Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres
|
|
|
- Kerry Fitzgerald
- 5 years ago
- Views:
Transcription
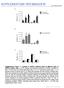
1 Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres generated from the CD133-positive cells infected with either Control- NeoR or KRAS-NeoR. (B) Transplanted pancreas with S1 KCST spheres. Yellow arrows indicate the cyst-like structures found during pancreas dissection. (C) Representative images of H&E and Alcian blue staining from S1 KCST cultured spheres (left) and transplanted mice (right). Note that no alcian blue-positive cytoplasm in cultured spheres. Scale Bars, 100µM. (D) Indel spectrum images of TIDE analysis for S1 KCST spheres. Red and black bars show p < 0.05 and p > 0.05, respectively. Orange bar shows the peak with no Indel mutations.
2 Supplementary Figure 2. Phenotype and genotype of cultured and transplanted S2 and S3 spheres (A and B) Indel spectrum images of TIDE analysis for S2 KECST (A) and S3 KECST (B) spheres. (C) Transplanted pancreas with S2 and S3 KECST spheres. Yellow arrows indicate the cyst-like structures found during pancreas dissection.
3 Supplementary Figure 3. Immunohistochemistry with antibodies against phospho-erk and phospho-akt Human PDA, PanIN, and pancreas transplanted with S1 KCST, S2 KECST, and S3 KECST spheres was stained with anti-phospho- ERK and phospho-akt antibodies. Note that phospho-akt signal is weak or absent in human native PanIN and all transplanted hipanin lesions. Scale Bars, 200µM.
4 Supplementary Figure 4. Phenotype and genotype of cultured and transplanted spheres (A-C) H&E and Alcian blue staining of the transplanted S2 KECST (A and B) and S3 KECST (B) spheres. (D) Pancreas transplanted with S2 KECST spheres was stained with human nuclear-specific antibody (HuNu, white) and mcherry fluorescence (red) along with DAPI nuclear staining (blue). Note that neither mcherry- nor HuNu-positive cells were found in mouse duct of the same pancreas. Magnified view of the boxed areas are shown to the right. Scale Bars, 200µM. (E) Genomic DNA PCR of cloned spheres for assessing the presence of lentiviral transgenes. Numbers on top denote clone numbers.
5 Supplementary Figure 5. Immunohistochemistry with anti-human mitochondria antibody Pancreas transplanted with S1 KCST, S2 KECST, and S3 KECST spheres was stained with anti-human mitochondria antibody. Scale Bars, 200µM.
6 Supplementary Figure 6. Hematoxylin and Eosin and anti-human mitochondria antibody staining on serial sections Serial sections of the pancreas transplanted with S2 KECST (ID 187) spheres were stained with H&E and anti-human mitochondria antibody. Scale Bars, 200µM.
7 Supplementary Figure 7. Genotype and phenotype of S2 KCT clone3 (A) TIDE Indel spectrum images of TIDE analysis for S2 KCT clone3. Note that Indel spectrum for CDKN2A is not available due to the large size of deletion. (B) Genomic DNA sequences of CRISPR-Cas9-targeted loci. The numbers next to the gene names indicate the total number of nucleotide deleted or inserted. Arrows indicate the expected cut site by Cas9 nuclease. (C) H&E staining of the human grafts found in transplanted pancreas with S2 KCT clone3. Scale Bars, 200µM.
8 Supplementary Figure 8. Genotype and phenotype of S1 clones (A and B) TIDE Indel spectrum images of TIDE analysis for S1 KCST clone3 (A) and S1 KCST clone3 (B). (C and D) Genomic DNA sequences of CRISPR-Cas9-targeted loci for S1 KCST clone3 (C) and S1 KCST clone3 (D). The numbers next to the gene names indicate the total number of nucleotide deleted or inserted. Arrows indicate the expected cut site by Cas9 nuclease.
9 Supplementary Figure 9. H&E images of S1 clones (A-B) H&E staining of PanIN structures found in transplanted pancreas with S1 KCST clone4. (A) and S1 KECST clone4 (B). Note that the PanIN structures show cribriforming (A, ID203), abnormal nuclei (yellow arrows), and necrotic cells in the lumen (red arrows), features of human PanIN2 and 3. Magnified images are shown on right. (C) Relative mrna expression level of ERBB2 transgene Error bars = S.D. n = 2. Scale Bars, 200µM.
10 Supplementary Figure 10. Genotype and phenotype of S2 KCST clone8 (A) TIDE Indel spectrum images of TIDE analysis for S2 KCST clone8. (B) Genomic DNA sequences of CRISPR-Cas9-targeted loci for S2 KCST clone8. The numbers next to the gene names indicate the total number of nucleotide deleted or inserted. Arrows indicate the expected cut site by Cas9 nuclease. (C) H&E staining of PanIN structures found in transplanted pancreas with S2 KCST clone8. Scale Bars, 200µM.
11 Supplementary Figure 11. Off-target analysis result of S1 KCST clone3 Off-target analysis result of S1 KCST clone3. See Supplementary Table 4.
12 Supplementary Figure 12. Off-target analysis result of S1 KCST clone4 Off-target analysis result of S1 KCST clone4. See Supplementary Table 4.
13 Supplementary Figure 13. Off-target analysis result of S2 KCT clone3 Off-target analysis result of S2 KCT clone3. See Supplementary Table 4.
