitates. Phosphorylation of RNAs was performed in the presence of 1 mm ATP with T4 PNK (NEB). Alkaline dephosphorylation was performed with calf intestinal phosphatase (NEB). Acidic dephosphorylation was carried out with T4 PNK (Takara) in 50 mm Tris-HCl (ph 6.5), 10 mm MgCl 2 and 5mM DTT. Enzymes were inactivated by phenol/chloroform extraction, followed by RNA precipitation. Precipitates were dissolved in H 2 O. Polyadenylation was performed with poly (A) polymerase (NEB) and 1 mm ATP in 1 poly (A) polymerase reaction buffer. RNAs were resolved on a 12% denaturing polyacrylamide gel and blotted on Hybond N+ (GE Healthcare) or Hybond N (GE Healthcare) membrane. To cross-link RNAs, UV irradiation or EDC reagent was used. 2
3 Drosophila strains Yellow white (y w) and Oregon R were employed as WT strains. The armi alleles used were armi 72.1 and armi 1 (Cook et al. 2004). All stocks were maintained at 25 C. myc-piwi expression and RNAi in OSCs To yield RNAi-resistant piwi cdna (Piwi-r), 5 and 3 fragments of piwi cdna were individually amplified by KOD plus DNA polymerase (Toyobo) using the following primers: 5 fragment, Piwi-F and Piwi-mt-R; 3 fragment, Piwi-mt-F and Piwi-R (primers are listed in Supplemental Table S2). To yield Piwi PAZ mutant cdna (myc-piwi-pazmt), 5 and 3 fragments of piwi cdna were individually amplified by KOD plus DNA polymerase (Toyobo) using the following primers: 5 fragment, Piwi-F and Piwi-PAZmt-R; 3 fragment, Piwi-PAZmt-F and Piwi-R (primers are shown in Table S2). The resulting DNA fragments were gel-purified, mixed and used as templates for PCR amplification. DNA fragments were amplified by PCR with Piwi-F and Piwi-R. The resulting PCR products were cloned into the pacm vector (Saito et al. 2009) and the sequence was validated by DNA sequence analysis. A short DNA fragment encoding a myc-tag was inserted between the Xho I and Bam HI sites of pacm to produce pacm-myc. Full-length zuc cdna was inserted between the Nhe I and Xho I sites of pacm-myc to yield pac-zuc-myc. For RNAi and expressing myc-tagged proteins in OSCs, trypsinized OSCs ( cells) were suspended in 100 µl of Solution V of the Cell Line Nucleofector Kit V (Amaxa Biosystems) together with 3
4 either 200 pmol of sirna duplex or 5 µg of plasmid vector. Transfection was conducted in electroporation cuvettes using a Nucleofector device (Amaxa Biosystems). The transfected cells were transferred to fresh OSC medium and incubated at 26 C for 2 d for further experiments. The sirnas utilized for RNAi are summarized in Table S2. The sirna was enhanced green fluorescent protein (EGFP) sirna (Saito et al. 2009). Quantitative RT-PCR was performed as previously described (Kawamura et al. 2008). Total RNA (0.5 µg) was used to reverse transcribe target sequences using oligo(dt) primer. The resulting cdna was analyzed by quantitative RT-PCR using a LightCycler real-time PCR system (Roche Diagnostics) using SYBR Premix Ex Taq (Takara). Relative steady-state mrna levels were determined from the threshold cycle for amplification. Table S2 lists the PCR primer sequences. Ribosomal protein 49 (Rp49) was used as an internal. Cloning of small RNAs and processing sequence tags was immunopurified from OSCs using a specific antibody. After immunoprecipitation, total RNAs were isolated from the anti- immunoprecipitates. Acidic dephosphorylation was carried out with T4 PNK (Takara) in 50 mm Tris-HCl (ph 6.5), 10 mm MgCl 2 and 5mM DTT. Enzymes were inactivated by phenol/chloroform extraction, followed by RNA precipitation. Precipitates were dissolved in H 2 O. RNAs were resolved on a 12% denaturing polyacrylamide gel with 5 -labelled Decade Marker (Ambion) as a size marker. RNAs of nt were excised from the gel. Cloning of RNAs was carried out as described previously (Saito et al. 4
5 2009). The sequencing was performed on a GS FLX system (Roche). Genome mapping, annotation and frequency mapping We followed a previously described procedure to map and annotate sequences (Saito et al. 2009). We mapped 116,680 pir-il sequences to the Drosophila melanogaster (dm3) genome. The annotation results are shown as a pie-chart in Supplemental Fig. S4A. Maps for pil-ils were generated (Supplemental Fig. S5 and S6A), representing the number of pir-ils that are 100% identical to the genomic sequence. RNA sequences of pir-ils have been deposited at the GEO database under accession number GSE Northern blotting Northern blotting was performed as previously described (Saito et al. 2006). DNA oligonucleotides used are summarized in Table S2. 5
6 Supplemental Figures A * 220 k- 160 k- 120 k- 100 k- 80 k- 70 k- 60 k- 50 k- 40 k- (Da) ovary S2 - - Tubulin 1188 Helicase domain B 220 k- 160 k- 120 k- 100 k- 80 k- 70 k- 60 k- 50 k- 40 k- (Da) n.i. anti- anti S2 lysate - Figure S1A-D Saito et al. C D OSC /DAPI / DAPI /DAPI egg chamber / DAPI RNAi /DAPI germarium
7 Figure S1E-G Saito et al. E G y w y w 1 armi / + 1 armi / armi / armi F 1 1 armi / armi / DAPI armi / armi (kb) long form short form - - Tubulin Supplemental Figure S1. (A) Western blot analyses on ovary and S2 cell lysates using anti- antibodies. Anti-tubulin was utilized as an internal. The antigen (the N-terminal end of ; aa 1-200; indicated as *) used for immunizing mice is shown at the bottom. (B) Immunoprecipitation was performed from S2 cells using anti- antibodies. Mass spectrometric analysis determined that the ~150 kda protein is. (C) Immunofluorescence of in OSCs and ovaries. DAPI staining (blue) shows the locations of the nuclei. (D) Immunostaining of OSCs before and after the treatment with RNAi for indicates that the dotted signals in the cytoplasm reflect the subcellular localization of in the cells. (E) Immunostaining pattern of in the armi transheterozygous mutant ovary. (F) Western blot analysis shows that is expressed even in the armi homozygous mutant ovaries. (G) Northern blotting shows that the armi transcripts (short form) are expressed in the armi homozygous mutant ovaries.
