Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte
|
|
|
- Shavonne Harmon
- 5 years ago
- Views:
Transcription
1 Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte Carlo algorithm for one million generations to obtain a majority-rule consensus tree. Full-length amino acid sequences of 16 AtHD-ZIP IV family and five AtHD-ZIP III (outgroup) were analyzed. Posterior probabilities of key nodes are shown at the respective nodes. The scale bar indicates the number of amino acid changes per branch length. HDG2 is highlighted in red; AtML1 and PDF2 are highlighted in blue to show their close phylogenetic relationship with HDG2. Colored dots indicate known expression/promoter activities within the leaf epidermis with the following cell types: pink, uniformly in the protoderm (L1-layer); green, in trichomes; cyan, in stomatal-lineage cells (Rerie et al.,1994; Lu et al., 1996; Session et al., 1999; Abe et al. 2003; Nakamura et al., 2006).
2 Fig. S2. T-DNA insertion knockout alleles of HDG2, PDF2 and AtML1 show no obvious growth phenotypes. (A) RT- PCR analysis of HDG2, PDF2 and AtML1 from 10-day-old seedlings of wild-type (wt) and T-DNA insertion alleles, hdg2-2 (SALK_127828C), pdf2-2 (SALK_109425C), atml1-3 (SALK_128172) and atml1-4 (SALK_033408). Corresponding transcripts are undetectable in these T-DNA insertion alleles. ACT2 transcripts serve as a control. (B) 10-day-old seedlings of the corresponding genotypes. Images were taken under the same magnification. Scale bar: 5 mm. (C) Schematic diagram indicating the T-DNA insertion sites for three mutant alleles within the HDG2 gene. This diagram includes HDG2 genomic region from translational initiation codon to termination codon. Light-gray boxes, homeodomain; dark-gray boxes, START domain, which also contains a transcriptional activation domain of maize OCL1 (black boxes). Color-coded arrows correspond to locations of primer pairs used for RT-PCR analysis (see D). (D) RT-PCR analysis of three hdg2 T-DNA insertion alleles reveals expressions of aberrant transcripts of partial size, which are not derived from genomic DNA contamination based on their size. Each color code corresponds to the following primer pairs used for analysis: green, primers HDG2_1_XhoI and LBa1; blue, primers HDG2_1_XhoI and HDG2_1750.rc; red, HDG2_1168 and HDG2_2359.rc. See C for locations of primers. For primer sequence, see supplementary materials Table S1.
3 Fig. S3. Inducible ectopic overexpressors of HDG2, HDG2-SRDX and AtML1. (A,C) RT-PCR analysis of transgenic Arabidopsis lines carrying inducible HDG2, HDG2-SRDX and AtML1 with (+) or without ( ) estradiol induction. eif4a transcripts serve as a control. (B,D) Seedlings of the corresponding transgenic lines, HDG2 (top; 2 weeks old) and HDG2-SRDX (10 days old) with or without induction. Scale bars: 5 mm.
4 Fig. S4. Induced ectopic overexpression of HDG2 and AtML1 in additional four transgenic lines for each construct, each showing ectopic stomatal differentiation in mesophyll layers. (A,B) mps-pi stained optical sections of mesophyll layer of 12-dayold cotyledons from four independent transgenic lines harboring inducible HDG2 transgene (A) and four independent transgenic lines harboring inducible AtML1 transgene (B). Ectopic differentiation of internal stomata (yellow) is evident. Images were taken under the same magnification. Scale bar: 50 mm. (C,D) RT-PCR analysis of additional transgenic Arabidopsis lines carrying inducible HDG2 or AtML1 with (+) or without ( ) estradiol induction. eif4 transcripts serve as a control. The line number corresponds to the lines examined for phenotypes (A,B).