14 Supplementary Figure 14. Off-target analysis result of S2 KCST clone8 Off-target analysis result of S2 KCST clone8. See Supplementary Table 4.
15 Supplementary Figure 15. Genetic modification of human ductal cell line HPDE induces invasive PDA development (A) Indel spectrum images of TIDE analysis for HPDE KECST (B) Schematics of lenticrisprv2 and new sets of sgrna sequences (KECST2). (C) Genomic DNA PCR for assessing the presence of lentiviral transgenes in HPDE cells. (D) Relative mrna expression level of oncogenic KRAS and ERBB2 transgene. Error bars = S.D. (E) Indel efficiency of each genomic loci assessed by TIDE analysis. (F) Stereoscopic and representative H&E and CK19 (green) staining images of the tumors formed in the transplanted pancreas with HPDE KECST2. Red arrows indicate tumor nodules. (G) Representative H&E staining of spleen with metastatic cells found in ID 215. The metastatic cells are CK19-positive (green, bottom). (H) Stereoscopic and representative H&E staining images of the tumors formed in the transplanted pancreas with HPDE KCST2. Scale Bars, 200µM.
16 Supplementary Table 1. Phenotypes of pancreas donors ANONYMOUS ID AGE (YEAR) GENDER BODY MASS INDEX S1 54 M 40.3 S2 40 F 25.2 S3 51 M 28.4
17 Supplementary Table 2. List of PanIN-like structures in transplanted mouse pancreas SAMPLE ID MOUSE ID HUMAN PATHOLOGY REVIEW GRAFT FOUND S1 KCST 192 YES PanIN1 193 YES PanIN1 194 YES PanIN1 S2 KECST 179 YES PanIN1, PanIN2 180 YES PanIN2 181 NO N/A 185 YES PanIN1 186 YES PanIN1 190 YES PanIN2 S3 KECST 187 YES PanIN1 188 YES PanIN1, PanIN2 191 YES PanIN1
18 Supplementary Table 3. List of isolated sphere clones and their genotypes Sample ID Clone ID Presence of Transgenes Indel mutation status K E C S T CDKN2A SMAD4 TP53 Note S1 KCST 3 Y N Y Y Y -32/-5-22/-20-4/-4 S1 KCST clone3 4 Y N Y Y Y -28/-10-65/-5-4/5 S1 KCST clone4 5 Y N Y Y Y -22/-22-2/-5-6/-85 6 Y N Y Y Y -12/-10-24/-2-21/-21 7 Y N Y Y Y -9/-9-5/-1-12/-6 9 Y N Y Y Y -29/-5-5/-1-12/-6 10 Y N Y Y Y -12/-5-5/-1-12/-6 11 Y N Y Y Y -12/-76-5/-5-6/-4 12 Y N Y Y Y -35/-35-5/-5-12/1 13 Y N Y Y Y N/D -5/-5-6/-4 14 Y N Y Y Y -5/-4 0/-13-11/4 15 Y N Y Y Y N/D -5/-5-6/-4 16 Y N Y Y Y N/D -5/-5-6/-4 17 Y N Y Y Y -36/-5 N/D N/D 18 Y N Y N Y -21/-28 0/0-12/4 S2 KECST 3 Y N Y N Y -95/-95 0/0 +1/+1 S2 KCT clone3 7 Y N Y Y Y -25/-5-2/-2 N/D 8 Y N Y Y Y -17/-17-2/+1-1/-1 S2 KCST clone8 S3 KECST 2 Y Y Y Y Y -21/-21-6/-6-6/-6 4 Y Y Y Y Y -21/-21-6/-6-6/-6 7 Y Y Y Y Y -21/-21-6/-6-6/-6
19 Supplementary Table 4. Off-target analysis result locus SMAD4#1 OFF1 sgrna human chromosome alignment(dots are matched bp) off-target score Note activity=1 when CDKN2A#1 Query_1 ACCGTAACTATTCGGTGCGTNGG PAM=NGG Ch9: T.. 1 CDKN2A locus Ch7: A...T.T CDKN2A#1 OFF1 activity=0.4 when Query_1 ACCGTAACTATTCGGTGCGTNaG PAM=NAG Ch2: A CDKN2A#1 OFF2 activity=1 when SMAD4#1 Query_1 ACAACTCGTTCGTAGTGATANGG PAM=NGG Ch18: SMAD T.. 1 Ch7: T..T...G activity=1 when TP53#2 Query_1 GGGCAGCTACGGTTTCCGTCNGG PAM=NGG Ch17: T.. 1 TP53 locus
20 Supplementary Table 5. Tumors found in mouse pancreas transplanted with transduced HPDE cells SAMPLE ID MOUSE METASTASIS PATHOLOGY REVIEW ID HPDE KECST 211 Lung poorly differentiated carcinoma 212 mid-to-poorly differentiated adenocarcinoma 214 poorly differentiated adenocarcinoma HPDE KCST 219 poorly differentiated adenocarcinoma 220 moderate-to poorly differentiated adenocarcinoma 221 poorly differentiated adenocarcinoma HPDE KECST2 215 Spleen poorly differentiated adenocarcinoma 217 poorly differentiated adenocarcinoma 218 moderate-to-poorly differentiated adenocarcinoma HPDE KCST2 228 poorly differentiated adenocarcinoma 229 Liver poorly differentiated adenocarcinoma 230 No tumors found
21 Supplementary Table 6. Gene Set Enrichment Analysis (GSEA) result HALLMARK NAME SIZE NES p-val FDR q-val EPITHELIAL_MESENCHYMAL_TRANSITION G2M_CHECKPOINT TNFA_SIGNALING_VIA_NFKB IL6_JAK_STAT3_SIGNALING COMPLEMENT UV_RESPONSE_DN SPERMATOGENESIS MITOTIC_SPINDLE APOPTOSIS HALLMARK NAME = name of the individual hallmark geneset, SIZE = number of genes in each geneset, NES = Normalized Enrichment Score, p-val = nominal p-value, FDR = False Discovery Rate