8 A DCP1 DCP1/Yb DCP1/Yb/DAPI Saito et al. Figure S2 B Yb Yb / DAPI Yb Yb / DAPI RNAi / DAPI / DAPI Yb RNAi Supplemental Figure S2. (A) Coimmunostaining of OSCs with anti-yb and anti-dcp1 antibodies shows that Yb bodies do not colocalize with P bodies. (B) Depletion of did not affect the cellular localization of Yb whereas depletion of Yb caused to be localized to the cytoplasm.
9 Saito et al. Figure S3A-D A Piwi RNAi Yb - Piwi C RNAi in OSC Piwi Yb ] 50 nt - - Yb - Tubulin 40 nt - 30 nt - 20 nt - - tj pirna - mir-310 B Piwi RNAi Piwi - sl-1 esirna D myc-piwi-pazmt myc-piwi-pazmt/dapi DAPI DAPI myc-piwi myc-piwi/dapi TJ TJ
10 Saito et al. Figure S3E-F E IP from OSC kda input n.i. anti-piwi anti-yb AGO2 AGO F IP n.i. anti-piwi anti /20 1/40 1/60 1 dilution - Piwi Supplemental Figure S3. (A) Western blotting was performed using anti-piwi, anti- and anti- Yb antibodies on OSC lysates in which Piwi, and Yb were depleted by RNAi. Anti-Tubulin was used as an internal. sirna for EGFP was employed as a negative (). (B) Depletion of caused Piwi (green) to be mislocalized to the cytoplasm. In contrast, the nuclear localization of TJ protein (red) was unaffected by depletion. (C) Yb depletion causes a severe reduction of the pirna level in OSCs (~25% of the signal). (D) The Piwi PAZ mutant (green) (myc-piwi-pazmt; containing two mutations, Y327A and Y328A) accumulates in the cytoplasm while myc-piwi wt (green) localizes to the nucleus, supporting the idea that pirna loading is required for the nuclear localization of Piwi. DAPI staining (blue) shows the locations of nuclei in the cells. (E) The complexes immunoprecipitated with anti-yb and anti-piwi antibodies contain neither AGO1 nor AGO2. (F) Western blot analysis shows that the amounts of Piwi are approximately equal in the complexes immunoisolated with anti-piwi and anti- antibodies only when the anti-piwi complex was diluted 1:40 before loading.
11 A no annotation 6.2% Saito et al. Figure S4A-B transposons 16.6% snorna 1.4% snrna 3.5% rrna 35.7% protein-coding gene 35.9% protein-coding genes rrna ncrna trna repeat snrna transposons snorna no annotation B IP total RNA n.i. anti- oligo antisense oligo IP total RNA n.i. anti- oligo antisense oligo IP total RNA n.i. anti- oligo antisense oligo IP total RNA n.i. anti- oligo antisense oligo actin 5c ORF probe 1 actin 5c ORF probe 2 tj ORF probe 1 tj ORF probe 2 Supplemental Figure S4. pir-il analyses. (A) The small RNA content (50-75 nt) in the - Piwi-Yb complex perfectly matches the Drosophila genome sequence. (B) Northern blotting analyses. RNAs isolated from OSCs (total RNAs) and the anti- immunoprecipitates (anti- IP) were probed with DNA oligos corresponding to actin 5c and tj ORF. n.i. was used as a negative for immunoprecipitation. The sequences of the probes used are; 5'-GCCTCGGTCAGCAGCACGGGGTGCT (actin 5c-probe1) 5'-CGCAGGATGGCATGGGGAAGGGCAT (actin 5c-probe2) 5'-TCAACGTGGTCAGCATGTCGTCATT (tj-probe1) 5'-ACCCGAAGCACTACCGTACGAGTTC (tj-probe2) RNAs isolated from the -IP complex appeared negative although oligos were - posi tive, suggesting that clones corresponding to actin 5c and tj ORF are background.
12 A Chr. X 21,500,000 21,550,000 21,600,000 21,650,000 Saito et al. Figure S5A OSC Piwi-associated pirnas Unique mappers pirnas (black shows pirnas overlapping with pir-ils) pir-ils (black shows pir-ils overlapping with pirnas) OSC pir-ils Unique mappers 21,502,000 21,504,000 21,506,000 21,508,000 21,510,000 pirnas Unique mappers pirnas (black shows pirnas overlapping with pir-ils) pir-ils Unique mappers pir-ils (black shows pir-ils overlapping with pirnas)
13 Saito et al. Figure S5B B chrx:21,508,080-21,508,550 21,508,100 21,508,200 21,508,300 21,508,400 21,508,500 OSC Piwi-associated pirnas Unique mappers (plus) OSC pir-ils Unique mappers (plus) pirnas (black shows pirnas overlapping with pir-ils) pir-ils (black shows pir-ils overlapping with pirnas) SINE LINE LTR DNA Simple Low Complexity Satellite RNA Other Unknown Repeating Elements by RepeatMasker Supplemental Figure S5. (A) Upper: The distribution patterns of pirnas (Saito et al. 2009) and pir-ils on the pirna locus flam sequence. Only uniquely mapped ones are shown. Black bars show pirnas/pir-ils overlapping with pir-ils/pirnas. Green bars show pirnas/pir-ils non-overlapping with pir-ils/pirnas. Lower: The magnification of the region specified in (A). (B) The distribution of uniquely mapped pir-ils and mature pirnas on the chromosome X, position 21,508,080-21,508,550. Black bars show pirnas/pir-ils overlapping with pir-ils/pirnas. Green bars show pirnas/pir-ils non-overlapping with pir-ils/pirnas.
