Supplemental Data. Borg et al. Plant Cell (2014) /tpc
|
|
|
- Duane McDaniel
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown below. Conserved amino acids are bold in dark shaded boxes and similar amino acids are in light shaded boxes. Black lines (below) show conserved regions and the single grey line indicates the unaligned second EAR motif of rice (OsEAR2). Gene names, a, b, denote paralogs, Zf1, Zf2, Zf3, zinc fingers 1-3, NLS, putative nuclear localization signal. Sb, Sorghum bicolor, Bd, Brachypodium distachyon, Os, Oryza sativa, Pt, Poplar trichocarpa, Br, Brassica rapa, Al, Arabidopsis lyrata, At, Arabidopsis thaliana.
2 Supplemental Figure 2 Analysis of DAZ1 and DAZ2 promoter activity in developing pollen Expression of DAZ1 and DAZ2 is restricted to the male germline. Images show typical examples of developing pollen from transgenic lines expressing ProDAZ1:H2B-GFP (A- E) and ProDAZ2:H2B-GFP (F-J). GFP expression is first detectable at early bicellular stage (A,F), increasing during mid-bicellular (B,G) and late bicellular stage (C,H). GFP fluorescence persists into tricellular stage (D,I) and mature pollen (E,J). Images from each marker line were captured under standard conditions. Scale bars = 10µm.
3 Supplemental Figure 3 DAZ1 and DAZ2 protein localisation in sperm cells Stable transformants were generated expressing DAZ1 and DAZ2 as mcherry fusions (ProDAZ1:DAZ1-mCherry and ProDAZ2:DAZ2-mCherry). Mature pollen was stained with DAPI and observed using fluorescence microscopy. Representative examples of the localisation of DAZ1 and DAZ2 protein in sperm cell nuclei are presented as pairs of images, DAPI fluorescence (A,C,E) and mcherry fluorescence (B,D,F). DAZ1 (B) and DAZ2 (D) co-localise with DAPI-stained sperm cell nuclei (A and C respectively). DAZ1 also shows nuclear and cytoplasmic localisation (E-F) in 63.1% of sperm cells, based on analysis of three independent single locus lines (G). Scale bars = 15µm.
4 Supplementary Figure 4 - RT-PCR expression analysis of DAZ1 and DAZ2 PCR amplifications were carried out on cdna reactions performed with total RNA samples from the tissues indicated and with genomic DNA (gdna), Reactions performed with (+) and without (-) reverse transcriptase to control for genomic DNA contamination. The primers used were specific for DAZ1 (upper panel), DAZ2 (middle panel) and HTR5 transcripts (lower panel).
5 Supplemental Figure 5 Characterisation of T-DNA insertion lines in DAZ1 and DAZ2. (A) Schematic diagram of the DAZ1 and DAZ2 loci indicating T-DNA insertion sites and primers used for RT-PCR analysis [qrt-f/qrt-r] (B) DAZ1 and DAZ2 transcripts in pollen from WT plants and homozygous T-DNA insertion mutants. Transcripts were undetectable in the daz1-2 and daz2-1 alleles, whereas residual expression of a likely aberrant transcript was observed in daz1-1. Amplifications were performed on cdna and genomic DNA (gdna) using primers specific for DAZ1 (upper panel), DAZ2 (middle panel) and HTR5 (lower panel). Contamination from gdna was not observed in reverse transcriptase minus cdna samples (not shown).
