SUPPLEMENTARY INFORMATION
|
|
|
- Julianna Owens
- 6 years ago
- Views:
Transcription
1 a Densitometry (AU) t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule expansion in wild-type ES cells after bistatin treatment does not involve changes in the protein levels of tubulin. Wild-type ES cells treated with 25 nm bistatin () for 3 minutes showed immediate expansion of microtubules, but no increase in the total levels of -tubulin. (b) RNA interference by myosin IIA sirna transfection in human foreskin fibroblasts reduced protein levels of myosin IIA by 6% at 1 nm and 65% at 1 nm sirna (densitometry calculations were corrected for GAPDH levels). Myosin IIB levels were not significantly altered (right-hand graph). Error bars indicate range Nature Publishing Group
2 a 6 5 Velocity ( m/h) WT IIB -/- fibroblasts WT IIB -/- ES cells b 12 1 IIA -/- ES cells c IIA -/- IIA -/- Velocity ( m/h) Figure S2 S1 Analyses of roles of myosin IIB. (a) Myosin IIB-deficient fibroblasts and ES cells exhibit slightly slower migration rates compared to wild-type cells, as quantified by time-lapse microscopy. (b, c) Inhibition of myosin IIB by bistatin () in myosin IIA-null ES cells. Blebbistatin (5 or 1 nm) treatment of myosin IIA-null ES cells (IIA-/-) to completely inhibit all myosin II isoforms (including IIB) did not significantly change migration rates (b), but 5 nm bistatin eliminated residual stress fibres (c) Nature Publishing Group
3 a 1 b D/T.6.4 D/T P =.31 WT IIA-/- P =.39 Figure S3 S2 Myosin II-deficient cells show decreased directionality. Human foreskin fibroblasts treated with bistatin (a) or myosin IIA-null ES cells (b) migrate with less directional persistence compared to their untreated or wild-type counterparts. Directionality was evaluated by time-lapse microscopy and calculated as D/T: the ratio between the total trajectory length (T) and the linear distance between the start and end points (D) Nature Publishing Group
4 Fluorescence intensity (AU) Fluorescence intensity (AU) Rac Distance (μm) Tiam Distance (μm) Figure S4 S3 Line scans of fluorescence intensity of the immunolocalization of Rac1 and Tiam1 at the leading edge. Fluorescence was measured by MetaMorph image analysis software starting at the cell periphery (left) along 7 lines drawn perpendicular to the leading edge of the myosin IIAnull ES cell in Fig. 4a. Fill colors designate paired scans for each protein. Note the strong, co-localized staining for Rac1 and Tiam1 near the leading edge Nature Publishing Group
5 Legends for Supplementary Videos The supplementary videos are best viewed with QuickTime 7 or a higher version. The viewer is available at no cost from Apple at (for Mac) or (for Windows). Supplementary videos 1-4: COS-7 cells, which lack the myosin IIA isoform but express myosin IIB, were co-transfected with mcherry- -tubulin and either EGFPmyosin IIA or EGFP-myosin IIB plasmids. Time-lapse spinning-disk focal fluorescent imaging was performed hours after transfection for ~6 minutes. Human foreskin fibroblasts, which express both myosin II isoforms, were transfected and imaged similarly. Video movies were compressed using QuickTime Pro and the H.264 codec; the original higher-resolution movies can be obtained from the authors upon request. Supplementary video 1: Myosin IIA-transfected COS-7 cells show substantial microtubule dynamics (left panel and red in the merged color image) in areas of active lamellar ruffling. EGFP-myosin IIA (middle, green) shows vigorous retrograde movements of myosin IIA clusters in lamellae. The scale bar represents 5 μm. Supplementary video 2: Myosin IIB-transfected COS-7 cells show relatively stable microtubules (left, red) with slow turnover. The EGFP-myosin IIB (middle, green) was primarily associated with stress fibers, and the low quantities in lamellae displayed slower retrograde flow than myosin IIA. Similar stability of microtubules was observed in cells without myosin II transfection. The scale bar represents 5 μm. Supplementary video 3: Human fibroblasts transfected with mcherry- -tubulin (left, red) and EGFP-myosin IIA (middle, green) show microtubule dynamics and vigorous retrograde flow of the EGFP-myosin IIA. The scale bar represents 5 μm. Supplementary video 4: Human fibroblasts were transfected with mcherry- tubulin (left, red) and EGFP-myosin IIB (middle, green) and show microtubule dynamics (these cells express endogenous myosin IIA) but only minimal retrograde flow of the EGFP-myosin IIB. The scale bar represents 5 μm. Supplementary video 5: Comparisons of microtubule dynamics in COS-7 cells transfected with myosin IIA (left panels), trol (center), or myosin IIB (right) plasmids. The scale bar represents 5 μm Nature Publishing Group