14 Saito et al. Figure S6 A chr2l 19,465,000 19,466,000 19,467,000 19,468,000 pirnas pirnas (black shows pirnas overlapping with pir-ils) pir-ils pir-ils (black shows pir-ils overlapping with pirnas) 5'UTR traffic jam ORF 3'UTR B 100% 80% C 60% G 40% U 20% 0% A Supplemental Figure S6. (A) The distribution patterns of pirnas and pir-ils on the tj gene sequence. Some pir-ils were mapped to the tj ORF. However, the signals for the clones were not detected in the -Piwi-Yb complex (Supplemental Fig. S4B) by Northern blotting, suggesting that they may be considered as 'background' of the small RNA library. pir-ils were detected in the complex by Northern blotting (Fig. 3C and 3D). (B) The 5' end of pir-ils mapped to transposons was not U-rich as oppose to that of mature transposon-pirnas. Only a small number of transposn-pir-ils (~4%) has G at their 5' end, although the reason for this appearance remains unknown.
15 Saito et al. Figure S7 A 1.2 B Yb / / DAPI Zuc RNAi Yb / / DAPI 1.0 relative Zuc mrna D 0 Mito Zuc C Zuc RNAi Mito / Zuc Mito / Zuc / DAPI n.i. ZucRNAi anti-myc n.i. anti-myc E 40 nt - 1/8 1/8 1 1 dilution Mito Yb 30 nt - Yb / Zuc / DAPI Mito / Yb Mito / Yb / DAPI 20 nt - < < Supplemental Figure S7. (A) The efficiency of Zuc depletion in OSC was determined by qrt-pcr. (B) Cellular localization of in OSC was not affected by Zuc depletion. (C) 32 P end-labeling showed that when Zuc was depleted, myc-piwi was loaded with far fewer pirnas compared with loading under normal conditions. (D) Zuc (green) localizes to mitochondria (red) when expressed in OSCs. (E) Left panel; Zuc signals (green) can be detected in close proximity to Yb bodies (red). Right four panels; Yb signals (green) are detected in close proximity to mitochondria (red).
16 Saito et al. Figure S A anti-piwi anti CIP T4 PNK anti-ago CIP T4 PNK B anti PAP T4 PNK (Acidic) 100 nt - 50 nt - 40 nt - 30 nt - 20 nt - EDC 30 nt - 20 nt - EDC 100 nt - 75 nt - 50 nt - 40 nt - mir nt nt - 75 nt - 50 nt - 40 nt - 30 nt - UV 20 nt - tj-pirna 20 nt - tj-pirna Supplemental Figure S8. (A) pir-ils are undetectable by northern blotting in which the RNAs were cross-linked by EDC. They become detectable after phosphorylation, suggesting that pir-ils are free from phosphate groups at both ends. It should be noted that mature tj-pirna and mir-310 are detectable by the EDC method even without phosphorylation. (B) pirna-ils are barely polyadenylated unless they were pre-treated with PNK under acidic conditions, suggesting that they contain a cyclic phosphate at their 3' end. From this result, together with those in (A), it is suggested that pir-ils are free from phosphate groups at 5' end but contain a cyclic phosphate at their 3' end.
17 A C Saito et al. Figure S siegfp sipiwi B D E Piwi RNAi Piwi RNAi Supplemental Figure S9. (A) Piwi depletion in OSCs causes derepression of various transposons. (B) Piwi and depletion by RNAi in OSCs caused derepression of mdg1 transposon. RNAi was performed for 48 hours. (C) Yb and Zuc depletion by RNAi in OSC caused derepression of mdg1. RNAi was performed for 96 hours. (D) Quantitative RT-PCR analysis for the mdg1 transposon. In OSCs where endogenous Piwi was depleted, myc-piwi-r (RNAi-insensitive Piwi), but not myc-piwi or myc-piwi-δn (Piwi lacking its nuclear localization signal), was able to rescue transposon derepression. (E) RNAi-insensitive Piwi-r and Piwi-DDAA-r rescued transposon silencing in Piwidepleted OSC.