6 Supplemental Table 1. Complementation of duo1-1 pollen with ProDUO1:DAZ1-mCherry No rescue (expect 1:1) Full rescue (expect 1:1) Line Total Tricellular % Bicellular % X 2 Significance Total %GFP + %GFP - X 2 Significance AA % 25.4% *** % 52.2% ns AB % 24.8% *** % 40.3% ** AC % 25.0% *** % 42.7% * BA % 26.8% *** % 46.6% ns BB % 31.0% *** % 47.5% ns BC % 42.1% * % 52.3% ns Chi-square analysis was used to test significant deviation from the ratios indicated, * p < 0.05, ** p < 0.01, *** p < 0.001, ns = not significant Supplemental Table 2. Complementation of daz1;daz2 pollen with ProDUO1:CYCB1;1 Chi-square analysis Genotype Line Total Tricellular (TC) Bicellular (BC) Ratio (TC : BC) X 2 Significance A : ns A : ns daz1-1 +/- ; daz2-1 -/- D : ns E : ns E : ns A : ns A : ns A : ns daz1-1 -/- ; daz2-1 +/- B : ns D : ns D : ns D : ns Chi-square analysis was used to test significant deviation from the ratio given no rescue (i.e. 1:1), ns = not significant
7 Supplemental Table 3. Segregation of F2 genotypes from daz1-1 +/- ;daz2-1 +/- F1 plants Normal transmission daz1;daz2 blocked transmission Genotypes Observed numbers Expected proportion X 2 Significance Expected proportion X 2 Significance DAZ1/DAZ1 ; DAZ2/DAZ *** ns DAZ1/daz1-1 ; DAZ2/DAZ DAZ1/DAZ1 ; DAZ2/daz DAZ1/daz1-1 ; DAZ2/daz DAZ1/DAZ1 ; daz2-1/daz daz1-1/daz1-1 ; DAZ2/DAZ daz1-1/daz1-1 ; DAZ2/daz DAZ1/daz1-1 ; daz2-1/daz daz1-1/daz1-1 ; daz2-1/daz Chi-square analysis was used to test significant deviation from the ratios indicated, *** p < ns = not significant Supplemental Table 4. Genetic transmission analysis of daz1;daz2 pollen Line Silique Total WT daz1;daz2 TE (%) A A Plants of F1 progeny from ms1 x daz1-1 +/- ;daz2-1 +/- cross were screened to identify a ~25% bicellular pollen phenotype
8 Supplemental Table 5 - Primers used in this study. The att sites and restriction sites are indicated in bold and mutagenised regions are underlined T-DNA genotyping primers DAZ1-TDNA-LP DAZ1-TDNA-RP DAZ2-TDNA-LP DAZ2-TDNA-RP DAZ1 and DAZ2 expression clones ProDAZ1-attB4-F ProDAZ1-attB1-R DAZ1-attB1-F DAZ1-attB2-R DAZ1-attB2-nsR ProDAZ2-attB4-F ProDAZ2-attB1-R DAZ2-attB1-F DAZ2-attB2-R DAZ2-attB2-nsR DAZ1 mear varants DAZ1Trunc1-attB2-nsR DAZ1-Trunc1-attB2-stopR mear1-fm mear1-rm mear2 Rs mear2 Rns TGATTTCGAAATGTGGAATGG CAACAACTTCCACCCTGAATC CAGATGCTTATGGCATTTTCTG CTCATGTGACCAAAGAGAGCC TGTATAGAAAAGTTGAAGTGGCACAAACCAACCC TTTTGTACAAACTTGTATTATTGAGTCTCTTACTAGAG ACAAAAAAGCAGGCTCTATGTCGAACACTTCAAACTCCG ACAAGAAAGCTGGGTCTCAAAGTCCTAACCTAAGATCC ACAAGAAAGCTGGGTCAAGTCCTAACCTAAGATCC TGTATAGAAAAGTTGATACTTGTATTTGGATTCC TTTTGTACAAACTTGTGTGAGCACGTAGAGGAAGTG ACAAAAAAGCAGGCTCTATGAACAACAATCATTCCTATG ACAAGAAAGCTGGGTCTTAGAGTCCTAACCTAAGATC ACAAGAAAGCTGGGTCGAGTCCTAACCTAAGATC ACAAGAAAGCTGGGTGTTCCTGATCTTTCTCCCAATGAC ACAAGAAAGCTGGGTGTCATTCCTGATCTTTCTCCCAATGAC GCATTGCTGCTGCTGTTGCTGCGGC GCCGCAGCAACAGCAGCAGCAATGC ACAAGAAAGCTGGGTCTCAAGCTCCTGCCCTAGCATCCGCGG ACAAGAAAGCTGGGTCAGCTCCTGCCCTAGCATCCGCGG MYB site mutagenesis for luciferase assays ProDAZ1-mMBS-A-F CAAAACAGGGAGAAGGAGCTCCTAAGAAACCGTCG ProDAZ1-mMBS-A-R CGACGGTTTCTTAGGAGCTCCTTCTCCCTGTTTTG ProDAZ1-mMBS-B-F GGAGAAGTAACTGCTAAGAGAGCTCCGATGAAAACTAACTTGTTC ProDAZ1-mMBS-B-R GAACAAGTTAGTTTTCATCGGAGCTCTCTTAGCAGTTACTTCTCC ProDAZ2-mMBS-A-F GTAAACCGCTCTGTTCCTGAGCTCAAGTTAACTGTCTCCCTG ProDAZ2-mMBS-A-R CAGGGAGACAGTTAACTTGAGCTCAGGAACAGAGCGGTTTAC ProDAZ2-mMBS-B-F GTTCCTCAACTGAAGTGAGCTCTCTCCCTGTTCCTCC ProDAZ2-mMBS-B-R GGAGGAACAGGGAGAGAGCTCACTTCAGTTGAGGAAC ProDAZ2-mMBS-C-F GTCTCTTCCTTACATAGTAGAGCTCTCTGTTCCTCAACTGAAG