6 Supplemental Information: Equipment and Settings Time-Lapse Fluorescence Microscopy: Dual wavelength time-lapse images of mcherry-tubulin with EGFP-myosin IIA or EGFP-myosin IIB were acquired using an Ultraview ERS spinning-disk focal scanning system (Perkin-Elmer) attached to a Zeiss Axiovert 2M microscope equipped with a 1X Plan-APOCHROMAT (N.A. 1.4) oil DIC objective (Zeiss). 24-hours after transfection cells were plated onto glass-bottom culture dishes (MatTek). Images were captured every 6 seds (4-5 sec. dead time) for total durations of approximately 6 minutes using a Hamamatsu EM CCD camera (Hamamatsu Corporation) in 1x1 binning mode with a center sub-pixel array of 496x496. A Melles Griot Kr-Ar laser was used for excitation of EGFP and mcherry chimeric proteins at 488 nm and 568 nm, respectively. Exposure times and laser power were ~5ms and 35% for the 488 nm line and ~75 ms and 55% for the 568 nm line. An environmental chamber mounted on the microscope maintained stant 37ºC temperature, ~1% CO2, and humidity during acquisition. Image filtering and quantification: No image filtering was performed on fixed samples. Sequential background subtraction, low pass filtering, Gaussian filtering and sharpening image processing was carried out for fluorescence time series using custom-made volution filters created on both ImageJ (NIH) and MetaMorph software (Molecular Devices). All volution filter settings were the same for all images collected. Images were acquired at a bit-depth of 16 and then verted to 8-bit format following filtering. For analysis of myosin IIA and IIB flow rates within the lamella, kymographs of raw data were generated with ImageJ, and the slopes of lines depicting myosin flow were calculated from X, Y coordinates and verted to micrometers/minute. Microtubule dwell time was calculated from the time-lapse movie sequences by counting the number of frames in which the tip of a microtubule stayed within a zone 1 micrometers from the leading edge of cells Nature Publishing Group
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Prospéri et al.,
 Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
The definitive version is available at www3.intersci. Supporting_Information.pdf (Supporting Information) Instructions for use
 Title Nonmuscle myosin II folds into a 10S form via two po Kiboku, Takayuki; Katoh, Tsuyoshi; Nakamura, Akio; K Author(s) Masayuki CitationGenes to Cells, 18(2): 90-109 Issue Date 2013-02 Doc URL http://hdl.handle.net/2115/53054
Title Nonmuscle myosin II folds into a 10S form via two po Kiboku, Takayuki; Katoh, Tsuyoshi; Nakamura, Akio; K Author(s) Masayuki CitationGenes to Cells, 18(2): 90-109 Issue Date 2013-02 Doc URL http://hdl.handle.net/2115/53054
MT minus end catastrophe
 DOI: 1.138/ncb3241 MT minus end catastrophe frequency [s -1 ].2.15.1.5 control mgfp- TPX2 * * mgfp- TPX2 mini Supplementary Figure 1. TPX2 reduces catastrophes at the microtubule minus ends. Modified box-and-whiskers
DOI: 1.138/ncb3241 MT minus end catastrophe frequency [s -1 ].2.15.1.5 control mgfp- TPX2 * * mgfp- TPX2 mini Supplementary Figure 1. TPX2 reduces catastrophes at the microtubule minus ends. Modified box-and-whiskers
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Paul et al.,
 Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplementary Figures
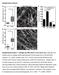 Supplementary Figures Supplementary Figure 1: Collagen gel fibril sizes in vitro and in vivo. (a-b) Box and whisker plots for collagen fibril/bundle diameter (A) and length (B) for HR and FB16 matrices.*
Supplementary Figures Supplementary Figure 1: Collagen gel fibril sizes in vitro and in vivo. (a-b) Box and whisker plots for collagen fibril/bundle diameter (A) and length (B) for HR and FB16 matrices.*
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
supplementary information
 DOI: 10.1038/ncb1992 Figure S1 (a-b) Verification of actin flow patter using photobleaching of actin:gfp and Lifeact:RFP in migrating dendritic cells (DCs). (a) Actin:GFP and Lifeact:RFP double-trafected
DOI: 10.1038/ncb1992 Figure S1 (a-b) Verification of actin flow patter using photobleaching of actin:gfp and Lifeact:RFP in migrating dendritic cells (DCs). (a) Actin:GFP and Lifeact:RFP double-trafected
Kinesin Walks on Microtubules
 Kinesin Walks on Microtubules Other Motor Proteins Question: Alone or in Groups? Our live-cell imaging: DIC (differential interference contrast) Normal DIC Microscopy Image 50x50 m Orca ER camera 125
Kinesin Walks on Microtubules Other Motor Proteins Question: Alone or in Groups? Our live-cell imaging: DIC (differential interference contrast) Normal DIC Microscopy Image 50x50 m Orca ER camera 125
Supplemental material JCB. Pitaval et al., Cell shape and ciliogenesis Pitaval et al.