18 Table S1. Saito et al. Supplemental Table 1. Genes depleted in OSCs. Gene Function Reference armi pirna pathway (Vagin et al., 2006; Malone et al., 2009) mael pirna pathway (Lim and Kai, 2007) spn-e pirna pathway (Vagin et al., 2006; Malone et al., 2009) zuc pirna pathway (Malone et al., 2009; Pane et al., 2007; Saito et al., 2009) dicer1 mirna pathway (Vagin et al., 2006) dicer2 sirna pathway (Vagin et al., 2006)
19 Table S2. Saito et al. Supplemental Table 2 Experiment Primer Name Primer sequence Vector construction Piwi-F Piwi-R Piwi-mt-F Piwi-mt-R Piwi-PAZmt-F Piwi-PAZmt-R Myc-tag-5 Myc-tag-3 Zuc-F Zuc-R ATAAGCTTGCTGATGATCAGGGACGTG ATAGATCTTTATAGATAATAAAACTTCTTTTCGAG CGAGGAGGCACCGCGCAGGGAGGGAGGCCCGCCAGAGCGAAAGC CG CGGGCCTCCCTCCCTGCGCGGTGCCTCCTCGAGAGCTCCTCTCTCT C GATATCAGTTTCGTGGAAGCCGCTCTCACTAAATATAATATACGCATTC GCG GCGTATATTATATTTAGTGAGAGCGGCTTCCACGAAACTGATATCTCT ACC TCGAGGGAGAGCAGAAACTGATCTCTGAAGAAGACCTGAACTAAG GATCCTTAGTTCAGGTCTTCTTCAGAGATCAGTTTCTGCTCTCCC TTGCTAGCCACCATGTTGATTACCCAAATAATTATGAAACAA ATCTCGAGCTTGAGCTGGATTTGGCTCC Experiment Gene Primer sequence (Forward, Reverse) RP CCGCTTCAAGGGACAGTATCTG, ATCTCGCCGCAGTAAACGC CTGGCAAAGGGATTTCATCA, TGCATTCCTAAGGCCAAATG TATGGGCTGAGGCGATAAAC, CAAGTGGCTCACTGCTGGTA I- element GACCAAATAAAAATAATACGACTTC, AACTAATTGCTGGCTTGTTATG qrt-pcr HeT-A CGCGCGGAACCCATCTTCAGA, CGCCGCAGTCGTTTGGTGAGT Mdg1 Roo ZAM Zucchini AACAGAAACGCCAGCAACAGC, CGTTCCCATGTCCGTTGTGAT CGTCTGCAATGTACTGGCTCT, CGGCACTCCACTAACTTCTCC ACTTGACCTGGATACACTCACAAC, GAGTATTACGGCGACTAGGGATAC TGATTTGGAAGCTGGTGCAG, GCGACACTTGTGGTTTCTGG Experiment Gene Probe sequence Northern Blot U6 snrna mir-310 sl-1 esirna Tj-piRNA Idefix-piRNA GGGCCATGCTAATCTTCTCTGTA AAAGGCCGGGAAGTGTGCAATA GGAGCGAACTTGTTGGAGTCAA GGTAATGGGAATGCACTTCTCTTGAA AAACTACTGGCAATCGTTTGGGAA Experiment sirna name sirna sense sirna antisense RNAi EGFP-si Piwi-si Zucchini si tage-si Yb-si Maelstrom-si Spindle-E-si Dicer1-si Dicer2-si GGCAAGCUGACCCUGAAGUTT GCUCCCAGGCGUGAAGGUGTT GCAUUGCCGUCAGCACUGUTT CAGCUUAAGGCGAAUCCAATT GGGGUACAUCAACCAAGCUTT CGCCAAGAUGUCCCAUGAUTT CUCGCGGCAUGUAAAGAUUTT GAUUGUCAUGACAGCGCCGTT CAGGGAGUGGAUGACUGGATT ACUUCAGGGUCAGCUUGCCTT CACCUUCACGCCUGGGAGCTT ACAGUGCUGACGGCAAUGCTT UUGGAUUCGCCUUAAGCUGTT AGCUUGGUUGAUGUACCCCTT AUCAUGGGACAUCUUGGCGTT AAUCUUUACAUGCCGCGAGTT CGGCGCUGUCAUGACAAUCTT UCCAGUCAUCCACUCCCUGTT
Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb
 Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
SiRNA transfection, northern blotting and RT-qPCR analysis were performed as
 Supplemental Material: Supplemental Materials and Methods Supplemental Figures S1 to S7 Supplemental Tables S1 and S Additional References Supplemental Materials and Methods Knockdown, northern blotting
Supplemental Material: Supplemental Materials and Methods Supplemental Figures S1 to S7 Supplemental Tables S1 and S Additional References Supplemental Materials and Methods Knockdown, northern blotting
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplemental Data. Noncoding Transcription by RNA Polymerase Pol IVb/Pol V Mediates Transcriptional Silencing of Overlapping and Adjacent Genes
 Cell, Volume 135 Supplemental Data Noncoding Transcription by RNA Polymerase Pol IVb/Pol V Mediates Transcriptional Silencing of Overlapping and Adjacent Genes Andrzej T. Wierzbicki, Jeremy R. Haag, and
Cell, Volume 135 Supplemental Data Noncoding Transcription by RNA Polymerase Pol IVb/Pol V Mediates Transcriptional Silencing of Overlapping and Adjacent Genes Andrzej T. Wierzbicki, Jeremy R. Haag, and
MeCP2. MeCP2/α-tubulin. GFP mir1-1 mir132
 Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
gypsy fold change in steady state RNA levels RnpS1 Acn mago tsu GFP armi sirna mediated depletion against indicated genes in OSCs
 Hayashi_FigS1 fold change in steady state RNA levels 1000 100 10 1 gypsy GFP armi mago tsu Acn RnpS1 btz sirna mediated depletion against indicated genes in OSCs Supplementary Figure S1. The EJC is required
Hayashi_FigS1 fold change in steady state RNA levels 1000 100 10 1 gypsy GFP armi mago tsu Acn RnpS1 btz sirna mediated depletion against indicated genes in OSCs Supplementary Figure S1. The EJC is required
Supplemental Figure legends Figure S1. (A) (B) (C) (D) Figure S2. Figure S3. (A-E) Figure S4. Figure S5. (A, C, E, G, I) (B, D, F, H, Figure S6.
 Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Supporting Information
 Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
Giardia RNAi Paper Discussion. Supplemental - VSG clonality. Variant-specific surface protein VSP9B10
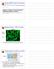 Giardia RNAi Paper Discussion Supplemental - VSG clonality VSP9B10 Green - VSP9B10 antibody Blue - DAPI Demonstration that cell line expresses a single VSP Variant-specific surface protein Outside Inside
Giardia RNAi Paper Discussion Supplemental - VSG clonality VSP9B10 Green - VSP9B10 antibody Blue - DAPI Demonstration that cell line expresses a single VSP Variant-specific surface protein Outside Inside
Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and
 SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
A direct role for HSP90 in pre-risc formation in Drosophila
 Supplementary Information for A direct role for HSP90 in pre-risc formation in Drosophila Tomohiro Miyoshi 1, Akiko Takeuchi 1, Haruhiko Siomi 1 and Mikiko C. Siomi 1,2 * 1 Keio University School of Medicine,
Supplementary Information for A direct role for HSP90 in pre-risc formation in Drosophila Tomohiro Miyoshi 1, Akiko Takeuchi 1, Haruhiko Siomi 1 and Mikiko C. Siomi 1,2 * 1 Keio University School of Medicine,
Mechanism of Induction and Suppression of Antiviral Immunity Directed by Virus-Derived Small RNAs in Drosophila
 Cell Host & Microbe, Volume 4 Supplemental Data Mechanism of Induction and Suppression of Antiviral Immunity Directed by Virus-Derived Small RNAs in Drosophila Roghiyh Aliyari, Qingfa Wu, Hong-Wei Li,
Cell Host & Microbe, Volume 4 Supplemental Data Mechanism of Induction and Suppression of Antiviral Immunity Directed by Virus-Derived Small RNAs in Drosophila Roghiyh Aliyari, Qingfa Wu, Hong-Wei Li,
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
SUPPLEMENTARY INFORMATION
 The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
SUPPLEMENTAL MATERIAL. Supplemental Methods. Cumate solution was from System Biosciences. Human complement C1q and complement
 SUPPLEMENTAL MATERIAL Supplemental Methods Reagents Cumate solution was from System Biosciences. Human complement Cq and complement C-esterase inhibitor (C-INH) were from Calbiochem. C-INH (Berinert) for
SUPPLEMENTAL MATERIAL Supplemental Methods Reagents Cumate solution was from System Biosciences. Human complement Cq and complement C-esterase inhibitor (C-INH) were from Calbiochem. C-INH (Berinert) for
Eric J. Wagner, Brandon D. Burch, Ashley C. Godfrey, Harmony R. Salzler, Robert J. Duronio, and William F. Marzluff
 Molecular Cell, Volume 28 Supplemental Data A Genome-Wide RNA Interference Screen Reveals that Variant Histones Are Necessary for Replication-Dependent Histone Pre-mRNA Processing Eric J. Wagner, Brandon
Molecular Cell, Volume 28 Supplemental Data A Genome-Wide RNA Interference Screen Reveals that Variant Histones Are Necessary for Replication-Dependent Histone Pre-mRNA Processing Eric J. Wagner, Brandon
RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the
 Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Information
 Journal : Nature Biotechnology Supplementary Information Targeted genome engineering in human cells with RNA-guided endonucleases Seung Woo Cho, Sojung Kim, Jong Min Kim, and Jin-Soo Kim* National Creative
Journal : Nature Biotechnology Supplementary Information Targeted genome engineering in human cells with RNA-guided endonucleases Seung Woo Cho, Sojung Kim, Jong Min Kim, and Jin-Soo Kim* National Creative
Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila
 Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Document S1. Supplemental Experimental Procedures and Three Figures (see next page)
 Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1. Roles of Armi, Zucchini, and Piwi in transposon silencing in OSS cells. (A) Knockdown of tage, chini and after dsrna treatment were assessed by qpcr. qpcr data was
Supplementary Figure Legends Figure S1. Roles of Armi, Zucchini, and Piwi in transposon silencing in OSS cells. (A) Knockdown of tage, chini and after dsrna treatment were assessed by qpcr. qpcr data was
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Learning Objectives. Define RNA interference. Define basic terminology. Describe molecular mechanism. Define VSP and relevance
 Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and
 1 Supplemental information 2 3 Materials and Methods 4 Reagents and animals 5 8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and 6 Silencer Select Pre-designed sirna
1 Supplemental information 2 3 Materials and Methods 4 Reagents and animals 5 8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and 6 Silencer Select Pre-designed sirna
Combining Techniques to Answer Molecular Questions
 Combining Techniques to Answer Molecular Questions UNIT FM02 How to cite this article: Curr. Protoc. Essential Lab. Tech. 9:FM02.1-FM02.5. doi: 10.1002/9780470089941.etfm02s9 INTRODUCTION This manual is
Combining Techniques to Answer Molecular Questions UNIT FM02 How to cite this article: Curr. Protoc. Essential Lab. Tech. 9:FM02.1-FM02.5. doi: 10.1002/9780470089941.etfm02s9 INTRODUCTION This manual is
SUPPLEMENTARY INFORMATION. LIN-28 co-transcriptionally binds primary let-7 to regulate mirna maturation in C. elegans
 SUPPLEMENTARY INFORMATION LIN-28 co-transcriptionally binds primary let-7 to regulate mirna maturation in C. elegans Priscilla M. Van Wynsberghe 1, Zoya S. Kai 1, Katlin B. Massirer 2-4, Victoria H. Burton
SUPPLEMENTARY INFORMATION LIN-28 co-transcriptionally binds primary let-7 to regulate mirna maturation in C. elegans Priscilla M. Van Wynsberghe 1, Zoya S. Kai 1, Katlin B. Massirer 2-4, Victoria H. Burton
Description Wako Catalog No. Page # microrna Purification Kits based on Immunoprecipitation method using antibody against Argonaute protein
 Wako microrna Products Wako Pure Chemical Industries, Ltd. microrna Cloning Ago IP microrna Purification Target mrna Screening micrornas (mirnas) are endogenous RNAs, approximately 22 nucleotides in length,
Wako microrna Products Wako Pure Chemical Industries, Ltd. microrna Cloning Ago IP microrna Purification Target mrna Screening micrornas (mirnas) are endogenous RNAs, approximately 22 nucleotides in length,
Premix Ex Taq (Probe qpcr)
 For Research Use Premix Ex Taq (Probe qpcr) Product Manual Table of Contents I. Description... 3 II. Principle... 4 III. Components... 5 IV. Materials Required but not Provided... 5 V. Storage... 5 VI.
For Research Use Premix Ex Taq (Probe qpcr) Product Manual Table of Contents I. Description... 3 II. Principle... 4 III. Components... 5 IV. Materials Required but not Provided... 5 V. Storage... 5 VI.