22 Supplementary Table 7. PCR primer sequences Purpose ID Fwd/Rev sequence lenticrispr cloning Control Forward caccggtagcgaacgtgtccggcgt lenticrispr cloning Control Reverse aaacacgccggacacgttcgctacc lenticrispr cloning CDKN2A#1 Forward caccgaccgtaactattcggtgcgt lenticrispr cloning CDKN2A#1 Reverse aaacacgcaccgaatagttacggtc lenticrispr cloning SMAD4#1 Forward caccgacaactcgttcgtagtgata lenticrispr cloning SMAD4#1 Reverse aaactatcactacgaacgagttgtc lenticrispr cloning TP53#2 Forward caccggggcagctacggtttccgtc lenticrispr cloning TP53#2 Reverse aaacgacggaaaccgtagctgcccc lenticrispr cloning CDKN2A#3 Forward caccggcatggagccttcggctgac lenticrispr cloning CDKN2A#3 Reverse aaacgtcagccgaaggctccatgcc lenticrispr cloning SMAD4#2 Forward caccgtacgaacgagttgtatcacc lenticrispr cloning SMAD4#2 Reverse aaacggtgatacaactcgttcgtac lenticrispr cloning TP53#3 Forward caccgaccagcagctcctacaccgg lenticrispr cloning TP53#3 Reverse aaacccggtgtaggagctgctggtc Transgene-specific PCR KRAS Forward ctccgagcggatgtaccc Transgene-specific PCR KRAS Reverse cattgcactgtactcctctt Transgene-specific PCR CDKN2A#1 Forward gagggcctatttcccatgatt Transgene-specific PCR CDKN2A#1 Reverse aaacacgcaccgaatagttacggtc Transgene-specific PCR SMAD4#1 Forward gagggcctatttcccatgatt Transgene-specific PCR SMAD4#1 Reverse aaactatcactacgaacgagttgtc Transgene-specific PCR TP53#2 Forward gagggcctatttcccatgatt Transgene-specific PCR TP53#2 Reverse aaacgacggaaaccgtagctgcccc Transgene-specific PCR ERBB2 Forward atgcggttttggcagtacat Transgene-specific PCR ERBB2 Reverse cgagatctgagtccggtagc Transgene-specific PCR CDKN2A#3 Forward gagggcctatttcccatgatt Transgene-specific PCR CDKN2A#3 Reverse aaacgtcagccgaaggctccatgcc Transgene-specific PCR SMAD4#2 Forward gagggcctatttcccatgatt Transgene-specific PCR SMAD4#2 Reverse aaacggtgatacaactcgttcgtac Transgene-specific PCR TP53#3 Forward gagggcctatttcccatgatt Transgene-specific PCR TP53#3 Reverse aaacccggtgtaggagctgctggtc TIDE PCR CDKN2A#1 and CDKN2A#3 Forward ggctcctcattcctcttcctt TIDE PCR CDKN2A#1 and CDKN2A#3 Reverse cttgcctggaaagataccgc TIDE PCR SMAD4#1 and SMAD4#2 Forward gcacaggccttgaaattatacc TIDE PCR SMAD4#1 and SMAD4#2 Reverse ccttatttaaagtcgcgggcta TIDE PCR TP53#2 and TP53#3 Forward gggtgtgatgggatggataaaag TIDE PCR TP53#2 and TP53#3 Reverse ctgctcttttcacccatctacag Off target analysis CDKN2A#1, OFF1 Forward gtgcacctgtaatccctgct Off target analysis CDKN2A#1, OFF1 Reverse agggcttgggacataaagga Off target analysis CDKN2A#1, OFF2 Forward attggtgaaccagggtgaaa Off target analysis CDKN2A#1, OFF2 Reverse ttcaatggcctgtaaaattgg Off target analysis SMAD4#1, OFF1 Forward ttttctcccccatctcagtg Off target analysis SMAD4#1, OFF1 Reverse tggcttttaaggtgcattcc
23 Supplementary Note 1. Full unedited gel image for Figure 1E and 3B
24 Supplementary Note 1. Full unedited gel image for Figure 1E and 3B (continued)
25 Supplementary Note 1. Full unedited gel image for Figure 1E and 3B (continued)
26 Supplementary Note 1. Full unedited gel image for Figure 1E and 3B (continued)
27 Supplementary Note 1. Full unedited gel image for Figure 1E and 3B (continued)
28 Supplementary Note 2. Full unedited gel image for Figure 5B and S1 sphere clones in Supplementary Figure 4E
29 Supplementary Note 2. Full unedited gel image for Figure 5B and S1 sphere clones in Supplementary Figure 4E (continued)
30 Supplementary Note 2. Full unedited gel image for Figure 5B and S1 sphere clones in Supplementary Figure 4E (continued)
31 Supplementary Note 2. Full unedited gel image for Figure 5B and S1 sphere clones in Supplementary Figure 4E (continued)
32 Supplementary Note 2. Full unedited gel image for Figure 5B and S1 sphere clones in Supplementary Figure 4E (continued)
33 Supplementary Note 2. Full unedited gel image for Figure 5B and S1 sphere clones in Supplementary Figure 4E (continued)
34 Supplementary Note 3. Full unedited gel image for S2 and S3 sphere clones in Supplementary Figure 4E
35 Supplementary Note 3. Full unedited gel image for S2 and S3 sphere clones in Supplementary Figure 4E (continued)
36 Supplementary Note 3. Full unedited gel image for S2 and S3 sphere clones in Supplementary Figure 4E (continued)
37 Supplementary Note 4. Full unedited gel image for Figure 6A
38 Supplementary Note 4. Full unedited gel image for Figure 6A (continued)
39 Supplementary Note 5. Full unedited gel image for Supplementary Figure 15C
40 Supplementary Note 5. Full unedited gel image for Supplementary Figure 15C (continued)
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Nature Biotechnology: doi: /nbt Supplementary Figure 1. In vitro validation of OTC sgrnas and donor template.
 Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supporting Information
 Supporting Information Park et al. 10.1073/pnas.1410555111 5 -TCAAGTCCATCTACATGGCC-3 5 -CAGCTGCCCGGCTACTACTA-3 5 -TGCAGCTGCCCGGCTACTAC-3 5 -AAGCTGGACATCACCTCCCA-3 5 -TGACAGGAACACCTACAAGT-3 5 -AAGGCACCTTTCTGTCTCCA-3
Supporting Information Park et al. 10.1073/pnas.1410555111 5 -TCAAGTCCATCTACATGGCC-3 5 -CAGCTGCCCGGCTACTACTA-3 5 -TGCAGCTGCCCGGCTACTAC-3 5 -AAGCTGGACATCACCTCCCA-3 5 -TGACAGGAACACCTACAAGT-3 5 -AAGGCACCTTTCTGTCTCCA-3
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
Supplemental Data. Borg et al. Plant Cell (2014) /tpc
 Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Nature Genetics: doi: /ng.2949
 Supplementary Figure 1. Abnormal differentiation in vivo and colony growth in vitro of B cells with triplication of chr.21 orthologs. (a) (Top) B220 and CD43 staining of bone marrow from Ts1Rhr and wild-type
Supplementary Figure 1. Abnormal differentiation in vivo and colony growth in vitro of B cells with triplication of chr.21 orthologs. (a) (Top) B220 and CD43 staining of bone marrow from Ts1Rhr and wild-type
Supporting Information
 Supporting Information Fujii et al. 10.1073/pnas.1217563110 A Human Cen chromosome 4q ART3 NUP54 SCARB2 FAM47E CCDC8 Tel B Bac clones RP11-54D17 RP11-628A4 Exon 1 2 34 5 6 7 8 9 10 11 12 5 3 PCR region
Supporting Information Fujii et al. 10.1073/pnas.1217563110 A Human Cen chromosome 4q ART3 NUP54 SCARB2 FAM47E CCDC8 Tel B Bac clones RP11-54D17 RP11-628A4 Exon 1 2 34 5 6 7 8 9 10 11 12 5 3 PCR region
somatic transition to was evident into airways in the
 Supplementary Fig. 1. A. Activation of an oncogenic K-Ras allele by recombination in somatic cells leads to spontaneous lung cancer in LA1 mice (ref. 20). In this mouse model, subpleural precancerous adenomas
Supplementary Fig. 1. A. Activation of an oncogenic K-Ras allele by recombination in somatic cells leads to spontaneous lung cancer in LA1 mice (ref. 20). In this mouse model, subpleural precancerous adenomas
Supplementary Information
 Supplementary Information Vidigal and Ventura a wt locus 5 region 3 region CCTCTGCCACTGCGAGGGCGTCCAATGGTGCTTG(...)AACAGGTGGAATATCCCTACTCTA predicted deletion clone 1 clone 2 clone 3 CCTCTGCCACTGCGAGGGCGTC-AGGTGGAATATCCCTACTCTA
Supplementary Information Vidigal and Ventura a wt locus 5 region 3 region CCTCTGCCACTGCGAGGGCGTCCAATGGTGCTTG(...)AACAGGTGGAATATCCCTACTCTA predicted deletion clone 1 clone 2 clone 3 CCTCTGCCACTGCGAGGGCGTC-AGGTGGAATATCCCTACTCTA
Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin
 Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin locus. The EcoRI restriction site between exons M1 and M2
Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin locus. The EcoRI restriction site between exons M1 and M2
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 )
 Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
SUPPLEMENT MATERIALS FOR CURTIN,
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 SUPPLEMENT MATERIALS FOR CURTIN, et al. Validating genome-wide association candidates: Selecting, testing, and characterizing
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 SUPPLEMENT MATERIALS FOR CURTIN, et al. Validating genome-wide association candidates: Selecting, testing, and characterizing
Supplementary Information
 Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
Pei et al. Supplementary Figure S1
 Pei et al. Supplementary Figure S1 C H-CUL9: + + + + + Myc-ROC1: - - + + + U2OS/pcDN3 U2OS/H-CUL9 U2OS/ + H-CUL9 IP: -H -myc input -H -myc 1 2 3 4 5 H-CUL9 Myc-ROC1 -H -H -H -H H-CUL9: wt RR myc-roc1:
Pei et al. Supplementary Figure S1 C H-CUL9: + + + + + Myc-ROC1: - - + + + U2OS/pcDN3 U2OS/H-CUL9 U2OS/ + H-CUL9 IP: -H -myc input -H -myc 1 2 3 4 5 H-CUL9 Myc-ROC1 -H -H -H -H H-CUL9: wt RR myc-roc1:
Supplementary Information
 Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 Sox2 localizes in neutrophils of mouse spleen. (a) Microscopy analysis of Sox2 and MPO in wild-type mouse spleen. Nucl, nucleus. (b) Immunohistochemistry staining of Sox2 and MPO
Supplementary Figure 1 Sox2 localizes in neutrophils of mouse spleen. (a) Microscopy analysis of Sox2 and MPO in wild-type mouse spleen. Nucl, nucleus. (b) Immunohistochemistry staining of Sox2 and MPO