 Supplemental material THE JOURNAL OF CELL BIOLOGY Pitaval et al., http://www.jcb.org/cgi/content/full/jcb.201004003/dc1 S1 Figure S1. Cell cycle exit analysis. (A) Experimental procedure. RPE1 cells were
Supplemental material THE JOURNAL OF CELL BIOLOGY Pitaval et al., http://www.jcb.org/cgi/content/full/jcb.201004003/dc1 S1 Figure S1. Cell cycle exit analysis. (A) Experimental procedure. RPE1 cells were
No wash 2 Washes 2 Days ** ** IgG-bead phagocytosis (%)
 Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Graphene oxide-enhanced cytoskeleton imaging and mitosis tracking
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Supplementary information for Graphene oxide-enhanced cytoskeleton imaging and mitosis tracking
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Supplementary information for Graphene oxide-enhanced cytoskeleton imaging and mitosis tracking
Supplementary methods Shoc2 In Vitro Ubiquitination Assay
 Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Supplementary Figures
 Supplementary Figures Supplementary Figure S1. Inhibition kinetics of p53/hdm2 binding induced by Nutlin 3 on live cells. Reproducibility of the p53/hdm2 binding disruption kinetics induced by Nutlin 3
Supplementary Figures Supplementary Figure S1. Inhibition kinetics of p53/hdm2 binding induced by Nutlin 3 on live cells. Reproducibility of the p53/hdm2 binding disruption kinetics induced by Nutlin 3
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y
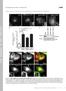 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
λ N -GFP: an RNA reporter system for live-cell imaging
 λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing
 Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
NOGO-A/RTN4A and NOGO-B/RTN4B are simultaneously expressed in. epithelial, fibroblast and neuronal cells and maintain ER morphology
 SREP-16-11884B Supplementary information NOGO-A/RTN4A and NOGO-B/RTN4B are simultaneously expressed in epithelial, fibroblast and neuronal cells and maintain ER morphology Olli Rämö a*, Darshan Kumar a*,
SREP-16-11884B Supplementary information NOGO-A/RTN4A and NOGO-B/RTN4B are simultaneously expressed in epithelial, fibroblast and neuronal cells and maintain ER morphology Olli Rämö a*, Darshan Kumar a*,
Supplementary Materials for
 www.sciencemag.org/cgi/content/full/science.1245533/dc1 Supplementary Materials for A Mechanism for Reorientation of Cortical Microtubule Arrays Driven by Microtubule Severing Jelmer J. Lindeboom, Masayoshi
www.sciencemag.org/cgi/content/full/science.1245533/dc1 Supplementary Materials for A Mechanism for Reorientation of Cortical Microtubule Arrays Driven by Microtubule Severing Jelmer J. Lindeboom, Masayoshi
Supplementary Figure 1
 Supplementary Figure a.5 fold of increase.5 plaa plaa with LY plaa /piaa plaa /piaa with LY b 5 5 5 plaa plaa /piaa No LY with LY No LY with LY c 6X fluorescence plaa plaa with LY plaa /piaa plaa /piaa
Supplementary Figure a.5 fold of increase.5 plaa plaa with LY plaa /piaa plaa /piaa with LY b 5 5 5 plaa plaa /piaa No LY with LY No LY with LY c 6X fluorescence plaa plaa with LY plaa /piaa plaa /piaa
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Fluorescence Imaging with One Nanometer Accuracy Lab
 I. Introduction. Fluorescence Imaging with One Nanometer Accuracy Lab Traditional light microscope is limited by the diffraction limit of light, typically around 250 nm. However, many biological processes
I. Introduction. Fluorescence Imaging with One Nanometer Accuracy Lab Traditional light microscope is limited by the diffraction limit of light, typically around 250 nm. However, many biological processes
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Multiplexed 3D FRET imaging in deep tissue of live embryos Ming Zhao, Xiaoyang Wan, Yu Li, Weibin Zhou and Leilei Peng
 Scientific Reports Multiplexed 3D FRET imaging in deep tissue of live embryos Ming Zhao, Xiaoyang Wan, Yu Li, Weibin Zhou and Leilei Peng 1 Supplementary figures and notes Supplementary Figure S1 Volumetric
Scientific Reports Multiplexed 3D FRET imaging in deep tissue of live embryos Ming Zhao, Xiaoyang Wan, Yu Li, Weibin Zhou and Leilei Peng 1 Supplementary figures and notes Supplementary Figure S1 Volumetric
SUPPLEMENTARY INFORMATION
 DOI: 1.18/ncb46 a Damage Fracture s s 7 5 16 1 9 26 5 µm 5 µm b Position of photo-damage site in cells 15 Counts 1 5-14 -12-1 -8-6 -4-2 Distance from damage site to tip (µm) Supplementary Figure 1 Effect
DOI: 1.18/ncb46 a Damage Fracture s s 7 5 16 1 9 26 5 µm 5 µm b Position of photo-damage site in cells 15 Counts 1 5-14 -12-1 -8-6 -4-2 Distance from damage site to tip (µm) Supplementary Figure 1 Effect
