Supplementary Materials for
|
|
|
- Frederick McDonald
- 5 years ago
- Views:
Transcription
1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells Anita K. Mehta, Kevin Hua, William Whipple, Minh-Thuy Nguyen, Ching-Ti Liu, Johannes Haybaeck, Joanne Weidhaas, Jeff Settleman, Anurag Singh* The PDF file includes: *Corresponding author. Published 12 September 2017, Sci. Signal. 10, eaam6291 (2017) DOI: /scisignal.aam6291 Fig. S1. Regulation of mesenchymal-epithelial transition by KE-associated mirnas. Fig. S2. Effects of primary MIR-124 lentiviral expression on cellular growth and viability. Fig. S3. Live-cell imaging of autophagic flux in H460 KM cells. Fig. S4. BECN1 expression is suppressed by mir-124. Fig. S5. Repression of p62 and downstream signaling by mir-124. Fig. S6. Control of inflammatory cytokine expression by mir-124. Legends for tables S1 and S2 Table S3. Primer and oligo sequences. Legends for movies S1 to S4 Other Supplementary Material for this manuscript includes the following: (available at Table S1 (Microsoft Excel format). TLDA raw data. Table S2 (Microsoft Excel format). TargetScan mir-124 predicted targets. Movie S1 (.avi format). Live-cell imaging of mcherry-gfp-lc3 in control H460 cells. Movie S2 (.avi format). Live-cell imaging of mcherry-gfp-lc3 in mir-124 transfected H460 cells. Movie S3 (.avi format). Live-cell imaging of mcherry-gfp-lc3 in chloroquinetreated H460 cells.
2 Movie S4 (.avi format). Live-cell imaging of mcherry-gfp-lc3 in mir chloroquine treated H460 cells.
3 Figure S1. Regulation of mesenchymal-epithelial transition by KE-associated mirnas. (A) Abundance of indicated mirnas following sirna-mediated KRAS depletion in representative KE H2009 and H358 cells. Western blot, inset indicates protein KRAS abundance following sikras transfection. Data are the mean of 3 replicates + SEM and are representative of 2 independent experiments. (B) Confocal immunofluorescence micrographs of H460 cells transfected with mir-205 and mir-200c. Cells are shown co-stained with E-cadherin (green channel) and vimentin (red channel). Scale bar, 25µM. (C) Western blot of A549 KM cells after mir-200c and mir-205 reconstitution, using 50nM or 10nM of the synthetic mimic oligonucleotides. (D) Western blot of Zeb1 and E-cadherin abundance in H460 cells after reconstitution with the indicated mirna. Data are representative of 3 independent experiments. (E) Pearson correlation analysis of primary MIR200C and MIR124 RNA transcript expression from a microarray dataset of primary NSCLCs.
4 Figure S2. Effects of primary MIR-124 lentiviral expression on cellular growth and viability. (A) 3D Matrigel assays with MCF7 cells expressing either Luciferase control vector or a vector encoding pri-mir124. Scale bar, 20µM. (B) Automated quantitation of data in (A) showing individual colony size and number. Data are in arbitrary pixel units. (C) Immunofluorescence micrographs showing cleaved caspase-3 amounts (red fluorescence) after mir-124 reconstitution in H460 cells. Nuclei were counterstained with DAPI (blue). Scale bar, 20µM. (D) Clonogenic growth assays of H460 cells expressing control or pri-mir124 expressing lentiviruses.
5 Figure S3. Live-cell imaging of autophagic flux in H460 KM cells. (A and B) Images of H460 cells 36 hours after stable transfection of the mcherry-gfp-lc3 reporter with mir-124 or control (NC) vector and treated with chloroquine. Zoomed-in images (B) demonstrate identification and quantitation of green/gfp puncta (phagophores/autophagosomes) and red/mcherry puncta (autolysosomes), using an integrated machine learning software tool (Biotek Gen5). Individual red and green puncta are outlined. GFP/mCherry double-positive puncta are excluded from being classified as autolysosomes using a thresholding option in the Gen5 software. Scale bars = 50µm (A), 10µM (B).
6 Figure S4. BECN1 expression is suppressed by mir-124. (A) Relative luminescence of luciferase-becn1-3 UTR constructs in A549 cells after control (NC), mir-200c or mir-124 transfections. (B) Cell proliferation in control or Beclin-1 overexpressing A549 cells after mir-124 reconstitution. (C) Western blot analysis of cleaved PARP and LC3-II amounts after mir-124 reconstitution in control (GFP) and Beclin-1 transfected A549 cells. (D) Densitometry-based quantitation of data in panel (C). (E) Correlation analysis of BECN1 and MIR124 expression in primary NSCLC samples from patients; r indicates Pearson correlation coefficient.