RNP-IP (Modified Method)-Getting Majority RNA from RNA Binding Protein in the Cytoplasm Fengzhi Liu *
 RNP-IP (Modified Method)-Getting Majority RNA from RNA Binding Protein in the Cytoplasm Fengzhi Liu * School of Biomedical Sciences, Thomas Jefferson University, Philadelphia, USA *For correspondence:
RNP-IP (Modified Method)-Getting Majority RNA from RNA Binding Protein in the Cytoplasm Fengzhi Liu * School of Biomedical Sciences, Thomas Jefferson University, Philadelphia, USA *For correspondence:
Supplemental Materials and Methods
 Supplemental Materials and Methods In situ hybridization In situ hybridization analysis of HFE2 and genin mrna in rat liver tissues was performed as previously described (1). Briefly, the digoxigenin-labeled
Supplemental Materials and Methods In situ hybridization In situ hybridization analysis of HFE2 and genin mrna in rat liver tissues was performed as previously described (1). Briefly, the digoxigenin-labeled
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1154040/dc1 Supporting Online Material for Selective Blockade of MicroRNA Processing by Lin-28 Srinivas R. Viswanathan, George Q. Daley,* Richard I. Gregory* *To whom
www.sciencemag.org/cgi/content/full/1154040/dc1 Supporting Online Material for Selective Blockade of MicroRNA Processing by Lin-28 Srinivas R. Viswanathan, George Q. Daley,* Richard I. Gregory* *To whom
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Mir-X mirna First-Strand Synthesis and SYBR qrt-pcr
 User Manual Mir-X mirna First-Strand Synthesis and SYBR qrt-pcr User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories,
User Manual Mir-X mirna First-Strand Synthesis and SYBR qrt-pcr User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories,
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Sequence, Biogenesis, and Function of Diverse Small RNA Classes Bound to the Piwifamily
 Sequence, Biogenesis, and Function of Diverse Small RNA Classes Bound to the Piwifamily Proteins of Tetrahymena thermophila Mary T. Couvillion, Suzanne R. Lee, Brandon Hogstad, Colin D. Malone, Leath A.
Sequence, Biogenesis, and Function of Diverse Small RNA Classes Bound to the Piwifamily Proteins of Tetrahymena thermophila Mary T. Couvillion, Suzanne R. Lee, Brandon Hogstad, Colin D. Malone, Leath A.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Supplemental Information
 Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
N173A. residue number
 a 0.3 Δω 1 ( 1 H Ν ) [ppm] 0-0.3 N173A -0.6 Y177A b Δω 3 ( 13 C α ) [ppm] 2 0-2 c Δω 3 ( 13 C β ) [ppm] 2 0-2 -4 Supplementary Figure 1 140 150 160 170 180 190 200 residue number Comparison of chemical
a 0.3 Δω 1 ( 1 H Ν ) [ppm] 0-0.3 N173A -0.6 Y177A b Δω 3 ( 13 C α ) [ppm] 2 0-2 c Δω 3 ( 13 C β ) [ppm] 2 0-2 -4 Supplementary Figure 1 140 150 160 170 180 190 200 residue number Comparison of chemical
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
Supporting Information
 Supporting Information He et al. 10.1073/pnas.1116302108 SI Methods Cell Culture. Mouse J774A.1 and RAW 264.7 macrophages were obtained from ATCC and were cultured in MEM supplemented with 10% FS (Sigma)
Supporting Information He et al. 10.1073/pnas.1116302108 SI Methods Cell Culture. Mouse J774A.1 and RAW 264.7 macrophages were obtained from ATCC and were cultured in MEM supplemented with 10% FS (Sigma)
Gene Forward Primer Reverse Primer GAPDH ATCATCCCTGCCTCTACTGG GTCAGGTCCACCACTGACAC SSB1 AACTTCAGTGAGCCAAACCC GTTCTCAGAGGCTGGAGAGG
 Supplemental Data EXPERIMENTAL PROCEDURES Plasmids and Antibodies- Full length cdna of INT11 or INT12 were cloned into ps- Flag-SBP vector respectively. Anti-RNA pol II (RPB1) was purchased from Santa
Supplemental Data EXPERIMENTAL PROCEDURES Plasmids and Antibodies- Full length cdna of INT11 or INT12 were cloned into ps- Flag-SBP vector respectively. Anti-RNA pol II (RPB1) was purchased from Santa
Reading Lecture 8: Lecture 9: Lecture 8. DNA Libraries. Definition Types Construction
 Lecture 8 Reading Lecture 8: 96-110 Lecture 9: 111-120 DNA Libraries Definition Types Construction 142 DNA Libraries A DNA library is a collection of clones of genomic fragments or cdnas from a certain
Lecture 8 Reading Lecture 8: 96-110 Lecture 9: 111-120 DNA Libraries Definition Types Construction 142 DNA Libraries A DNA library is a collection of clones of genomic fragments or cdnas from a certain
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Fatchiyah
 Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
Human Cell-Free Protein Expression System
 Cat. # 3281 For Research Use Human Cell-Free Protein Expression System Product Manual Table of Contents I. Description... 3 II. Components... 3 III. Materials Required but not Provided... 4 IV. Storage...
Cat. # 3281 For Research Use Human Cell-Free Protein Expression System Product Manual Table of Contents I. Description... 3 II. Components... 3 III. Materials Required but not Provided... 4 IV. Storage...
Table of Contents. I. Kit Components...2. Storage...2. Principle...2. IV. Precautions for operation...3. V. Protocol : reverse transcription...
 Table of Contents I. Kit Components...2 II. III. Storage...2 Principle...2 IV. Precautions for operation...3 V. Protocol : reverse transcription...3 VI. Protocol : Real-time PCR...5 VII. Appendix...7 VIII.
Table of Contents I. Kit Components...2 II. III. Storage...2 Principle...2 IV. Precautions for operation...3 V. Protocol : reverse transcription...3 VI. Protocol : Real-time PCR...5 VII. Appendix...7 VIII.