Flag-Rac Vector V12 V12 N17 C40. Vector C40 pakt (T308) Akt1. Myc-DN-PAK1 (N-SP)
 a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
Michal Reichman-Fried
 Zebrafish Development and Genetics course 2013 Cell labeling and migration Imaging primordial germ cell migration Instructors: Erez Raz (erez.raz@uni-muenster.de) Michal Reichman-Fried (mreichm@uni-muenster.de)
Zebrafish Development and Genetics course 2013 Cell labeling and migration Imaging primordial germ cell migration Instructors: Erez Raz (erez.raz@uni-muenster.de) Michal Reichman-Fried (mreichm@uni-muenster.de)
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Prashar et al., http://www.jcb.org/cgi/content/full/jcb.201304095/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. FBT phagocytosis in cells expressing PM-GFP. (A) T-PC
Supplemental material Prashar et al., http://www.jcb.org/cgi/content/full/jcb.201304095/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. FBT phagocytosis in cells expressing PM-GFP. (A) T-PC
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
MEFs were treated with the indicated concentrations of LLOMe for three hours, washed
 Supplementary Materials and Methods Cell Fractionation MEFs were treated with the indicated concentrations of LLOMe for three hours, washed with ice-cold PBS, collected by centrifugation, and then homogenized
Supplementary Materials and Methods Cell Fractionation MEFs were treated with the indicated concentrations of LLOMe for three hours, washed with ice-cold PBS, collected by centrifugation, and then homogenized
SUPPLEMENTAL INFORMATION: 1. Supplemental methods 2. Supplemental figure legends 3. Supplemental figures
 Supplementary Material (ESI) for Lab on a Chip This journal is The Royal Society of Chemistry 2008 A microfluidics-based turning assay reveals complex growth cone responses to integrated gradients of substrate-bound
Supplementary Material (ESI) for Lab on a Chip This journal is The Royal Society of Chemistry 2008 A microfluidics-based turning assay reveals complex growth cone responses to integrated gradients of substrate-bound
T H E J O U R N A L O F C E L L B I O L O G Y
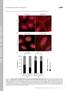 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Feng et al., http://www.jcb.org/cgi/content/full/jcb.201408079/dc1 Figure S1. A modest elevation of disulfide-bonded K14 in primary mouse
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Feng et al., http://www.jcb.org/cgi/content/full/jcb.201408079/dc1 Figure S1. A modest elevation of disulfide-bonded K14 in primary mouse
Supplemental Data includes Supplemental Experimental Procedures, 4 Figures and 5 Movie Legends.
 SUPPLEMENTAL DATA Supplemental Data includes Supplemental Experimental Procedures, 4 Figures and 5 Movie Legends. Supplemental Experimental Procedures Dominant negative nesprin KASH (DN KASH) plasmid construction-
SUPPLEMENTAL DATA Supplemental Data includes Supplemental Experimental Procedures, 4 Figures and 5 Movie Legends. Supplemental Experimental Procedures Dominant negative nesprin KASH (DN KASH) plasmid construction-
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
SUPPLEMENTARY INFORMATION
 Nanomechanical mapping of first binding steps of a virus to animal cells David Alsteens, Richard Newton, Rajib Schubert, David Martinez-Martin, Martin Delguste, Botond Roska and Daniel J. Müller This PDF
Nanomechanical mapping of first binding steps of a virus to animal cells David Alsteens, Richard Newton, Rajib Schubert, David Martinez-Martin, Martin Delguste, Botond Roska and Daniel J. Müller This PDF
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 1 SUPPLEMENTARY INFORMATION 2 WWW.NATURE.COM/NATURECELLBIOLOGY SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 3 SUPPLEMENTARY INFORMATION
SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 1 SUPPLEMENTARY INFORMATION 2 WWW.NATURE.COM/NATURECELLBIOLOGY SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 3 SUPPLEMENTARY INFORMATION
Supporting Online Material Material and Methods
 Supporting Online Material Material and Methods Live-cell DIC recordings PtK 1 cells (ATCC, Manassas, VA) were filmed on a Nikon (Nikon Instruments, Melville, NY) inverted microscope equipped with 60x
Supporting Online Material Material and Methods Live-cell DIC recordings PtK 1 cells (ATCC, Manassas, VA) were filmed on a Nikon (Nikon Instruments, Melville, NY) inverted microscope equipped with 60x
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
42 fl organelles = 34.5 fl (1) 3.5X X 0.93 = 78,000 (2)
 SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Quantitative real-time RT-PCR analysis of the expression levels of E-cadherin