Supplementary Figures and supplementary figure legends
 Supplementary Figures and supplementary figure legends Figure S1. Effect of different percentage of FGF signaling knockdown on TGF signaling and EndMT marker gene expression. HUVECs were subjected to different
Supplementary Figures and supplementary figure legends Figure S1. Effect of different percentage of FGF signaling knockdown on TGF signaling and EndMT marker gene expression. HUVECs were subjected to different
Supplementary Figure 1. Quantitative RT-PCR experimental validation of CRISPR/Cas9 and sgrnas expression in HEK293A transfected cells.
 Supplementary Figure 1. Quantitative RT-PCR experimental validation of CRISPR/Cas9 and sgrnas expression in HEK293A transfected cells. HEK293A cells were transfected with the indicated combinations of
Supplementary Figure 1. Quantitative RT-PCR experimental validation of CRISPR/Cas9 and sgrnas expression in HEK293A transfected cells. HEK293A cells were transfected with the indicated combinations of
Supplementary Figure 1.
 Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Sperm cells are passive cargo of the pollen tube in plant fertilization
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Before starting, write your name on the top of each page Make sure you have all pages
 Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Comparative assessment of vaccine vectors encoding ten. malaria antigens identifies two protective liver-stage
 Supplementary Information Comparative assessment of vaccine vectors encoding ten malaria antigens identifies two protective liver-stage candidates Rhea J. Longley 1,,,*, Ahmed M. Salman 1,2,, Matthew G.
Supplementary Information Comparative assessment of vaccine vectors encoding ten malaria antigens identifies two protective liver-stage candidates Rhea J. Longley 1,,,*, Ahmed M. Salman 1,2,, Matthew G.
To investigate the heredity of the WFP gene, we selected plants that were homozygous
 Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
SUPPLEMENTARY INFORMATION
 AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
C57BL/6N Female. C57BL/6N Male. Prl2 WT Allele. Prl2 Targeted Allele **** **** WT Het KO PRL bp. 230 bp CNX. C57BL/6N Male 30.
 A Prl Allele Prl Targeted Allele 631 bp 1 3 4 5 6 bp 1 SA β-geo pa 3 4 5 6 Het B Het 631 bp bp PRL CNX C 1 C57BL/6N Male (14) Het (7) (1) 1 C57BL/6N Female (19) Het (31) (1) % survival 5 % survival 5 ****
A Prl Allele Prl Targeted Allele 631 bp 1 3 4 5 6 bp 1 SA β-geo pa 3 4 5 6 Het B Het 631 bp bp PRL CNX C 1 C57BL/6N Male (14) Het (7) (1) 1 C57BL/6N Female (19) Het (31) (1) % survival 5 % survival 5 ****
SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector
 SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Richard J. Wall 1,#, Magali Roques 1,+, Nicholas J. Katris
SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Richard J. Wall 1,#, Magali Roques 1,+, Nicholas J. Katris
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Supplementary Figure 1. Characterization of the POP2 transcriptional and post-transcriptional regulatory elements. (A) POP2 nucleotide sequence
 1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
Growth factor, augmenter of liver regeneration
 Supplemental Table 1: Human and mouse PC1 sequence equivalencies Human Mouse Domain Clinical significance; Score* PolyPhen prediction; PSIC score difference C210G C210G WSC Highly likely pathogenic; 15
Supplemental Table 1: Human and mouse PC1 sequence equivalencies Human Mouse Domain Clinical significance; Score* PolyPhen prediction; PSIC score difference C210G C210G WSC Highly likely pathogenic; 15
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Allele-specific locus binding and genome editing by CRISPR at the
 Supplementary Information Allele-specific locus binding and genome editing by CRISPR at the p6ink4a locus Toshitsugu Fujita, Miyuki Yuno, and Hodaka Fujii Supplementary Figure Legends Supplementary Figure
Supplementary Information Allele-specific locus binding and genome editing by CRISPR at the p6ink4a locus Toshitsugu Fujita, Miyuki Yuno, and Hodaka Fujii Supplementary Figure Legends Supplementary Figure