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
Supporting Information. SI Material and Methods
 Supporting Information SI Material and Methods RNA extraction and qrt-pcr RNA extractions were done using the classical phenol-chloroform method. Total RNA samples were treated with Turbo DNA-free (Ambion)
Supporting Information SI Material and Methods RNA extraction and qrt-pcr RNA extractions were done using the classical phenol-chloroform method. Total RNA samples were treated with Turbo DNA-free (Ambion)
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Methods (detailed). for all the experiments. Cells were grown in 50 ml YEA. The cells were harvested and
 Methods (detailed). Purification of yeast chromosomal DNA. Strain JZ105 (mat1m Δmat2,3::LEU2, ade6-210, leu1-32, ura4-d18, his2) was used for all the experiments. Cells were grown in 50 ml YEA. The cells
Methods (detailed). Purification of yeast chromosomal DNA. Strain JZ105 (mat1m Δmat2,3::LEU2, ade6-210, leu1-32, ura4-d18, his2) was used for all the experiments. Cells were grown in 50 ml YEA. The cells
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Supplementary Material
 Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplementary Information: Materials and Methods. Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered
 Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE.
 Instruction manual RNA-direct SYBR Green Realtime PCR Master Mix 0810 F0930K RNA-direct SYBR Green Realtime PCR Master Mix Contents QRT-201T QRT-201 0.5mLx2 0.5mLx5 Store at -20 C, protected from light
Instruction manual RNA-direct SYBR Green Realtime PCR Master Mix 0810 F0930K RNA-direct SYBR Green Realtime PCR Master Mix Contents QRT-201T QRT-201 0.5mLx2 0.5mLx5 Store at -20 C, protected from light
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
SUPPLEMENTARY INFORMATION FILE
 SUPPLEMENTARY INFORMATION FILE Existence of a microrna pathway in anucleate platelets Patricia Landry, Isabelle Plante, Dominique L. Ouellet, Marjorie P. Perron, Guy Rousseau & Patrick Provost 1. SUPPLEMENTARY
SUPPLEMENTARY INFORMATION FILE Existence of a microrna pathway in anucleate platelets Patricia Landry, Isabelle Plante, Dominique L. Ouellet, Marjorie P. Perron, Guy Rousseau & Patrick Provost 1. SUPPLEMENTARY
Cat. # RR391A. For Research Use. Probe qpcr Mix. Product Manual. v201610da
 Cat. # RR391A For Research Use Probe qpcr Mix Product Manual Table of Contents I. Introduction... 3 II. Principle... 3 III. Components... 5 IV. Materials Required but not Provided... 5 V. Storage... 5
Cat. # RR391A For Research Use Probe qpcr Mix Product Manual Table of Contents I. Introduction... 3 II. Principle... 3 III. Components... 5 IV. Materials Required but not Provided... 5 V. Storage... 5
Premix Ex Taq (Probe qpcr)
 For Research Use Premix Ex Taq (Probe qpcr) Product Manual Table of Contents I. Description... 3 II. Principle... 4 III. Components... 5 IV. Materials Required but not Provided... 5 V. Storage... 5 VI.
For Research Use Premix Ex Taq (Probe qpcr) Product Manual Table of Contents I. Description... 3 II. Principle... 4 III. Components... 5 IV. Materials Required but not Provided... 5 V. Storage... 5 VI.
Supplementary information
 Supplementary information Supplementary Figure 1 (a) EM image of the pyramidal layer of wt mice. CA3 pyramidal neurons were selected according to their typical alignment, size and shape for subsequent
Supplementary information Supplementary Figure 1 (a) EM image of the pyramidal layer of wt mice. CA3 pyramidal neurons were selected according to their typical alignment, size and shape for subsequent
Supplementary Methods
 Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
hnrnp D/AUF1 Rabbit IgG hnrnp M
 Mouse IgG Goat IgG Rabbit IgG Mouse IgG hnrnp F Goat IgG Mouse IgG Kb 6 4 3 2 15 5 Supplementary Figure S1. In vivo binding of TERRA-bound RBPs to target RNAs. Immunoprecipitation (IP) assay using 3 mg
Mouse IgG Goat IgG Rabbit IgG Mouse IgG hnrnp F Goat IgG Mouse IgG Kb 6 4 3 2 15 5 Supplementary Figure S1. In vivo binding of TERRA-bound RBPs to target RNAs. Immunoprecipitation (IP) assay using 3 mg
The poly(a) tail blocks RDR6 from converting self mrnas into substrates for gene silencing
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17036 The poly(a) tail blocks RDR6 from converting self mrnas into substrates for gene silencing
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17036 The poly(a) tail blocks RDR6 from converting self mrnas into substrates for gene silencing
Supplemental Material Igreja and Izaurralde 1. CUP promotes deadenylation and inhibits decapping of mrna targets. Catia Igreja and Elisa Izaurralde
 Supplemental Material Igreja and Izaurralde 1 CUP promotes deadenylation and inhibits decapping of mrna targets Catia Igreja and Elisa Izaurralde Supplemental Materials and methods Functional assays and
Supplemental Material Igreja and Izaurralde 1 CUP promotes deadenylation and inhibits decapping of mrna targets Catia Igreja and Elisa Izaurralde Supplemental Materials and methods Functional assays and
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Motivation From Protein to Gene
 MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
Bootcamp: Molecular Biology Techniques and Interpretation
 Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Cat. # RR430S RR430A. For Research Use. SYBR Fast qpcr Mix. Product Manual. v201610da
 For Research Use SYBR Fast qpcr Mix Product Manual Table of Contents I. Description... 3 II. Principle... 4 III. Components... 5 IV. Storage... 5 V. Features... 6 VI. Precautions... 6 VII. Protocol...
For Research Use SYBR Fast qpcr Mix Product Manual Table of Contents I. Description... 3 II. Principle... 4 III. Components... 5 IV. Storage... 5 V. Features... 6 VI. Precautions... 6 VII. Protocol...