 Supplementary Information 1 Quantitative real-time RT-PCR analysis of the expression levels of E-cadherin and ribosomal protein L19 (RPL19) mrna in cleft and bud epithelial cells of embryonic salivary
Supplementary Information 1 Quantitative real-time RT-PCR analysis of the expression levels of E-cadherin and ribosomal protein L19 (RPL19) mrna in cleft and bud epithelial cells of embryonic salivary
Fluorescence Light Microscopy for Cell Biology
 Fluorescence Light Microscopy for Cell Biology Why use light microscopy? Traditional questions that light microscopy has addressed: Structure within a cell Locations of specific molecules within a cell
Fluorescence Light Microscopy for Cell Biology Why use light microscopy? Traditional questions that light microscopy has addressed: Structure within a cell Locations of specific molecules within a cell
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
Artificial targeting of misfolded cytosolic proteins to endoplasmic reticulum as a mechanism for clearance
 Supplementary Information Artificial targeting of misfolded cytosolic proteins to endoplasmic reticulum as a mechanism for clearance Fen Liu 1, Deanna M. Koepp 2, and Kylie J. Walters 1* 1 Protein Processing
Supplementary Information Artificial targeting of misfolded cytosolic proteins to endoplasmic reticulum as a mechanism for clearance Fen Liu 1, Deanna M. Koepp 2, and Kylie J. Walters 1* 1 Protein Processing
T H E J O U R N A L O F C E L L B I O L O G Y
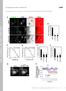 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Yamaguchi et al., http://www.jcb.org/cgi/content/full/jcb.201009126/dc1 S1 Figure S2. The expression levels of GFP-Akt1-PH and localization
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Yamaguchi et al., http://www.jcb.org/cgi/content/full/jcb.201009126/dc1 S1 Figure S2. The expression levels of GFP-Akt1-PH and localization
Fast, three-dimensional super-resolution imaging of live cells
 Nature Methods Fast, three-dimensional super-resolution imaging of live cells Sara A Jones, Sang-Hee Shim, Jiang He & Xiaowei Zhuang Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3
Nature Methods Fast, three-dimensional super-resolution imaging of live cells Sara A Jones, Sang-Hee Shim, Jiang He & Xiaowei Zhuang Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3
Innovations To Meet Your Needs
 Innovations To Meet Your Needs Cooled CCD Camera 1340 x 1037 pixel resolution for greatest image quality 12-bit precision provides 3 orders of linear dynamic range Windows and Power Macintosh Software
Innovations To Meet Your Needs Cooled CCD Camera 1340 x 1037 pixel resolution for greatest image quality 12-bit precision provides 3 orders of linear dynamic range Windows and Power Macintosh Software
Intercalation-based single-molecule fluorescence assay to study DNA supercoil
 Intercalation-based single-molecule fluorescence assay to study DNA supercoil dynamics Mahipal Ganji, Sung Hyun Kim, Jaco van der Torre, Elio Abbondanzieri*, Cees Dekker* Supplementary information Supplementary
Intercalation-based single-molecule fluorescence assay to study DNA supercoil dynamics Mahipal Ganji, Sung Hyun Kim, Jaco van der Torre, Elio Abbondanzieri*, Cees Dekker* Supplementary information Supplementary
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Hong et al.,
 Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 215 Electronic Supplementary Information Selective cell elimination in vitro and in vivo cell elimination
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 215 Electronic Supplementary Information Selective cell elimination in vitro and in vivo cell elimination
Plutoni et al., http :// /cgi /content /full /jcb /DC1
 Supplemental material JCB Plutoni et al., http ://www.jcb.org /cgi /content /full /jcb.201505105 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. Cell morphology during migration. (a) Protein extracts (20
Supplemental material JCB Plutoni et al., http ://www.jcb.org /cgi /content /full /jcb.201505105 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. Cell morphology during migration. (a) Protein extracts (20
Supplementary Figure S1
 Supplementary Figure S1 Supplementary Figure S1 Subcellular localization of Rab6a and LIS1 in the DRG neurons. (a) Exogenous expression of EGFP-Rab6a(wild-type) in the DRG neurons by the Neon transfection
Supplementary Figure S1 Supplementary Figure S1 Subcellular localization of Rab6a and LIS1 in the DRG neurons. (a) Exogenous expression of EGFP-Rab6a(wild-type) in the DRG neurons by the Neon transfection