5 Fig. S5. Stomatal development defects in rosette leaves of hdg2 mutants and higher-order mutants in closely related HD-ZIP IV genes. (A) SI of 6-week-old abaxial rosette leaf 4 from wild type, atml1, pdf2, hdg2, hdg2 atml1 and hdg2 pdf2. hdg2-2 and atml1-3 alleles were used for the analysis. Bars indicate means (n=16-21); error bars indicate s.e.m. Total numbers of stomata counted: 1875 (wild type), 1224 (atml1), 1424 (pdf2), 1931 (hdg2), 1608 (hdg2 atml1) and 1749 (hdg2 pdf2). (B) MI of the six genotypes described above. Bars indicates means; error bars indicate s.e.m. Total numbers of meristemoids counted: 12 (wild type), 5 (atml1), 22 (pdf2), 40 (hdg2), 58 (hdg2 atml1) and 31 (hdg2 pdf2). (C) SLI of the six genotypes described above. Bars indicate means; error bars indicate s.e.m. Total numbers of cells counted: 7272 (wild type), 4795 (atml1), 6229 (pdf2), 8010 (hdg2), 7585 (hdg2 atml1) and 7123 (hdg2 pdf2). For B,C, genotypes with non-significant phenotypes were grouped together with a letter (Tukey s HSD test after One-way ANOVA). For C, only one genotype was significantly different from others (Tukey s HSD test; P<0.01). (D) Representative confocal images of 6-week-old abaxial rosette leaf 4 epidermis from six different genotypes. Images were taken under the same magnification. Scale bar: 20 mm. hdg2 mutant rosette leaf epidermis shows characteristic delayed stomatal differentiation, a phenotype exaggerated in hdg2 atml1 double mutant (brackets).
6 POLAR SCRM/ICE1 Fig. S6. Co-expression network of genes controlling stomatal development and/or protodermal differentiation. The network diagram was generated using ATTED II network drawer ( HDG2 is highlighted in red; known regulators of stomatal development are in green; known genes with protodermal expression are in blue; hexagons represent transcription factors. Thickness of lines (edges) represents mutual rank (MR) of co-expression; the higher the rank, the thicker the line. Dotted red line indicates known protein-protein interactions. Fig. S7. Expression of transcriptional and translational reporters of HDG2 in developing trichomes. (A,B) Confocal images of abaxial rosette leaf epidermis expressing HDG2pro::nls-3xGFP (A) and HDG2pro::HDG-GFP (B). Each developing trichome has very large nucleus (fluorescing green). Scale bars: 50 mm. Table S1. List of primer DNA sequences for real-time quantitative RT-PCR and RT-PCR. Download Table S1 Table S2. List of plasmid constructs generated in this study. Download Table S2 Table S3. DNA sequences used for yeast one-hybrid analysis. Download Table S3 Table S4. Bioinformatic analysis of promoter motifs in stomatal development genes. Overrepresentation analysis of promoter elements in 18 genes involved in stomatal development (EPF1, EPF2, ERECTA, ERL1, ERL2, TMM, SPCH, MUTE, FAMA, SCRM, SCRM2, HDG2, AtML1, POLAR, BASL, MYB88, FLP and SDD1) were performed with the Athena tool ( wsu.edu/athena/). Download Table S4
7 Movie 1. Complete optical sectioning of a wild-type 10-day-old cotyledon. Z stacks were made from mps-pi stained confocal optical sections in 1 mm intervals. Movie 2. Complete optical sectioning of a 10-day-old cotyledon from HDG2 induced ectopic overexpression. Z stacks were made from mps-pi stained confocal optical sections in 1 mm intervals. Movie 3. Complete optical sectioning of a 10-day-old cotyledon from AtML1 induced ectopic overexpression. Z stacks were made from mps-pi stained confocal optical sections in 1 mm intervals.
Supplementary Fig. 1. Microscopic image of cotyledon adaxial epidermis of 3-dayold Col, epf2-1, ros1-4 and rdd. Scale bar, 100 m.