7 Figure S5. Repression of p62 and downstream signaling by mir-124. (A) H460 KM cells transfected with mir-124 or control vector (NC; 50nM). Endogenous p62 aggregates visualized in red (AlexaFluor 594) and nuclei with DAPI. Scale bar, 50µm. (B) Quantification of p62 aggregates per cell using software-based image analysis (CellProfiler). (C) Western blot of total and phosphorylated S6 (ps6) in mir-124 reconstituted KM cells. (D) Expression of SQSTM1 mrna in mir-124-transfected H460 cells relative to controls, as determined by Taqman qpcr assays. (E) Bright field micrographs of H358 KE cells and KM-derivative H358M cells with pri- MIR-124. Images are representative of 2 independent experiments. (F) Western blot of E-cadherin, vimentin and p62 in mir-124-reconstituted PDAC KM cells. GAPDH, loading control. (G) Western blotting of p62 and cleaved PARP abundance upon depletion of p62 by sirna (10nM) in H460 and H358 cells. (H) Abundance of p62 in H460 cells expressing control vector, GFP-tagged-p62 or mir-124 vectors in the indicated combinations.
8 Figure S6. Control of inflammatory cytokine expression by mir-124. (A) Inverse correlation between TRAF6 and MIR124 RNA expression in primary NSCLCs as determined by Pearson correlation coefficient (r). (B) MiR- 124 reconstitution promotes altered cytokine expression as determined by qpcr. Data are representative of two independent experiments.
9 Table S1. TLDA raw data. Probe information, raw fluorescence Rn and threshold cycle data for qpcr mirna profiling in 3 KE cell lines (A549, H460 and SW1573) and 3 KM cell lines (H2009, H358 and H441). Table is provided in the auxiliary online materials (.xls). Table S2. TargetScan mir-124 predicted targets. Predicted seed sequences for mir-124 in 3 UTR regions for indicated gene transcripts, as documented by the Targetscan web portal ( Table is provided in the auxiliary online materials (.xls). Table S3. Primer and oligo sequences. DNA sequences used for molecular cloning and qpcr experiments (FW = forward primer, REV = reverse primer). Primer Name PRI-MIR attb1 PRI-MIR attb2 p62 3'UTR WT_1 sense p62 3'UTR WT_1 antisense p62 3'UTR WT_2 sense p62 3'UTR WT_2 antisense p62 3'UTR mutant 1 sense p62 3'UTR mutant 1 antisense p62 3'UTR mutant 2 sense p62 3'UTR mutant 2 antisense BECN1 3 UTR WT attb1 BECN1 3 UTR WT attb2 IL6-FW IL6-REV IL7-FW IL7-REV TNFA-FW TNFA-REV IL1B-FW IL1B-REV IL11-FW IL11-REV Primer Sequence (5' to 3') TTTTTCTCGAGTGTTTTTAGGCGTGT (XhoI site) TTTTTGCGGCCGCGGTACCATTTCCGCGG (NotI site) GTCTCTGTACGGGCCAGTTT CCTGACAGCTTCCAGCAGAA AGAAGCCATTTAGGGCAGCA GGCCTGACATGGAAGGTGAA GGGACAGGGTTCCCCGAAACGCTCGTTTTAAAGATACTCTGTAAG CTTACAGAGTATCTTTAAAACGAGCGTTTCGGGGAACCCTGTCCC GGACAGGGTTCCCCGTTAGCCAGGTTTTAAAGATACTCTGTAAGAA TTCTTACAGAGTATCTTTAAAACCTGGCTAACGGGGAACCCTGTCC GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGATATCTTTCCTTAGGGGGAGGTTTG GGGGACCACTTTGTACAAGAAAGCTGGGTCCAGTTTTCAGACTGCAGCAAA AGAGTAGTGAGGAACAAGCCA TCAGGGGTGGTTATTGCATCT CGCAAGTTGAGGCAATTTCT TTCCTGGCCAGTGCAGTT AGCCCATGTTGTAGCAAACC GAGGTACAGGCCCTCTGATG TCTTCAGCCAATCTTCATTGCTC TGGAAGGAGCACTTCATCTG GCGGACAGGGAAGGGTTAAAG CAGGCGGCAAACACAGTTC
10 Movie S1. Live-cell imaging of mcherry-gfp-lc3 in control H460 cells. Time lapse movie showing H460 cells transduced to express mcherry-gfp-lc3 autophagy flux reporter under stable conditions, transfected with NC control oligonucleotide. Movie of compiled still images is shown at 15 frames per sec. with each frame representing 10 min in real time. Time is shown as 00h:00m:00s. Movie is provided in the auxiliary online materials. Movie S2. Live-cell imaging of mcherry-gfp-lc3 in mir-124 transfected H460 cells. As described for Movie S1, in cells transfected with mir-124. Movie is provided in the auxiliary online materials. Movie S3. Live-cell imaging of mcherry-gfp-lc3 in chloroquine-treated H460 cells. As described for Movie S1, in cells treated with chloroquine (50µM). Movie is provided in the auxiliary online materials. Movie S4. Live-cell imaging of mcherry-gfp-lc3 in mir chloroquine treated H460 cells. As described for Movie S1, in cells transfected with mir-124 and treated with chloroquine. Movie is provided in the auxiliary online materials.