Supplementary Figure 1: Two modes of low concentration of BsSMC on a DNA (a) Protein staining (left) and fluorescent imaging of Cy3 (right) confirm
 Supplementary Figure 1: Two modes of low concentration of BsSMC on a DNA (a) Protein staining (left) and fluorescent imaging of Cy3 (right) confirm that BsSMC was labeled with Cy3 NHS-Ester. In each panel,
Supplementary Figure 1: Two modes of low concentration of BsSMC on a DNA (a) Protein staining (left) and fluorescent imaging of Cy3 (right) confirm that BsSMC was labeled with Cy3 NHS-Ester. In each panel,
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Live cell microscopy
 Live cell microscopy 1. Why do live cell microscopy? 2. Maintaining living cells on a microscope stage. 3. Considerations for imaging living cells. 4. Fluorescence labeling of living cells. 5. Imaging
Live cell microscopy 1. Why do live cell microscopy? 2. Maintaining living cells on a microscope stage. 3. Considerations for imaging living cells. 4. Fluorescence labeling of living cells. 5. Imaging
T H E J O U R N A L O F C E L L B I O L O G Y
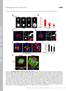 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
Supplementary Information
 Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
Supplemental Data Length Control of the Metaphase Spindle
 Supplemental Data Length Control of the Metaphase Spindle S1 Gohta Goshima, Roy Wollman, Nico Stuurman, Jonathan M. Scholey, and Ronald D. Vale Supplemental Experimental Procedures Cell Culture, RNA Interference,
Supplemental Data Length Control of the Metaphase Spindle S1 Gohta Goshima, Roy Wollman, Nico Stuurman, Jonathan M. Scholey, and Ronald D. Vale Supplemental Experimental Procedures Cell Culture, RNA Interference,
J. Cell Sci. 128: doi: /jcs : Supplementary Material. Supplemental Figures. Journal of Cell Science Supplementary Material
 Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Supplementary Figure S1. Hoechst. cells/field. Myogenin. % of Myogenin+ cells. 0 h chase CONTROL. BrdU. 72 h chase. CONTROL BrdU CONTROL BrdU
 Hoechst Myogenin amyhc 24 h 48 h 72 h b cells/field 50 40 30 20 10 0 24h 48h 72h h after plating (4 days growth+ t) c e CONTROL 0 h chase % of Myogenin+ cells h after plating (4 days growth+ t) CONTROL
Hoechst Myogenin amyhc 24 h 48 h 72 h b cells/field 50 40 30 20 10 0 24h 48h 72h h after plating (4 days growth+ t) c e CONTROL 0 h chase % of Myogenin+ cells h after plating (4 days growth+ t) CONTROL
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Egan et al., http://www.jcb.org/cgi/content/full/jcb.201112101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Localization of dynein to microtubule plus ends requires
Supplemental material Egan et al., http://www.jcb.org/cgi/content/full/jcb.201112101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Localization of dynein to microtubule plus ends requires
SI Appendix. Supporting Materials (references cited here can be found in the reference list in the main text):
 SI Appendix Supporting Materials (references cited here can be found in the reference list in the main text): SupplementaryText Figs. S1 to S13 Table S1 Captions for Supporting Movies S1 to S5 Other Supporting
SI Appendix Supporting Materials (references cited here can be found in the reference list in the main text): SupplementaryText Figs. S1 to S13 Table S1 Captions for Supporting Movies S1 to S5 Other Supporting
Supporting Online Material
 Supporting Online Material Two Distinct Actin Networks Drive the Protrusion of Migrating Cells A. Ponti, M. Machacek, S. L. Gupton, C. M. Waterman-Storer, G. Danuser This document contains: Experimental
Supporting Online Material Two Distinct Actin Networks Drive the Protrusion of Migrating Cells A. Ponti, M. Machacek, S. L. Gupton, C. M. Waterman-Storer, G. Danuser This document contains: Experimental
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
Chemotaxis assay using µ-slide Chemotaxis
 Chemotaxis assay using µ-slide Chemotaxis 1. General information The µ-slide Chemotaxis is a tool for observing chemotactical responses of adherent migrating cells over extended periods of time. The linear
Chemotaxis assay using µ-slide Chemotaxis 1. General information The µ-slide Chemotaxis is a tool for observing chemotactical responses of adherent migrating cells over extended periods of time. The linear
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence
 Supplementary Information Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Yoshinori Kousoku, Aya Ohkuni,
Supplementary Information Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Yoshinori Kousoku, Aya Ohkuni,
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators.
 Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Department of Chemistry, Center for Photochemical Sciences, Bowling Green State University,
 Supporting Information for Revealing Abrupt and Spontaneous Ruptures of Protein Native Structure under PicoNewton Compressive Force Manipulation S. Roy Chowdhury, Jin Cao, Yufan He, H. Peter Lu * Department
Supporting Information for Revealing Abrupt and Spontaneous Ruptures of Protein Native Structure under PicoNewton Compressive Force Manipulation S. Roy Chowdhury, Jin Cao, Yufan He, H. Peter Lu * Department
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
supplementary information Normalized Men ε/β ncrnas level 1.0 Ratio of Men ε/men β expression 0.5 DOI: /ncb2140
 DOI: 10.1038/ncb2140 2.5 10 Normalized Men ε/β ncrnas level 2.0 1.5 1.0 0.5 0 -DOX +DOX Ratio of Men ε/men β expression 5 0 -DOX +DOX Figure S1 Men ε/β reporter gene expression level by qrt-pcr. Left,
DOI: 10.1038/ncb2140 2.5 10 Normalized Men ε/β ncrnas level 2.0 1.5 1.0 0.5 0 -DOX +DOX Ratio of Men ε/men β expression 5 0 -DOX +DOX Figure S1 Men ε/β reporter gene expression level by qrt-pcr. Left,
Visualizing Cells Molecular Biology of the Cell - Chapter 9
 Visualizing Cells Molecular Biology of the Cell - Chapter 9 Resolution, Detection Magnification Interaction of Light with matter: Absorbtion, Refraction, Reflection, Fluorescence Light Microscopy Absorbtion
Visualizing Cells Molecular Biology of the Cell - Chapter 9 Resolution, Detection Magnification Interaction of Light with matter: Absorbtion, Refraction, Reflection, Fluorescence Light Microscopy Absorbtion
STORM/PALM. Super Resolution Microscopy 10/31/2011. Looking into microscopic world of life
 Super Resolution Microscopy STORM/PALM Bo Huang Department of Pharmaceutical Chemistry, UCSF CSHL Quantitative Microscopy, 1/31/211 Looking into microscopic world of life 1 µm 1 µm 1 nm 1 nm 1 nm 1 Å Naked
Super Resolution Microscopy STORM/PALM Bo Huang Department of Pharmaceutical Chemistry, UCSF CSHL Quantitative Microscopy, 1/31/211 Looking into microscopic world of life 1 µm 1 µm 1 nm 1 nm 1 nm 1 Å Naked
Fluorescence Microscopy: A Biological Perspective
 Fluorescence Microscopy: A Biological Perspective From nanometre to metre: the scale of life Instrumentation and accessible scale limits the questions that can be addressed in biology Why are there limits?
Fluorescence Microscopy: A Biological Perspective From nanometre to metre: the scale of life Instrumentation and accessible scale limits the questions that can be addressed in biology Why are there limits?
Localization Microscopy
 Localization Microscopy Theory, Sample Prep & Practical Considerations Patrina Pellett & Ann McEvoy Applications Scientist GE Healthcare, Cell Technologies May 27 th, 2015 Localization Microscopy Talk
Localization Microscopy Theory, Sample Prep & Practical Considerations Patrina Pellett & Ann McEvoy Applications Scientist GE Healthcare, Cell Technologies May 27 th, 2015 Localization Microscopy Talk
Fluorescence Nanoscopy
 Fluorescence Nanoscopy Keith A. Lidke University of New Mexico panda3.phys.unm.edu/~klidke/index.html Optical Microscopy http://en.wikipedia.org/wiki/k%c3%b6hler_illumination 30 µm Fluorescent Probes Michalet
Fluorescence Nanoscopy Keith A. Lidke University of New Mexico panda3.phys.unm.edu/~klidke/index.html Optical Microscopy http://en.wikipedia.org/wiki/k%c3%b6hler_illumination 30 µm Fluorescent Probes Michalet
Supplementary Table 1. Components of an FCS setup (1PE and 2PE)
 Supplementary Table 1. Components of an FCS setup (1PE and 2PE) Component and function Laser source Excitation of fluorophores Microscope with xy-translation stage mounted on vibration isolated optical
Supplementary Table 1. Components of an FCS setup (1PE and 2PE) Component and function Laser source Excitation of fluorophores Microscope with xy-translation stage mounted on vibration isolated optical
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/496/eaam6291/dc1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells
www.sciencesignaling.org/cgi/content/full/10/496/eaam6291/dc1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells
Supplementary Data. Supplementary Materials and Methods Quantification of delivered sirnas. Fluorescence microscopy analysis
 Supplementary Data Supplementary Materials and Methods Quantification of delivered sirnas After transfection, cells were washed in three times with phosphate buffered saline and total RNA was extracted
Supplementary Data Supplementary Materials and Methods Quantification of delivered sirnas After transfection, cells were washed in three times with phosphate buffered saline and total RNA was extracted
-RT RT RT -RT XBMPRII
 Base pairs 2000 1650 1000 850 600 500 400 Marker Primer set 1 Primer set 2 -RT RT RT -RT XBMPRII kda 100 75 50 37 25 20 XAC p-xac A 300 B Supplemental figure 1. PCR analysis of BMPRII expression and western
Base pairs 2000 1650 1000 850 600 500 400 Marker Primer set 1 Primer set 2 -RT RT RT -RT XBMPRII kda 100 75 50 37 25 20 XAC p-xac A 300 B Supplemental figure 1. PCR analysis of BMPRII expression and western
Nature Methods: doi: /nmeth Supplementary Figure 1. Retention of RNA with LabelX.