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna
 Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Table S1
 Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Genome research in eukaryotes
 Functional Genomics Genome and EST sequencing can tell us how many POTENTIAL genes are present in the genome Proteomics can tell us about proteins and their interactions The goal of functional genomics
Functional Genomics Genome and EST sequencing can tell us how many POTENTIAL genes are present in the genome Proteomics can tell us about proteins and their interactions The goal of functional genomics
b number of motif occurrences
 supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
CRISPR RNA-guided activation of endogenous human genes
 CRISPR RNA-guided activation of endogenous human genes Morgan L Maeder, Samantha J Linder, Vincent M Cascio, Yanfang Fu, Quan H Ho, J Keith Joung Supplementary Figure 1 Comparison of VEGF activation induced
CRISPR RNA-guided activation of endogenous human genes Morgan L Maeder, Samantha J Linder, Vincent M Cascio, Yanfang Fu, Quan H Ho, J Keith Joung Supplementary Figure 1 Comparison of VEGF activation induced
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Table S1. Primers used in the study
 Table S1. Primers used in the study Primer name Application Sequence I1F16 Genotyping GGCAAGTGAGTGAGTGCCTA I1R11 Genotyping CCCACTCGTATTGACGCTCT V19 Genotyping GGGTCTCAAAGTCAGGGTCA D18Mit184-F Genotyping
Table S1. Primers used in the study Primer name Application Sequence I1F16 Genotyping GGCAAGTGAGTGAGTGCCTA I1R11 Genotyping CCCACTCGTATTGACGCTCT V19 Genotyping GGGTCTCAAAGTCAGGGTCA D18Mit184-F Genotyping
Supplementary Figure 1. Generation of B2M -/- ESCs. Nature Biotechnology: doi: /nbt.3860
 Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
Supplemental Figure 1
 Supplemental Figure 1 A P1 NOD - SCID(X*X*)(0/0) NOD - SCID(X*Y)(A*0201/A*0201) B F1 (X*X)(A*0201/0) (X*X*)(0/0) F2 (X*X*)(0/0) (X*Y)(0/0) (X*X*)(A*0201/0) (X*X*)(A*0201/0) F3 (X*X*)(0/0) (X*X)(A*0201/0)
Supplemental Figure 1 A P1 NOD - SCID(X*X*)(0/0) NOD - SCID(X*Y)(A*0201/A*0201) B F1 (X*X)(A*0201/0) (X*X*)(0/0) F2 (X*X*)(0/0) (X*Y)(0/0) (X*X*)(A*0201/0) (X*X*)(A*0201/0) F3 (X*X*)(0/0) (X*X)(A*0201/0)
F4/80, CD11b, Gr-1, NK1.1, CD3, CD4, CD8 and CD19. A-antigen was detected with FITCconjugated
 SDC MATERIALS AND METHODS Flow Cytometric Detection of A-Antigen Expression Single cell suspensions were prepared from bone marrow, lymph node and spleen. Peripheral blood was obtained and erythrocytes
SDC MATERIALS AND METHODS Flow Cytometric Detection of A-Antigen Expression Single cell suspensions were prepared from bone marrow, lymph node and spleen. Peripheral blood was obtained and erythrocytes
Genome Editing: Cas9 Stable Cell Lines for CRISPR sgrna Validation, Library Screening, and More. Ed Davis, Ph.D.
 TECHNICAL NOTE Genome Editing: Cas9 Stable Cell Lines for CRISPR sgrna Validation, Library Screening, and More Introduction Ed Davis, Ph.D. The CRISPR-Cas9 system has become greatly popular for genome
TECHNICAL NOTE Genome Editing: Cas9 Stable Cell Lines for CRISPR sgrna Validation, Library Screening, and More Introduction Ed Davis, Ph.D. The CRISPR-Cas9 system has become greatly popular for genome
University of Dundee. Published in: Scientific Reports. DOI: /srep Publication date: 2016
 University of Dundee SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Wall, Richard J.; Roques, Magali; Katris,
University of Dundee SAS6-like protein in Plasmodium indicates that conoid-associated apical complex proteins persist in invasive stages within the mosquito vector Wall, Richard J.; Roques, Magali; Katris,
SUPPLEMENTARY INFORMATION
 VOLUME: 1 ARTICLE NUMBER: 0066 In the format provided by the authors and unedited. Engineering CRISPR-Cpf1 crrnas and mrnas to maximize genome editing efficiency Bin Li 1, Weiyu Zhao 1, Xiao Luo 1, Xinfu
VOLUME: 1 ARTICLE NUMBER: 0066 In the format provided by the authors and unedited. Engineering CRISPR-Cpf1 crrnas and mrnas to maximize genome editing efficiency Bin Li 1, Weiyu Zhao 1, Xiao Luo 1, Xinfu
Supplementary Figure 1. CRISPR/Cas9-induced targeted mutations in TaGASR7, TaDEP1, TaNAC2, TaPIN1, TaLOX2 and TaGW2 genes in wheat protoplasts.