ProDAZ2-mMBS-C-R CTTCAGTTGAGGAACAGAGAGCTCTACTATGTAAGGAAGAGAC GAL4DB fusions for repression assay DAZ1-EAR-SmaI-F DAZ1-EAR-SalI-R DAZ1-smEAR2-SalI-R AGGACCCGGGCCAAGTCAGGGGCATTG AAGCGTCGACTCAAAGTCCTAACCTAAG AAGCGTCGACTCAAGCTCCTGCCCTAGCATCCGCGG qrt-pcr primers for developmental stages and sporophytic tissues HTR5-qRT-F AGCTCCCTTTCCAGAGGCTA HTR5-qRT-R TCCAAGTCTCCTACACCCAAA DUO1-qRT-F AACGTCAAACCAATCCGTCAATCC DUO1-qRT-R CGAACAATGGCTCAGAAGAATCAGC DAZ1-qRT-F TGATGCTTCACAAATCGCACG DAZ1-qRT-R CAGGAGGCTATGTTATGCTCTTCC DAZ2-qRT-F GAAAGTTTTGGTCTTGGAAGGCTC DAZ2-qRT-R ACTCGCTCTGTGTCCTCCTAAAGCC
9 qrt-pcr primers for expression differences in mutant pollen HTR5-qF AGATGGCTCGTACTAAGCAAACAG HTR5-qR GCAGACTTACGTGCAGCCTTTG VCK-qF TCGCAATTTCAACAAGAGGTCGAG VCK-qR AAGTAGAACCATGTTCGGATGCC CYCB1;1-qF AGTTGCTGCTCGAGAGAAGAAGG CYCB1;1-qR TCTAAACCACAAGCAGCCTTGC CYCB1;2-qF GCCACCTCAGGTTAACGATCTG CYCB1;2-qR CCAAGAATTGCCTTCTCCATCACC MYB101-qF TCATTCATCATTGGGAGCAAACCC MYB101-qR GCAAAGGTGTTGTGGTTGAAGGG HTR10-qF AAACTGCGAGGAAATCTACTGGTG HTR10-qR ATTTGCGGATCTCACGAAGAGC HAP2-qF TCTCCTCGCTCTCTTCCCAATTAC HAP2-qR CGGTTAACAACGTCTGCTCTGG TIP5;1-qF TGCCTCTGTTATGGCTTGCCTTG TIP5;1-qR ATCGGTACGTGCTGTTCCATGAC OPT8-qF ACCATGTTCATGGCACAGGTTG OPT8-qR TCCCGGTGGAAGCAGATTAGTG PCR11-qF TGTGTTGCCTTTGGACGCATCG PCR11-qR ATGTACATCGCTCCGCTCACAC DUO1-qF ATGGATGCAAGTTCTCGGCTGAC DUO1-qR ACGTAGCGATTCTCGCCCATTTG Reporter constructs tdtomato-attb2-f tdtomato-attb3-r MDB-attB1-F MDB-mCherry-Fm MDB-mCherry-Rm mcherry-attb2-rstop TCTTGTACAAAGTGGCGATGGTGAGCAAGGGCGAGGAGG TGTATAATAAAGTTGTTTACTTGTACAGCTCGTCCATGC AAAAAAGCAGGCTTCATGATGACTTCTCGTTCGATTGTTC TCGAGAGAAGATGGTGAGCAAGGGCGAGGAGG TGCTCACCATCTTCTCTCGAGCAGCAACTAAACC ACAAGAAAGCTGGGTTTTACTTGTACAGCTCGTCCATGC
Sperm cells are passive cargo of the pollen tube in plant fertilization
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
SUPPLEMENTARY INFORMATION
 Materials and Methods Transgenic Plant Materials and DNA Constructs. VEX1::H2B-GFP, ACA3::H2B-GFP, KRP6::H2B-GFP, KRP6::mock21ts-GFP, KRP6::TE21ts- GFP and KRP6::miR161ts-GFP constructs were generated
Materials and Methods Transgenic Plant Materials and DNA Constructs. VEX1::H2B-GFP, ACA3::H2B-GFP, KRP6::H2B-GFP, KRP6::mock21ts-GFP, KRP6::TE21ts- GFP and KRP6::miR161ts-GFP constructs were generated
To investigate the heredity of the WFP gene, we selected plants that were homozygous
 Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary Information. Supplementary Figure S1. Phenotypic comparison of the wild type and mutants.
 Supplementary Information Supplementary Figure S1. Phenotypic comparison of the wild type and mutants. Supplementary Figure S2. Transverse sections of anthers. Supplementary Figure S3. DAPI staining and
Supplementary Information Supplementary Figure S1. Phenotypic comparison of the wild type and mutants. Supplementary Figure S2. Transverse sections of anthers. Supplementary Figure S3. DAPI staining and
Supplementary Information
 Supplementary Information Supplementary Fig. 1. Seed dormancy and germination responses of RVE1 and PIF1. (a) Diagram of RVE1 and the T-DNA insertion of the rve1-2 mutant (SAIL_326_A01). Black boxes represent
Supplementary Information Supplementary Fig. 1. Seed dormancy and germination responses of RVE1 and PIF1. (a) Diagram of RVE1 and the T-DNA insertion of the rve1-2 mutant (SAIL_326_A01). Black boxes represent
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Supplemental Data. Ko et al. (2014). Plant Cell /tpc Supplemental Figure 1. Evidence of T-DNA Insertion in ms142 Mutant.