 Supplementary Figure 1 Retention of RNA with LabelX. (a) Epi-fluorescence image of single molecule FISH (smfish) against GAPDH on HeLa cells expanded without LabelX treatment. (b) Epi-fluorescence image
Supplementary Figure 1 Retention of RNA with LabelX. (a) Epi-fluorescence image of single molecule FISH (smfish) against GAPDH on HeLa cells expanded without LabelX treatment. (b) Epi-fluorescence image
Supplementary Material. The MIAPE-GE glossary (MIAPE-GE version 1.4).
 Supplementary Material. The MIAPE-GE glossary (MIAPE-GE version 1.4). Classification Definition 1. General features 1.1.1 Date stamp The date on which the work described was initiated; given in the standard
Supplementary Material. The MIAPE-GE glossary (MIAPE-GE version 1.4). Classification Definition 1. General features 1.1.1 Date stamp The date on which the work described was initiated; given in the standard
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO monoclonal antibody. (a) LC-MS analyses of tryptic
 Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
Aoki et al.,
 JCB: SUPPLEMENTAL MATERIAL Aoki et al., http://www.jcb.org/cgi/content/full/jcb.200609017/dc1 Supplemental materials and methods Calculation of the raw number of translocated proteins First, the average
JCB: SUPPLEMENTAL MATERIAL Aoki et al., http://www.jcb.org/cgi/content/full/jcb.200609017/dc1 Supplemental materials and methods Calculation of the raw number of translocated proteins First, the average
Synonymous Modification Results in High Fidelity Gene Expression of Repetitive Protein and Nucleotide Sequences
 Synonymous Modification Results in High Fidelity Gene Expression of Repetitive Protein and Nucleotide Sequences Bin Wu *,1,2, Veronika Miskolci *,1, Hanae Sato 1, Evelina Tutucci 1, Charles A. Kenworthy
Synonymous Modification Results in High Fidelity Gene Expression of Repetitive Protein and Nucleotide Sequences Bin Wu *,1,2, Veronika Miskolci *,1, Hanae Sato 1, Evelina Tutucci 1, Charles A. Kenworthy
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
SUPPLEMENTARY INFORMATION
 Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Supplementary information for:
 Supplementary information for: Generation of brilliant green fluorescent petunia plants by using a new and potent fluorescent protein transgene Dong Poh Chin 1,, Ikuo Shiratori 2,, *, Akihisa Shimizu 2,
Supplementary information for: Generation of brilliant green fluorescent petunia plants by using a new and potent fluorescent protein transgene Dong Poh Chin 1,, Ikuo Shiratori 2,, *, Akihisa Shimizu 2,
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide,
 Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
< Supporting Information >
 SUPPORTING INFORMATION 1 < Supporting Information > Discovery of autophagy modulators through the construction of high-content screening platform via monitoring of lipid droplets Sanghee Lee, Eunha Kim,
SUPPORTING INFORMATION 1 < Supporting Information > Discovery of autophagy modulators through the construction of high-content screening platform via monitoring of lipid droplets Sanghee Lee, Eunha Kim,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION a b Supplementary Figure 1. Cellular viability in reformulated culture media cells were cultured in serum containing DMEM culture medium supplemented with 4%PEG and 5%DEX for
SUPPLEMENTARY INFORMATION a b Supplementary Figure 1. Cellular viability in reformulated culture media cells were cultured in serum containing DMEM culture medium supplemented with 4%PEG and 5%DEX for
Brightfield and Fluorescence Imaging using 3D PrimeSurface Ultra-Low Attachment Microplates
 A p p l i c a t i o n N o t e Brightfield and Fluorescence Imaging using 3D PrimeSurface Ultra-Low Attachment Microplates Brad Larson, BioTek Instruments, Inc., Winooski, VT USA Anju Dang, S-BIO, Hudson,
A p p l i c a t i o n N o t e Brightfield and Fluorescence Imaging using 3D PrimeSurface Ultra-Low Attachment Microplates Brad Larson, BioTek Instruments, Inc., Winooski, VT USA Anju Dang, S-BIO, Hudson,