 Supplementary Figure 1. CRISPR/Cas9-induced targeted mutations in TaGASR7, TaDEP1, TaNAC2, TaPIN1, TaLOX2 and TaGW2 genes in wheat protoplasts. Lanes 1 and 2: digested CRISPR/Cas9-transformed protoplasts;
Supplementary Figure 1. CRISPR/Cas9-induced targeted mutations in TaGASR7, TaDEP1, TaNAC2, TaPIN1, TaLOX2 and TaGW2 genes in wheat protoplasts. Lanes 1 and 2: digested CRISPR/Cas9-transformed protoplasts;
Targeted complete next generation sequencing and quality control of transgenes and integration sites in CHO cell line development
 Targeted Locus Amplification Technology Targeted complete next generation sequencing and quality control of transgenes and integration sites in CHO cell line development Cergentis B.V. Yalelaan 62 3584
Targeted Locus Amplification Technology Targeted complete next generation sequencing and quality control of transgenes and integration sites in CHO cell line development Cergentis B.V. Yalelaan 62 3584
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
Genome-wide genetic screening with chemically-mutagenized haploid embryonic stem cells
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 Supplementary Information Genome-wide genetic screening with chemically-mutagenized haploid embryonic stem cells Josep V. Forment 1,2, Mareike Herzog
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 Supplementary Information Genome-wide genetic screening with chemically-mutagenized haploid embryonic stem cells Josep V. Forment 1,2, Mareike Herzog
Measurement of peritoneal macrophage apoptosis by Celigo plate imaging cytometer
 SUPPLEMENTAL METHODS Measurement of peritoneal macrophage apoptosis by Celigo plate imaging cytometer For Celigo experiments, 0.1 ml containing 5 x 10 4 cells was seeded into 96 well plates for 30 min
SUPPLEMENTAL METHODS Measurement of peritoneal macrophage apoptosis by Celigo plate imaging cytometer For Celigo experiments, 0.1 ml containing 5 x 10 4 cells was seeded into 96 well plates for 30 min
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1
 Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nucleaseinduced
 Supplementary Data Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nucleaseinduced mutations Suresh Ramakrishna 1, Seung Woo Cho 2, Sojung Kim 2, Myungjae Song 1, Ramu Gopalappa
Supplementary Data Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nucleaseinduced mutations Suresh Ramakrishna 1, Seung Woo Cho 2, Sojung Kim 2, Myungjae Song 1, Ramu Gopalappa
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/4/e1602814/dc1 Supplementary Materials for CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice Yu Zhang, Chengzu Long, Hui
advances.sciencemag.org/cgi/content/full/3/4/e1602814/dc1 Supplementary Materials for CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice Yu Zhang, Chengzu Long, Hui
Supplementary Information. Optimization of the production of knock-in alleles by CRISPR/Cas9 microinjection into the mouse zygote
 Supplementary Information Supplementary Figures S1 to S5 and Tables S1 to S3. Optimization of the production of knock-in alleles by CRISPR/Cas9 microinjection into the mouse zygote Aurélien Raveux, Sandrine
Supplementary Information Supplementary Figures S1 to S5 and Tables S1 to S3. Optimization of the production of knock-in alleles by CRISPR/Cas9 microinjection into the mouse zygote Aurélien Raveux, Sandrine
Supplementary Materials. China
 Supplementary Materials An Efficient Genotyping Method for Genome-modified Animals and Human Cells Generated with CRISPR/Cas9 System Xiaoxiao Zhu 1,2 *, Yajie Xu 1 *, Shanshan Yu 1 *, Lu Lu 1,2, Mingqin
Supplementary Materials An Efficient Genotyping Method for Genome-modified Animals and Human Cells Generated with CRISPR/Cas9 System Xiaoxiao Zhu 1,2 *, Yajie Xu 1 *, Shanshan Yu 1 *, Lu Lu 1,2, Mingqin
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Biology 4361 Developmental Biology Lecture 4. The Genetic Core of Development
 Biology 4361 Developmental Biology Lecture 4. The Genetic Core of Development The only way to get from genotype to phenotype is through developmental processes. - Remember the analogy that the zygote contains
Biology 4361 Developmental Biology Lecture 4. The Genetic Core of Development The only way to get from genotype to phenotype is through developmental processes. - Remember the analogy that the zygote contains
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1.
 Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplemental Figure 1 (Figure S1), related to Figure 1 Figure S1 provides evidence to demonstrate Nfatc1Cre is a mouse line that directed gene
 Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Information
 Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
PV92 PCR Bio Informatics
 Purpose of PCR Chromosome 16 PV92 PV92 PCR Bio Informatics Alu insert, PV92 locus, chromosome 16 Introduce the polymerase chain reaction (PCR) technique Apply PCR to population genetics Directly measure
Purpose of PCR Chromosome 16 PV92 PV92 PCR Bio Informatics Alu insert, PV92 locus, chromosome 16 Introduce the polymerase chain reaction (PCR) technique Apply PCR to population genetics Directly measure
The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells
 Supplementary Information The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells Bing Zhao 1,3, Zhen Qi 1,3, Yehua Li 1,3, Chongkai Wang 2,
Supplementary Information The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells Bing Zhao 1,3, Zhen Qi 1,3, Yehua Li 1,3, Chongkai Wang 2,
Supplementary Materials
 Supplementary Materials Supplementary Methods Supplementary Discussion Supplementary Figure 1 Calculated frequencies of embryo cells bearing bi-allelic alterations. Targeted indel mutations induced by
Supplementary Materials Supplementary Methods Supplementary Discussion Supplementary Figure 1 Calculated frequencies of embryo cells bearing bi-allelic alterations. Targeted indel mutations induced by
SUPPLEMENTARY FIG. S2. Expression of single HLA loci in shns- and shb 2 m-transduced MKs. Expression of HLA class I antigens (HLA-ABC) as well as
 Supplementary Data Supplementary Methods Flow cytometric analysis of HLA class I single locus expression Expression of single HLA loci (HLA-A and HLA-B) by shns- and shb 2 m-transduced megakaryocytes (MKs)
Supplementary Data Supplementary Methods Flow cytometric analysis of HLA class I single locus expression Expression of single HLA loci (HLA-A and HLA-B) by shns- and shb 2 m-transduced megakaryocytes (MKs)
Nature Immunology: doi: /ni Supplementary Figure 1. Construction of lnckdm2b RFP reporter and lnckdm2b-knockout mice.