 Supplemental Figure 1. Evidence of T-DNA Insertion in ms142 Mutant. (A) DNA gel blotting to hptii probe confirmed single T-DNA insertion (marked with arrow) in ms142 mutant. (B) T-DNA tagged construction
Supplemental Figure 1. Evidence of T-DNA Insertion in ms142 Mutant. (A) DNA gel blotting to hptii probe confirmed single T-DNA insertion (marked with arrow) in ms142 mutant. (B) T-DNA tagged construction
Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte
 Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte Carlo algorithm for one million generations to obtain
Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte Carlo algorithm for one million generations to obtain
Supplementary Information
 Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
Supplemental Figure 1
 Supplemental Figure 1 A LK sls1 lks1-2 F 1 sls1 B LK sls1 lks1-2 F 1 lks1-2 sls1 F 1 lks1-2 sls1 F 2 Col lks1-2 Col+LKS1 Col Col+LKS1 Supplemental Figure 1. Genetic analysis of sls1 mutant. (A) and (B)
Supplemental Figure 1 A LK sls1 lks1-2 F 1 sls1 B LK sls1 lks1-2 F 1 lks1-2 sls1 F 1 lks1-2 sls1 F 2 Col lks1-2 Col+LKS1 Col Col+LKS1 Supplemental Figure 1. Genetic analysis of sls1 mutant. (A) and (B)
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis
 Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis seedlings expressing ERF109pro-GUS. The GUS staining
Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis seedlings expressing ERF109pro-GUS. The GUS staining
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplemental Data Supplemental Figure 1.
 Supplemental Data Supplemental Figure 1. Silique arrangement in the wild-type, jhs, and complemented lines. Wild-type (WT) (A), the jhs1 mutant (B,C), and the jhs1 mutant complemented with JHS1 (Com) (D)
Supplemental Data Supplemental Figure 1. Silique arrangement in the wild-type, jhs, and complemented lines. Wild-type (WT) (A), the jhs1 mutant (B,C), and the jhs1 mutant complemented with JHS1 (Com) (D)
Supporting Information
 Supporting Information Park et al. 10.1073/pnas.1410555111 5 -TCAAGTCCATCTACATGGCC-3 5 -CAGCTGCCCGGCTACTACTA-3 5 -TGCAGCTGCCCGGCTACTAC-3 5 -AAGCTGGACATCACCTCCCA-3 5 -TGACAGGAACACCTACAAGT-3 5 -AAGGCACCTTTCTGTCTCCA-3
Supporting Information Park et al. 10.1073/pnas.1410555111 5 -TCAAGTCCATCTACATGGCC-3 5 -CAGCTGCCCGGCTACTACTA-3 5 -TGCAGCTGCCCGGCTACTAC-3 5 -AAGCTGGACATCACCTCCCA-3 5 -TGACAGGAACACCTACAAGT-3 5 -AAGGCACCTTTCTGTCTCCA-3
Supplemental Data. Zhou et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. Confirmation of mutant mapping results. (A) Complementation assay with stably transformed genomic fragments (ComN-N) (2 kb upstream of TSS and 1.5 kb downstream of TES) and CaMV
Supplemental Figure 1. Confirmation of mutant mapping results. (A) Complementation assay with stably transformed genomic fragments (ComN-N) (2 kb upstream of TSS and 1.5 kb downstream of TES) and CaMV
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
Supplemental Figure legends Figure S1. (A) (B) (C) (D) Figure S2. Figure S3. (A-E) Figure S4. Figure S5. (A, C, E, G, I) (B, D, F, H, Figure S6.
 Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
Supplemental Data. Wang et al. (2017). Plant Cell /tpc NIP1;2 NIP7;1
 Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
Barley as a model for cereal engineering and genome editing. Wendy Harwood
 Barley as a model for cereal engineering and genome editing Wendy Harwood MonoGram 29 th April 2015 www.bract.org BRACT Transformation Platform Over-expression of single genes RNAi based silencing Promoter
Barley as a model for cereal engineering and genome editing Wendy Harwood MonoGram 29 th April 2015 www.bract.org BRACT Transformation Platform Over-expression of single genes RNAi based silencing Promoter
Supplemental Figure 1. VLN5 retains conserved residues at both type 1 and type 2 Ca 2+ -binding
 Supplemental Figure 1. VLN5 retains conserved residues at both type 1 and type 2 Ca 2+ -binding sites in the G1 domain. Multiple sequence alignment was performed with DNAMAN6.0.40. Secondary structural
Supplemental Figure 1. VLN5 retains conserved residues at both type 1 and type 2 Ca 2+ -binding sites in the G1 domain. Multiple sequence alignment was performed with DNAMAN6.0.40. Secondary structural
SUPPLEMENTARY INFORMATION
 Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Supplemental Data. Wang et al. Plant Cell. (2013) /tpc
 SNL1 SNL Supplemental Figure 1. The Expression Patterns of SNL1 and SNL in Different Tissues of Arabidopsis from Genevestigator Web Site (https://www.genevestigator.com/gv/index.jsp). 1 A 1. 1..8.6.4..