supplementary information
 Figure S1 ZEB1 full length mrna. (a) Analysis of the ZEB1 mrna using the UCSC genome browser (http://genome.ucsc.edu) revealed truncation of the annotated Refseq sequence (NM_030751). The probable terminus
Figure S1 ZEB1 full length mrna. (a) Analysis of the ZEB1 mrna using the UCSC genome browser (http://genome.ucsc.edu) revealed truncation of the annotated Refseq sequence (NM_030751). The probable terminus
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary materials
 Supplementary materials NADPH oxidase promotes Parkinsonian phenotypes by impairing autophagic flux in an mtorc1- independent fashion in a cellular model of Parkinson s disease Rituraj Pal 1, Lakshya Bajaj
Supplementary materials NADPH oxidase promotes Parkinsonian phenotypes by impairing autophagic flux in an mtorc1- independent fashion in a cellular model of Parkinson s disease Rituraj Pal 1, Lakshya Bajaj
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in
 Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or
Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Supplementary Figure 1. ips_3y+mir-302b. ips_3y+mir-372. ips_3y+mir-302b + mir-372. ips_4y. ips_4y+mir-302b. ips_4y + mir-372
 Supplementary Figure 1 ips_3y+mir-32b ips_3y+ ips_3y+mir-32b + ips_4y ips_4y+mir-32b ips_4y + Nature Biotechnology: doi:1.138/nb.t.1862 TRA-1-6 DAPI Oct3/4 Supplementary Figure 1: ips cells derived from
Supplementary Figure 1 ips_3y+mir-32b ips_3y+ ips_3y+mir-32b + ips_4y ips_4y+mir-32b ips_4y + Nature Biotechnology: doi:1.138/nb.t.1862 TRA-1-6 DAPI Oct3/4 Supplementary Figure 1: ips cells derived from
a KYSE270-CON KYSE270-Id1
 a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
SUPPLEMENTARY FIGURES AND TABLES
 SUPPLEMENTARY FIGURES AND TABLES Suppl. Table 1 Oligonucleotides used in the study The table shows the structure and nucleotide position of the oligonucleotides used in the study along with their application
SUPPLEMENTARY FIGURES AND TABLES Suppl. Table 1 Oligonucleotides used in the study The table shows the structure and nucleotide position of the oligonucleotides used in the study along with their application
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/483/eaak9557/dc1 Supplementary Materials for The lncrna H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging
www.sciencesignaling.org/cgi/content/full/10/483/eaak9557/dc1 Supplementary Materials for The lncrna H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction
 Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern
 Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern blot. Northern blot analysis of mir-302b expression following infection with PAO1, PAK and Kp in (A) lung
Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern blot. Northern blot analysis of mir-302b expression following infection with PAO1, PAK and Kp in (A) lung
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Kimura et al.,
 Supplemental material JCB Kimura et al., http://www.jcb.org/cgi/content/full/jcb.201503023/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. TRIMs regulate IFN-γ induced autophagy. (A and B) HC image analysis
Supplemental material JCB Kimura et al., http://www.jcb.org/cgi/content/full/jcb.201503023/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. TRIMs regulate IFN-γ induced autophagy. (A and B) HC image analysis
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figure 1: List of primers/oligonucleotides used in this study. 1 Supplementary figure 2: Sequences and mirna-targets of i) mcherry expresses in transgenic fish used
SUPPLEMENTARY INFORMATION Supplementary figure 1: List of primers/oligonucleotides used in this study. 1 Supplementary figure 2: Sequences and mirna-targets of i) mcherry expresses in transgenic fish used
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/429/ra54/dc1 Supplementary Materials for Dephosphorylation of the adaptor LAT and phospholipase C by SHP-1 inhibits natural killer cell cytotoxicity Omri Matalon,
www.sciencesignaling.org/cgi/content/full/9/429/ra54/dc1 Supplementary Materials for Dephosphorylation of the adaptor LAT and phospholipase C by SHP-1 inhibits natural killer cell cytotoxicity Omri Matalon,
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr
 Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p53 activity
 IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p5 activity H6 5 5 H58 A59 H6 H58 A59 anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p5 activity H6 5 5 H58 A59 H6 H58 A59 anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification.
 Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3164 Supplementary Figure 1 Validation of effective Gnas deletion and epithelial thickness. a, Representative genotyping in mice treated or not with tamoxifen to show Gnas deletion. To
DOI: 10.1038/ncb3164 Supplementary Figure 1 Validation of effective Gnas deletion and epithelial thickness. a, Representative genotyping in mice treated or not with tamoxifen to show Gnas deletion. To
Supplementary Information. A superfolding Spinach2 reveals the dynamic nature of. trinucleotide repeat RNA
 Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Mariana Pehar, Mary Cabell Jonas, Theresa M. Hare and Luigi Puglielli. Primer pair Sequence Annealing temperature
 SLC33A1/AT-1 REGULATES THE INDUCTION OF AUTOPHAGY DOWN-STREAM OF IRE1/XBP1 Mariana Pehar, Mary Cabell Jonas, Theresa M. Hare and Luigi Puglielli SUPPLEMENTAL DATA Supplemental Table S1. Primers used in
SLC33A1/AT-1 REGULATES THE INDUCTION OF AUTOPHAGY DOWN-STREAM OF IRE1/XBP1 Mariana Pehar, Mary Cabell Jonas, Theresa M. Hare and Luigi Puglielli SUPPLEMENTAL DATA Supplemental Table S1. Primers used in
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction)
 Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting
 MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4 Hui Li 1, #, Ruoyun Ouyang 2, #, Zi Wang 1, #, Weihua Zhou 1, Huiyong Chen 1, Yawen Jiang 1, Yibin Zhang 1, Hui Li
MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4 Hui Li 1, #, Ruoyun Ouyang 2, #, Zi Wang 1, #, Weihua Zhou 1, Huiyong Chen 1, Yawen Jiang 1, Yibin Zhang 1, Hui Li
Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera
 SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3562 In the format provided by the authors and unedited. Supplementary Figure 1 Glucose deficiency induced FH-ATF2 interaction. In b-m, immunoblotting or immunoprecipitation analyses were
DOI: 10.1038/ncb3562 In the format provided by the authors and unedited. Supplementary Figure 1 Glucose deficiency induced FH-ATF2 interaction. In b-m, immunoblotting or immunoprecipitation analyses were
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of
 Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
SUPPLEMENTARY INFORMATION
 Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Appendix Figures Page 2 Appendix Figure Legends Page 9 Appendix Tables Page 13 Appendix Supplementary Methods Page 15 Appendix References Page 17
 APPENDIX Appendix Figures Page 2 Appendix Figure Legends Page 9 Appendix Tables Page 13 Appendix Supplementary Methods Page 15 Appendix References Page 17 Appendix Figure Legends Appendix Figure
APPENDIX Appendix Figures Page 2 Appendix Figure Legends Page 9 Appendix Tables Page 13 Appendix Supplementary Methods Page 15 Appendix References Page 17 Appendix Figure Legends Appendix Figure
SUPPLEMENTARY INFORMATION. Chemical modulation of Chaperone-mediated autophagy by novel
 SUPPLEMENTARY INFORMATION Chemical modulation of Chaperone-mediated autophagy by novel retinoic acid derivatives Jaime Anguiano 1, Thomas P Garner 2, Murugesan Mahalingam 1, Bhaskar C. Das 1, Evripidis
SUPPLEMENTARY INFORMATION Chemical modulation of Chaperone-mediated autophagy by novel retinoic acid derivatives Jaime Anguiano 1, Thomas P Garner 2, Murugesan Mahalingam 1, Bhaskar C. Das 1, Evripidis
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/4/198/ra74/dc1 Supplementary Materials for Short RNA Duplexes Elicit RIG-I Mediated Apoptosis in a Cell Type and Length-Dependent Manner Osamu Ishibashi, Md. Moksed
www.sciencesignaling.org/cgi/content/full/4/198/ra74/dc1 Supplementary Materials for Short RNA Duplexes Elicit RIG-I Mediated Apoptosis in a Cell Type and Length-Dependent Manner Osamu Ishibashi, Md. Moksed
0.9 5 H M L E R -C tr l in T w is t1 C M
 a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
Supplementary information for: Mutant p53 gain-of-function induces epithelial-mesenchymal transition. through modulation of the mir-130b-zeb1 axis
 Supplementary information for: Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the mir-3b-zeb axis AUTHORS: Peixin Dong, Mihriban Karaayvaz, Nan Jia, Masanori
Supplementary information for: Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the mir-3b-zeb axis AUTHORS: Peixin Dong, Mihriban Karaayvaz, Nan Jia, Masanori
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
Supplementary Materials for
 www.advances.sciencemag.org/cgi/content/full/1/7/e1500454/dc1 Supplementary Materials for CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation Xin Han, Zongbin Liu, Myeong chan Jo,