2.5. Equipment and materials supplied by user PCR based template preparation Influence of temperature on in vitro EGFP synthesis 11
 Manual 15 Reactions LEXSY in vitro Translation Cell-free protein expression kit based on Leishmania tarentolae for PCR-based template generation Cat. No. EGE-2010-15 FOR RESEARCH USE ONLY. NOT INTENDED
Manual 15 Reactions LEXSY in vitro Translation Cell-free protein expression kit based on Leishmania tarentolae for PCR-based template generation Cat. No. EGE-2010-15 FOR RESEARCH USE ONLY. NOT INTENDED
Transcriptional regulation of BRCA1 expression by a metabolic switch: Di, Fernandez, De Siervi, Longo, and Gardner. H3K4Me3
 ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
Supporting Information
 Supporting Information Tal et al. 10.1073/pnas.0807694106 SI Materials and Methods VSV Infection and Quantification. Infection was carried out by seeding 5 10 5 MEF cells per well in a 6-well plate and
Supporting Information Tal et al. 10.1073/pnas.0807694106 SI Materials and Methods VSV Infection and Quantification. Infection was carried out by seeding 5 10 5 MEF cells per well in a 6-well plate and
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Materials
 Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplemental Material
 Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
Product Name : Simple mirna Detection Kit
 Product Name : Simple mirna Detection Kit Code No. : DS700 This product is for research use only Kit Contents This kit provides sufficient reagents to perform 20 reactions for detecting microrna. Components
Product Name : Simple mirna Detection Kit Code No. : DS700 This product is for research use only Kit Contents This kit provides sufficient reagents to perform 20 reactions for detecting microrna. Components
1. Cross-linking and cell harvesting
 ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
SYBR Premix Ex Taq II (Tli RNaseH Plus), ROX plus
 Cat. # RR82LR For Research Use SYBR Premix Ex Taq II (Tli RNaseH Plus), ROX plus Product Manual The long-term storage temperature of this product has been changed to -20 since Lot. A1901A. See section
Cat. # RR82LR For Research Use SYBR Premix Ex Taq II (Tli RNaseH Plus), ROX plus Product Manual The long-term storage temperature of this product has been changed to -20 since Lot. A1901A. See section
Solid Phase cdna Synthesis Kit
 #6123 v.02.09 Table of Contents I. Description... 2 II. Kit components... 2 III. Storage... 2 IV. Principle... 3 V. Protocol V-1. Preparation of immobilized mrna... 4 Protocol A: Starting from Tissue or
#6123 v.02.09 Table of Contents I. Description... 2 II. Kit components... 2 III. Storage... 2 IV. Principle... 3 V. Protocol V-1. Preparation of immobilized mrna... 4 Protocol A: Starting from Tissue or
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Rapid Method for the Purification of Total RNA from Formalin- Fixed Paraffin-Embedded (FFPE) Tissue Samples
 Application Note 17 RNA Sample Preparation Rapid Method for the Purification of Total RNA from Formalin- Fixed Paraffin-Embedded (FFPE) Tissue Samples M. Melmogy 1, V. Misic 1, B. Lam, PhD 1, C. Dobbin,
Application Note 17 RNA Sample Preparation Rapid Method for the Purification of Total RNA from Formalin- Fixed Paraffin-Embedded (FFPE) Tissue Samples M. Melmogy 1, V. Misic 1, B. Lam, PhD 1, C. Dobbin,
Table of Contents. i. Insertion of DNA fragments into plasmid vector...4. ii. Self-circularization of linear blunt-ended DNA...4
 Table of Contents I. Description...2 II. Kit Components...2 III. Storage...2 IIV. Notes...2 V. Reference...3 VI. PROCEDURES A. Dephosphorylation of vector DNA...3 B. Blunting reaction...3 C. Ligation reaction
Table of Contents I. Description...2 II. Kit Components...2 III. Storage...2 IIV. Notes...2 V. Reference...3 VI. PROCEDURES A. Dephosphorylation of vector DNA...3 B. Blunting reaction...3 C. Ligation reaction
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
SUPPLEMENTAL MATERIALS. Chromatin immunoprecipitation assays. Single-cell suspensions of pituitary
 SUPPLEMENTAL MATERIALS METHODS Chromatin immunoprecipitation assays. Single-cell suspensions of pituitary and liver cells were prepared from the indicated transgenic mice, as described (Ho et al, 2002).
SUPPLEMENTAL MATERIALS METHODS Chromatin immunoprecipitation assays. Single-cell suspensions of pituitary and liver cells were prepared from the indicated transgenic mice, as described (Ho et al, 2002).
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
Protocol Immunprecipitation
 1 Protocol Immunprecipitation IP Juni 08, S.L. Before starting an IP on should be certain, that one can clearly detect the Protein to precipitate in a standard Western Blot with the lysis conditions used
1 Protocol Immunprecipitation IP Juni 08, S.L. Before starting an IP on should be certain, that one can clearly detect the Protein to precipitate in a standard Western Blot with the lysis conditions used
What is a proteome and how is the link between an organisms genome and a proteome.
 Answer to exam questions Bi2014 autumn 2013. Question 1a. What is a proteome and how is the link between an organisms genome and a proteome. The answer should discuss the following topics: What is a genome,
Answer to exam questions Bi2014 autumn 2013. Question 1a. What is a proteome and how is the link between an organisms genome and a proteome. The answer should discuss the following topics: What is a genome,
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of
 Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
CRISPR/Cas9 Gene Editing Tools
 CRISPR/Cas9 Gene Editing Tools - Guide-it Products for Successful CRISPR/Cas9 Gene Editing - Why choose Guide-it products? Optimized methods designed for speed and ease of use Complete kits that don t
CRISPR/Cas9 Gene Editing Tools - Guide-it Products for Successful CRISPR/Cas9 Gene Editing - Why choose Guide-it products? Optimized methods designed for speed and ease of use Complete kits that don t