SNL1 SNL Supplemental Figure 1. The Expression Patterns of SNL1 and SNL in Different Tissues of Arabidopsis from Genevestigator Web Site (https://www.genevestigator.com/gv/index.jsp). 1 A 1. 1..8.6.4..
- 1 - Supplemental Data
 - 1-1 Supplemental Data 2 3 4 5 6 7 8 9 Supplemental Figure S1. Differential expression of AtPIP Genes in DC3000-inoculated plants. Gene expression in leaves was analyzed by real-time RT-PCR and expression
- 1-1 Supplemental Data 2 3 4 5 6 7 8 9 Supplemental Figure S1. Differential expression of AtPIP Genes in DC3000-inoculated plants. Gene expression in leaves was analyzed by real-time RT-PCR and expression
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
0.5% Sucrose. Supplemental Data. Chao et al. (2011). Plant Cell /tpc Col-0 tsc10a-2. Root length (mm)
 A 0% 0.5% 1% Col-0 tsc10a-2 B Root length (mm) 35 30 25 20 15 10 5 0 0% 0.50% 1% 2% 4% Sucrose concentration 2% 4% Sucrose Col-0 tsc10a-2 Supplemental Figure 1. The effect of sucrose on root growth of
A 0% 0.5% 1% Col-0 tsc10a-2 B Root length (mm) 35 30 25 20 15 10 5 0 0% 0.50% 1% 2% 4% Sucrose concentration 2% 4% Sucrose Col-0 tsc10a-2 Supplemental Figure 1. The effect of sucrose on root growth of
Erhard et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. c1-hbr allele structure. Diagram of the c1-hbr allele found in stocks segregating 1:1 for rpd1-1 and rpd1-2 homozygous mutants showing the presence of a 363 base pair (bp) Heartbreaker
Supplemental Figure 1. c1-hbr allele structure. Diagram of the c1-hbr allele found in stocks segregating 1:1 for rpd1-1 and rpd1-2 homozygous mutants showing the presence of a 363 base pair (bp) Heartbreaker
Supplemental Data. Guo et al. (2015). Plant Cell /tpc
 Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Trasposable elements: Uses of P elements Problem set B at the end
 Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Nature Genetics: doi: /ng Supplementary Figure 1. ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets.
 Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Two classes of silencing RNAs move between Caenorhabditis elegans tissues.
 Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
SUPPLEMENTARY INFORMATION
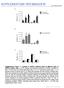 SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
Using mutants to clone genes
 Using mutants to clone genes Objectives: 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype
Using mutants to clone genes Objectives: 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype
Using mutants to clone genes
 Using mutants to clone genes Objectives 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype
Using mutants to clone genes Objectives 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
amplification of the 5 flanking region of CAT1 with a tail for the geneticin resistance gene cassette fusion CAT1-3F
 Supplementary materials Table S1. Primer list. Primer Sequence a (5-3 ) Description CAT1-5F CAT1-5R ATACGGATAAGGAAGCGATAGCAGCA gcacaggtacacttgtttagagagcgatccggattttaagtgaacg amplification of the 5 flanking
Supplementary materials Table S1. Primer list. Primer Sequence a (5-3 ) Description CAT1-5F CAT1-5R ATACGGATAAGGAAGCGATAGCAGCA gcacaggtacacttgtttagagagcgatccggattttaagtgaacg amplification of the 5 flanking
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
b number of motif occurrences
 supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
SUPPLEMENTAL FILES. Supplemental Figure 1. Expression domains of Arabidopsis HAM orthologs in both shoot meristem and root tissues.