 Supplementary Figure 1 Construction of lnckdm2b RFP reporter and lnckdm2b-knockout mice. (a) ILC3s (Lin CD45 lo CD90 hi ) were isolated from small intestines, followed by immunofluorescence staining. LncKdm2b
Supplementary Figure 1 Construction of lnckdm2b RFP reporter and lnckdm2b-knockout mice. (a) ILC3s (Lin CD45 lo CD90 hi ) were isolated from small intestines, followed by immunofluorescence staining. LncKdm2b
Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus α-hemolysin-mediated toxicity
 Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus α-hemolysin-mediated toxicity Sebastian Virreira Winter, Arturo Zychlinsky and Bart W. Bardoel Department of Cellular
Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus α-hemolysin-mediated toxicity Sebastian Virreira Winter, Arturo Zychlinsky and Bart W. Bardoel Department of Cellular
Figure S1 is related to Figure 1B, showing more details of outer segment of
 Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Incorporating SeqStudio Genetic Analyzer and Sanger sequencing into genome editing workflows
 Incorporating SeqStudio Genetic Analyzer and Sanger sequencing into genome editing workflows Stephen Jackson, Ph.D 27 May 2017 The world leader in serving science Key Applications for Genome Editing Research
Incorporating SeqStudio Genetic Analyzer and Sanger sequencing into genome editing workflows Stephen Jackson, Ph.D 27 May 2017 The world leader in serving science Key Applications for Genome Editing Research
a KYSE270-CON KYSE270-Id1
 a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
A Survey of Genetic Methods
 IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
Supplemental Data. Sethi et al. (2014). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
TheraLin. Universal Tissue Fixative Enabling Molecular Pathology
 TheraLin Universal Tissue Fixative Enabling Molecular Pathology TheraLin Universal Tissue Fixative Enabling Molecular Pathology Contents Page # TheraLin Universal Tissue Fixative 3 Introduction 5 Easy
TheraLin Universal Tissue Fixative Enabling Molecular Pathology TheraLin Universal Tissue Fixative Enabling Molecular Pathology Contents Page # TheraLin Universal Tissue Fixative 3 Introduction 5 Easy
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
SUPPLEMENTARY INFORMATION
 A diphtheria toxin resistance marker for in vitro and in vivo selection of stably transduced human cells Gabriele Picco, Consalvo Petti, Livio Trusolino, Andrea Bertotti and Enzo Medico SUPPLEMENTARY INFORMATION
A diphtheria toxin resistance marker for in vitro and in vivo selection of stably transduced human cells Gabriele Picco, Consalvo Petti, Livio Trusolino, Andrea Bertotti and Enzo Medico SUPPLEMENTARY INFORMATION
LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation
 LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation Mi Young Park, 1# Raki Sudan, 1# Neetu Srivastava, 1 Sudha Neelam, 1 Christie Youngs, 1 Jia- Wang Wang, 4 Robert W. Engelman, 5,6,7
LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation Mi Young Park, 1# Raki Sudan, 1# Neetu Srivastava, 1 Sudha Neelam, 1 Christie Youngs, 1 Jia- Wang Wang, 4 Robert W. Engelman, 5,6,7
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Zhang et al., RepID facilitates replication Initiation. Supplemental Information:
 Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Easi CRISPR for conditional and insertional alleles
 Easi CRISPR for conditional and insertional alleles C.B Gurumurthy, University Of Nebraska Medical Center Omaha, NE cgurumurthy@unmc.edu Types of Genome edits Gene disruption/inactivation Types of Genome
Easi CRISPR for conditional and insertional alleles C.B Gurumurthy, University Of Nebraska Medical Center Omaha, NE cgurumurthy@unmc.edu Types of Genome edits Gene disruption/inactivation Types of Genome
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb37 a mrna relative expression 1.3 1..8.5.3. CTR NGPS OCT4 SOX2 KLF4 c-myc b BANF1 mrna levels 1.2 1..8.6.4.2. plko.1 shbanf1 c Tra-1-6+ colonies 3 1 plko.1 shbanf1 d BANF1 locus Plasmid donor
DOI: 1.138/ncb37 a mrna relative expression 1.3 1..8.5.3. CTR NGPS OCT4 SOX2 KLF4 c-myc b BANF1 mrna levels 1.2 1..8.6.4.2. plko.1 shbanf1 c Tra-1-6+ colonies 3 1 plko.1 shbanf1 d BANF1 locus Plasmid donor
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
Supplementary Figures and Figure legends
 Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