www.advances.sciencemag.org/cgi/content/full/1/7/e1500454/dc1 Supplementary Materials for CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation Xin Han, Zongbin Liu, Myeong chan Jo,
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum. Blood-stage Development
 Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
SUPPORTING INFORMATION:
 SUPPORTING INFORMATION: Targeted m 6 A reader proteins to study epitranscriptomic regulation of single RNAs Simone Rauch, Chuan He,,, and Bryan C. Dickinson *, Department of Biochemistry and Molecular
SUPPORTING INFORMATION: Targeted m 6 A reader proteins to study epitranscriptomic regulation of single RNAs Simone Rauch, Chuan He,,, and Bryan C. Dickinson *, Department of Biochemistry and Molecular
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Supplementary Figure legends. Supplementary Methods
 Supplementary Methods Transmission electron microscopy. For transmission electron microscopy (TEM), the cell populations were rinsed with 0.1 Sorensen s buffer (ph 7.5), fixed in 2.5% glutaraldehyde for
Supplementary Methods Transmission electron microscopy. For transmission electron microscopy (TEM), the cell populations were rinsed with 0.1 Sorensen s buffer (ph 7.5), fixed in 2.5% glutaraldehyde for
USP19 modulates autophagy and antiviral immune responses by. deubiquitinating Beclin-1
 USP19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1 Shouheng Jin 1,2,, Shuo Tian 1,, Yamei Chen 1,, Chuanxia Zhang 1,2, Weihong Xie, 1 Xiaojun Xia 3,4, Jun Cui 1,3* &
USP19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1 Shouheng Jin 1,2,, Shuo Tian 1,, Yamei Chen 1,, Chuanxia Zhang 1,2, Weihong Xie, 1 Xiaojun Xia 3,4, Jun Cui 1,3* &
Supplementary Figure 1, Wiel et al
 Supplementary Figure 1, Wiel et al Supplementary Figure 1 ITPR2 increases in benign tumors and decreases in aggressive ones (a-b) According to the Oncomine database, expression of ITPR2 increases in renal
Supplementary Figure 1, Wiel et al Supplementary Figure 1 ITPR2 increases in benign tumors and decreases in aggressive ones (a-b) According to the Oncomine database, expression of ITPR2 increases in renal
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
Total RNA was isolated using Trizol reagent (Invitrogen) and reverse transcribed using
 Supplementary Methods RNA Isolation and Quantitative RT-PCR Total RNA was isolated using Trizol reagent (Invitrogen) and reverse transcribed using random hexamers and superscript II reverse transcriptase
Supplementary Methods RNA Isolation and Quantitative RT-PCR Total RNA was isolated using Trizol reagent (Invitrogen) and reverse transcribed using random hexamers and superscript II reverse transcriptase
Supplementary Data. Supplementary Materials and Methods Quantification of delivered sirnas. Fluorescence microscopy analysis
 Supplementary Data Supplementary Materials and Methods Quantification of delivered sirnas After transfection, cells were washed in three times with phosphate buffered saline and total RNA was extracted
Supplementary Data Supplementary Materials and Methods Quantification of delivered sirnas After transfection, cells were washed in three times with phosphate buffered saline and total RNA was extracted
Functional analysis reveals that RBM10 mutations. contribute to lung adenocarcinoma pathogenesis by. deregulating splicing
 Supplementary Information Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing Jiawei Zhao 1,+, Yue Sun 2,3,+, Yin Huang 6, Fan Song
Supplementary Information Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing Jiawei Zhao 1,+, Yue Sun 2,3,+, Yin Huang 6, Fan Song
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
HCT116 SW48 Nutlin: p53
 Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Supplementary Figure 1. Generation of B2M -/- ESCs. Nature Biotechnology: doi: /nbt.3860
 Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
University of Dundee. Published in: Scientific Reports. DOI: /srep Publication date: 2016
 University of Dundee SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Wall, Richard J.; Roques, Magali; Katris,
University of Dundee SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Wall, Richard J.; Roques, Magali; Katris,
Supplementary information. Supplementary Figures
 Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
Galina Gabriely, Ph.D. BWH/HMS
 Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