 SUPPLEMENTAL FILES SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Expression domains of Arabidopsis HAM orthologs in both shoot meristem and root tissues. RT-PCR amplification of AtHAM1, AtHAM2, AtHAM3,
SUPPLEMENTAL FILES SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Expression domains of Arabidopsis HAM orthologs in both shoot meristem and root tissues. RT-PCR amplification of AtHAM1, AtHAM2, AtHAM3,
Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres
 Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres generated from the CD133-positive cells infected with
Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres generated from the CD133-positive cells infected with
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
Fertilization Recovery after Defective. Sperm Cell Release in Arabidopsis. Current Biology, Volume 22 Supplemental Information
 Current Biology, Volume 22 Supplemental Information Fertilization Recovery after Defective Sperm Cell Release in Arabidopsis Ryushiro D. Kasahara, Daisuke Maruyama, Yuki Hamamura, Takashi Sakakibara, David
Current Biology, Volume 22 Supplemental Information Fertilization Recovery after Defective Sperm Cell Release in Arabidopsis Ryushiro D. Kasahara, Daisuke Maruyama, Yuki Hamamura, Takashi Sakakibara, David
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Supplemental Data. Benstein et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Supplemental Figure 1 A
 Supplemental Figure 1 A Supplemental Data. Han et al. (2016). Plant Cell 10.1105/tpc.15.00997 BamHI -1616 bp SINE repeats SalI ATG SacI +1653 bp HindIII LB HYG p35s pfwa LUC Tnos RB pfwa-bamhi-f: CGGGATCCCGCCTTTCTCTTCCTCATCTGC
Supplemental Figure 1 A Supplemental Data. Han et al. (2016). Plant Cell 10.1105/tpc.15.00997 BamHI -1616 bp SINE repeats SalI ATG SacI +1653 bp HindIII LB HYG p35s pfwa LUC Tnos RB pfwa-bamhi-f: CGGGATCCCGCCTTTCTCTTCCTCATCTGC
Candidate region (0.74 Mb) ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC ATC TCT GGG ACT CAT TAG CAG GAG GCT AGC
 A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
SUPPLEMENTARY INFORMATION
 AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the
 CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations Changtian Pan 1,2, Lei Ye 1,2, Li Qin 2, Xue Liu 2, Yanjun He 2, Jie wang 2, Lifei
CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations Changtian Pan 1,2, Lei Ye 1,2, Li Qin 2, Xue Liu 2, Yanjun He 2, Jie wang 2, Lifei
Supplementary information
 Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplemental Data. Li et al. (2015). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1: Characterization of asr3 T-DNA knockout lines and complementation transgenic lines. (A) The scheme of At2G33550 (ASR3) with gray boxes indicating exons and dash
Supplemental Data Supplemental Figure 1: Characterization of asr3 T-DNA knockout lines and complementation transgenic lines. (A) The scheme of At2G33550 (ASR3) with gray boxes indicating exons and dash
An antibiotic selection marker for nematode transgenesis
 nature methods An antibiotic selection marker for nematode transgenesis Rosina Giordano-Santini, Stuart Milstein, Nenad Svrzikapa, Domena Tu, Robert Johnsen, David Baillie, Marc Vidal & Denis Dupuy Supplementary
nature methods An antibiotic selection marker for nematode transgenesis Rosina Giordano-Santini, Stuart Milstein, Nenad Svrzikapa, Domena Tu, Robert Johnsen, David Baillie, Marc Vidal & Denis Dupuy Supplementary
Supplemental Data. Jing et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Characterization of epp1 Mutants. (A) Cotyledon angles of 5-d-old Col wild-type (gray bars) and epp1-1 (black bars) seedlings under red (R), far-red (FR) and blue (BL) light conditions,
Supplemental Figure 1. Characterization of epp1 Mutants. (A) Cotyledon angles of 5-d-old Col wild-type (gray bars) and epp1-1 (black bars) seedlings under red (R), far-red (FR) and blue (BL) light conditions,
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector
 SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Richard J. Wall 1,#, Magali Roques 1,+, Nicholas J. Katris
SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Richard J. Wall 1,#, Magali Roques 1,+, Nicholas J. Katris
Supplemental Figure S1. Validation of auxin-related gene expression from Fig. 1A. Relative gene expression in 2x and 3x seeds 6 days after
 Supplemental Figure S1. Validation of auxin-related gene expression from Fig. 1A. Relative gene expression in 2x and 3x seeds 6 days after pollination (6 DAP), as determined by RT-qPCR for YUC10 (A), YUC11
Supplemental Figure S1. Validation of auxin-related gene expression from Fig. 1A. Relative gene expression in 2x and 3x seeds 6 days after pollination (6 DAP), as determined by RT-qPCR for YUC10 (A), YUC11
Lecture 2: Using Mutants to study Biological processes
 Lecture 2: Using Mutants to study Biological processes Objectives: 1. Why use mutants? 2. How are mutants isolated? 3. What important genetic analyses must be done immediately after a genetic screen for
Lecture 2: Using Mutants to study Biological processes Objectives: 1. Why use mutants? 2. How are mutants isolated? 3. What important genetic analyses must be done immediately after a genetic screen for
Supplemental Data. Seo et al. (2014). Plant Cell /tpc
 Supplemental Figure 1. Protein alignment of ABD1 from other model organisms. The alignment was performed with H. sapiens DCAF8, M. musculus DCAF8 and O. sativa Os10g0544500. The WD40 domains are underlined.
Supplemental Figure 1. Protein alignment of ABD1 from other model organisms. The alignment was performed with H. sapiens DCAF8, M. musculus DCAF8 and O. sativa Os10g0544500. The WD40 domains are underlined.
Supplemental Data. Zhang et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. uvs90 gene cloning The T-DNA insertion in uvs90 was identified using thermal asymmetric interlaced (TAIL)-PCR. Three rounds of amplification were performed; the second (2 nd ) and
Supplemental Figure 1. uvs90 gene cloning The T-DNA insertion in uvs90 was identified using thermal asymmetric interlaced (TAIL)-PCR. Three rounds of amplification were performed; the second (2 nd ) and
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 )
 Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Experimental Tools and Resources Available in Arabidopsis. Manish Raizada, University of Guelph, Canada
 Experimental Tools and Resources Available in Arabidopsis Manish Raizada, University of Guelph, Canada Community website: The Arabidopsis Information Resource (TAIR) at http://www.arabidopsis.org Can order
Experimental Tools and Resources Available in Arabidopsis Manish Raizada, University of Guelph, Canada Community website: The Arabidopsis Information Resource (TAIR) at http://www.arabidopsis.org Can order
Intron distance to transcription start (nt)*
 Schwab et al. SOM: Enhanced microrna accumulation through stemloop-adjacent introns 1 Supplemental Table 1. Characteristics of introns used in this study Intron name Length intron (nt) Length cloned fragment
Schwab et al. SOM: Enhanced microrna accumulation through stemloop-adjacent introns 1 Supplemental Table 1. Characteristics of introns used in this study Intron name Length intron (nt) Length cloned fragment
Supplementary Information. A novel human endogenous retroviral protein inhibits cell-cell fusion. Supplementary Figures:
 Supplementary Information A novel human endogenous retroviral protein inhibits cell-cell fusion Jun Sugimoto, Makiko Sugimoto, Helene Bernstein, Yoshihiro Jinno and Danny J. Schust Supplementary Figures:
Supplementary Information A novel human endogenous retroviral protein inhibits cell-cell fusion Jun Sugimoto, Makiko Sugimoto, Helene Bernstein, Yoshihiro Jinno and Danny J. Schust Supplementary Figures:
Supplementary Information. c d e
 Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
Before starting, write your name on the top of each page Make sure you have all pages
 Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
SUPPLEMENT MATERIALS FOR CURTIN,
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 SUPPLEMENT MATERIALS FOR CURTIN, et al. Validating genome-wide association candidates: Selecting, testing, and characterizing
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 SUPPLEMENT MATERIALS FOR CURTIN, et al. Validating genome-wide association candidates: Selecting, testing, and characterizing
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast. Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice
 A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
*** *** * *** *** *** * *** ** ***
 Figure S: OsMADS6 over- or down-expression is stable across generations A, OsMADS6 expression in overexpressing (OX, OX, dark bars) and corresponding control (OX, WT, white bars) T4 plants. B, OsMADS6
Figure S: OsMADS6 over- or down-expression is stable across generations A, OsMADS6 expression in overexpressing (OX, OX, dark bars) and corresponding control (OX, WT, white bars) T4 plants. B, OsMADS6
gypsy fold change in steady state RNA levels RnpS1 Acn mago tsu GFP armi sirna mediated depletion against indicated genes in OSCs
 Hayashi_FigS1 fold change in steady state RNA levels 1000 100 10 1 gypsy GFP armi mago tsu Acn RnpS1 btz sirna mediated depletion against indicated genes in OSCs Supplementary Figure S1. The EJC is required
Hayashi_FigS1 fold change in steady state RNA levels 1000 100 10 1 gypsy GFP armi mago tsu Acn RnpS1 btz sirna mediated depletion against indicated genes in OSCs Supplementary Figure S1. The EJC is required
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for
 Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
 http://fire.biol.wwu.edu/trent/trent/direct_detection_of_genotype.html 1 Like most other model organism Arabidopsis thaliana has a sequenced genome? What do we mean by sequenced genome? What sort of info
http://fire.biol.wwu.edu/trent/trent/direct_detection_of_genotype.html 1 Like most other model organism Arabidopsis thaliana has a sequenced genome? What do we mean by sequenced genome? What sort of info
Abcam.com. hutton.ac.uk. Ipmdss.dk. Bo Gong and Eva Chou
 Abcam.com Bo Gong and Eva Chou Ipmdss.dk hutton.ac.uk What is a homeotic gene? A gene which regulates the developmental fate of anatomical structures in an organism Why study them? Understand the underlying
Abcam.com Bo Gong and Eva Chou Ipmdss.dk hutton.ac.uk What is a homeotic gene? A gene which regulates the developmental fate of anatomical structures in an organism Why study them? Understand the underlying
previously. co and fca-9 alleles were isolated from the Col background. Plants were
 Supporting Online Material Materials and Methods Plant Growth Conditions The flc-3, FRI Sf2 -Col (S1), fve-4 (S2), ld-1 (S3) and fpa (S4) lines were described previously. co and fca-9 alleles were isolated
Supporting Online Material Materials and Methods Plant Growth Conditions The flc-3, FRI Sf2 -Col (S1), fve-4 (S2), ld-1 (S3) and fpa (S4) lines were described previously. co and fca-9 alleles were isolated
Supplemental data. Zhao et al. (2009). The Wuschel-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice.