TOOLS sirna and mirna. User guide
 TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
Li et al., Supplemental Figures
 Li et al., Supplemental Figures Fig. S1. Suppressing TGM2 expression with TGM2 sirnas inhibits migration and invasion in A549-TR cells. A, A549-TR cells transfected with negative control sirna (NC sirna)
Li et al., Supplemental Figures Fig. S1. Suppressing TGM2 expression with TGM2 sirnas inhibits migration and invasion in A549-TR cells. A, A549-TR cells transfected with negative control sirna (NC sirna)
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators.
 Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative
 Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative RT-PCR For CD8-1 transcript, forward primer CD8-1F 5 -TAGTAACCAGAGGCCGCAAGA-3 reverse primer CD8-1R 5 -TCTACTAAGGTGTCCCATAGCATGAT-3
Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative RT-PCR For CD8-1 transcript, forward primer CD8-1F 5 -TAGTAACCAGAGGCCGCAAGA-3 reverse primer CD8-1R 5 -TCTACTAAGGTGTCCCATAGCATGAT-3
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2743 Figure S1 stabilizes cellular protein level, post-transcriptionally. (a, b) and DDR1 were RNAi-depleted from HEK.293.-CBG cells. Western blots with indicated antibodies (a). RT-PCRs
DOI: 10.1038/ncb2743 Figure S1 stabilizes cellular protein level, post-transcriptionally. (a, b) and DDR1 were RNAi-depleted from HEK.293.-CBG cells. Western blots with indicated antibodies (a). RT-PCRs
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplemental Table/Figure Legends
 MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
SUPPLEMENTARY EXPEMENTAL PROCEDURES
 SUPPLEMENTARY EXPEMENTAL PROCEDURES Plasmids- Total RNAs were extracted from HeLaS3 cells and reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) to obtain DNA template for the
SUPPLEMENTARY EXPEMENTAL PROCEDURES Plasmids- Total RNAs were extracted from HeLaS3 cells and reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) to obtain DNA template for the
Figure S1. GST-MDA-7 toxicity in GBM cells is dependent on cathepsin proteases, and weakly
 Supplemental Figure Legends. Figure S1. GST-MDA-7 toxicity in GBM cells is dependent on cathepsin proteases, and weakly dependent on caspase 8. GBM6 and GBM12 cells were treated 24 h after plating with
Supplemental Figure Legends. Figure S1. GST-MDA-7 toxicity in GBM cells is dependent on cathepsin proteases, and weakly dependent on caspase 8. GBM6 and GBM12 cells were treated 24 h after plating with
A) B) Ladder. Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical
 A) B) Ladder C) r4 r4 Nt- -Ct 78 kda Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical calpains is shown. The protease core consists of
A) B) Ladder C) r4 r4 Nt- -Ct 78 kda Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical calpains is shown. The protease core consists of
Supporting Information
 Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence
 Supplementary Information Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Yoshinori Kousoku, Aya Ohkuni,
Supplementary Information Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Yoshinori Kousoku, Aya Ohkuni,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
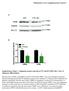 Merkestein et al. Supplementary Figure 1 A PLIN WT FTO KO 72 kda FABP4 17 kda HSC70 72 kda B Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
Merkestein et al. Supplementary Figure 1 A PLIN WT FTO KO 72 kda FABP4 17 kda HSC70 72 kda B Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
b alternative classical none
 Supplementary Figure. 1: Related to Figure.1 a d e b alternative classical none NIK P-IkBa Total IkBa Tubulin P52 (Lighter) P52 (Darker) RelB (Lighter) RelB (Darker) HDAC1 Control-Sh RelB-Sh NF-kB2-Sh
Supplementary Figure. 1: Related to Figure.1 a d e b alternative classical none NIK P-IkBa Total IkBa Tubulin P52 (Lighter) P52 (Darker) RelB (Lighter) RelB (Darker) HDAC1 Control-Sh RelB-Sh NF-kB2-Sh
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/258/rs3/dc1 Supplementary Materials for A Genome-Wide sirna Screen Reveals Positive and Negative Regulators of the NOD2 and NF-κB Signaling Pathways Neil Warner,
www.sciencesignaling.org/cgi/content/full/6/258/rs3/dc1 Supplementary Materials for A Genome-Wide sirna Screen Reveals Positive and Negative Regulators of the NOD2 and NF-κB Signaling Pathways Neil Warner,
Nature Methods: doi: /nmeth Supplementary Figure 1. Retention of RNA with LabelX.