 Supplemental data. Zhao et al. (2009). The Wuschel-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. A B Supplemental Figure 1. Expression of WOX11p-GUS WOX11-GFP
Supplemental data. Zhao et al. (2009). The Wuschel-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. A B Supplemental Figure 1. Expression of WOX11p-GUS WOX11-GFP
Supplemental Figure 1
 Supplemental Figure 1 A gta2-1 gta2-2 1kb AT4G08350 B Col-0 gta2-1 LP+RP+LBb1.3 C Col-0 gta2-2 LP+RP+LB1 D Col-0 gta2-2 gta2-1 GTA2 TUBLIN E Col0 gta2-1 gta2-2 Supplemental Figure 1. Phenotypic analysis
Supplemental Figure 1 A gta2-1 gta2-2 1kb AT4G08350 B Col-0 gta2-1 LP+RP+LBb1.3 C Col-0 gta2-2 LP+RP+LB1 D Col-0 gta2-2 gta2-1 GTA2 TUBLIN E Col0 gta2-1 gta2-2 Supplemental Figure 1. Phenotypic analysis
Supplementary Information. A superfolding Spinach2 reveals the dynamic nature of. trinucleotide repeat RNA
 Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
Supplementary Data 1.
 Supplementary Data 1. Evaluation of the effects of number of F2 progeny to be bulked (n) and average sequencing coverage (depth) of the genome (G) on the levels of false positive SNPs (SNP index = 1).
Supplementary Data 1. Evaluation of the effects of number of F2 progeny to be bulked (n) and average sequencing coverage (depth) of the genome (G) on the levels of false positive SNPs (SNP index = 1).
PIE1 ARP6 SWC6 KU70 ARP6 PIE1. HSA SNF2_N HELICc SANT. pie1-3 A1,A2 K1,K2 K1,K3 K3,LB2 A3, A4 A3,LB1 A1,A2 K1,K2 K1,K3. swc6-1 A3,A4.
 A B N-terminal SWC2 H2A.Z SWC6 ARP6 PIE1 HSA SNF2_N HELICc SANT C pie1-3 D PIE1 ARP6 5 Kb A1 200 bp A3 A2 LB1 arp6-3 A4 E A1,A2 A3, A4 A3,LB1 K1,K2 K1,K3 K3,LB2 SWC6 swc6-1 A1,A2 A3,A4 K1,K2 K1,K3 100
A B N-terminal SWC2 H2A.Z SWC6 ARP6 PIE1 HSA SNF2_N HELICc SANT C pie1-3 D PIE1 ARP6 5 Kb A1 200 bp A3 A2 LB1 arp6-3 A4 E A1,A2 A3, A4 A3,LB1 K1,K2 K1,K3 K3,LB2 SWC6 swc6-1 A1,A2 A3,A4 K1,K2 K1,K3 100
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
supplementary information
 DOI: 10.1038/ncb1979 Figure S1 Alignment and domain composition of HsTSN and PaTSN. Since both HsTSN (accession number AAA80488) and PaTSN (accession number CAL38976) have five staphylococcal nuclease
DOI: 10.1038/ncb1979 Figure S1 Alignment and domain composition of HsTSN and PaTSN. Since both HsTSN (accession number AAA80488) and PaTSN (accession number CAL38976) have five staphylococcal nuclease
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Data Supplementary Figures
 Supplementary Data Supplementary Figures Supplementary Figure 1. Pi04314 is expressed during infection, each GFP-Pi04314 fusion is stable and myr GFP-Pi04314 is removed from the nucleus while NLS GFP-Pi04314
Supplementary Data Supplementary Figures Supplementary Figure 1. Pi04314 is expressed during infection, each GFP-Pi04314 fusion is stable and myr GFP-Pi04314 is removed from the nucleus while NLS GFP-Pi04314
What are the Properties of the Protein EMB2777?
 Of EMB2777 and F14J22.20 - exons & introns by Hanbee O What are the Properties of the Protein EMB2777?! Embryo defective / seed lethal transcriptional factor o Sas10 / U3 ribonucleoprotein (Utp) family
Of EMB2777 and F14J22.20 - exons & introns by Hanbee O What are the Properties of the Protein EMB2777?! Embryo defective / seed lethal transcriptional factor o Sas10 / U3 ribonucleoprotein (Utp) family