 Supplementary Figure 1 Retention of RNA with LabelX. (a) Epi-fluorescence image of single molecule FISH (smfish) against GAPDH on HeLa cells expanded without LabelX treatment. (b) Epi-fluorescence image
Supplementary Figure 1 Retention of RNA with LabelX. (a) Epi-fluorescence image of single molecule FISH (smfish) against GAPDH on HeLa cells expanded without LabelX treatment. (b) Epi-fluorescence image
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
TRIM31 is recruited to mitochondria after infection with SeV.
 Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Long Noncoding RNA LOC Suppresses Apoptosis by. Targeting mir p and mir-4767 in Vascular Endothelial Cells
 Long Noncoding RNA LOC100129973 Suppresses Apoptosis by Targeting mir-4707-5p and mir-4767 in Vascular Endothelial Cells Wei Lu 1, ShuYa Huang 1, Le Su 1, BaoXiang Zhao 2, *, JunYing Miao 1, 3, * 1 Shandong
Long Noncoding RNA LOC100129973 Suppresses Apoptosis by Targeting mir-4707-5p and mir-4767 in Vascular Endothelial Cells Wei Lu 1, ShuYa Huang 1, Le Su 1, BaoXiang Zhao 2, *, JunYing Miao 1, 3, * 1 Shandong
MeCP2. MeCP2/α-tubulin. GFP mir1-1 mir132
 Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector
 SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Richard J. Wall 1,#, Magali Roques 1,+, Nicholas J. Katris
SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Richard J. Wall 1,#, Magali Roques 1,+, Nicholas J. Katris
Analyze microrna Activity in Cells Using Luciferase Reporters. Trista Schagat, Ph.D. December 2013
 Analyze microrna Activity in Cells Using Luciferase Reporters Trista Schagat, Ph.D. December 2013 agenda microrna Overview 3 5 3 5 3 5 How to and Tips Design Transfect Assay Analyze Case Studies 2 micrornas
Analyze microrna Activity in Cells Using Luciferase Reporters Trista Schagat, Ph.D. December 2013 agenda microrna Overview 3 5 3 5 3 5 How to and Tips Design Transfect Assay Analyze Case Studies 2 micrornas
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1154040/dc1 Supporting Online Material for Selective Blockade of MicroRNA Processing by Lin-28 Srinivas R. Viswanathan, George Q. Daley,* Richard I. Gregory* *To whom
www.sciencemag.org/cgi/content/full/1154040/dc1 Supporting Online Material for Selective Blockade of MicroRNA Processing by Lin-28 Srinivas R. Viswanathan, George Q. Daley,* Richard I. Gregory* *To whom
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
Document S1. Supplemental Experimental Procedures and Three Figures (see next page)
 Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
LysoTracker Red DND-99 (Invitrogen) was used as a marker of lysosome or acidic
 information MATERIAL AND METHODS Lysosome staining LysoTracker Red DND-99 (Invitrogen) was used as a marker of lysosome or acidic compartments, according to the manufacturer s protocol. Plasmid independent
information MATERIAL AND METHODS Lysosome staining LysoTracker Red DND-99 (Invitrogen) was used as a marker of lysosome or acidic compartments, according to the manufacturer s protocol. Plasmid independent