 Supplementary Fig. 1. Microscopic image of cotyledon adaxial epidermis of 3-dayold Col, epf2-1, ros1-4 and rdd. Scale bar, 100 m. Supplementary Fig. 2. The small-cell-cluster phenotype of different ros1
Supplementary Fig. 1. Microscopic image of cotyledon adaxial epidermis of 3-dayold Col, epf2-1, ros1-4 and rdd. Scale bar, 100 m. Supplementary Fig. 2. The small-cell-cluster phenotype of different ros1
Supplemental Data. Guo et al. (2015). Plant Cell /tpc
 Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
scrm-d/+ scrm-d Supplemental Fig. 1.
 Kanaoka, Pillitteri, Fujii, Yoshida, Bogenschutz, Takabayashi, Zhu, and Torii (2008) SCRM/ICE1 and SCRM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation WT scrm-d/+
Kanaoka, Pillitteri, Fujii, Yoshida, Bogenschutz, Takabayashi, Zhu, and Torii (2008) SCRM/ICE1 and SCRM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation WT scrm-d/+
SUPPLEMENTARY INFORMATION
 AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Information
 Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
sides of the aleurone (Al) but it is excluded from the basal endosperm transfer layer
 Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Supplemental Data. Borg et al. Plant Cell (2014) /tpc
 Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
SUPPLEMENTARY INFORMATION
 Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 )
 Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Supplemental Information. Boundary Formation through a Direct. Threshold-Based Readout. of Mobile Small RNA Gradients
 Developmental Cell, Volume 43 Supplemental Information Boundary Formation through a Direct Threshold-Based Readout of Mobile Small RNA Gradients Damianos S. Skopelitis, Anna H. Benkovics, Aman Y. Husbands,
Developmental Cell, Volume 43 Supplemental Information Boundary Formation through a Direct Threshold-Based Readout of Mobile Small RNA Gradients Damianos S. Skopelitis, Anna H. Benkovics, Aman Y. Husbands,
Nature Genetics: doi: /ng.3556 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE. Supplementary Figure 1
 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
To investigate the heredity of the WFP gene, we selected plants that were homozygous
 Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
embryos. Asterisk represents loss of or reduced expression. Brackets represent
 Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Chemical hijacking of auxin signaling with an engineered auxin-tir1
 1 SUPPLEMENTARY INFORMATION Chemical hijacking of auxin signaling with an engineered auxin-tir1 pair Naoyuki Uchida 1,2*, Koji Takahashi 2*, Rie Iwasaki 1, Ryotaro Yamada 2, Masahiko Yoshimura 2, Takaho
1 SUPPLEMENTARY INFORMATION Chemical hijacking of auxin signaling with an engineered auxin-tir1 pair Naoyuki Uchida 1,2*, Koji Takahashi 2*, Rie Iwasaki 1, Ryotaro Yamada 2, Masahiko Yoshimura 2, Takaho
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
- 1 - Supplemental Data
 - 1-1 Supplemental Data 2 3 4 5 6 7 8 9 Supplemental Figure S1. Differential expression of AtPIP Genes in DC3000-inoculated plants. Gene expression in leaves was analyzed by real-time RT-PCR and expression
- 1-1 Supplemental Data 2 3 4 5 6 7 8 9 Supplemental Figure S1. Differential expression of AtPIP Genes in DC3000-inoculated plants. Gene expression in leaves was analyzed by real-time RT-PCR and expression
WiscDsLox485 ATG < > //----- E1 E2 E3 E4 E bp. Col-0 arr7 ARR7 ACTIN7. s of mrna/ng total RNA (x10 3 ) ARR7.
 A WiscDsLox8 ATG < > -9 +0 > ---------//----- E E E E E UTR +9 UTR +0 > 00bp B S D ol-0 arr7 ol-0 arr7 ARR7 ATIN7 D s of mrna/ng total RNA opie 0. ol-0 (x0 ) arr7 ARR7 Supplemental Fig.. Genotyping and
A WiscDsLox8 ATG < > -9 +0 > ---------//----- E E E E E UTR +9 UTR +0 > 00bp B S D ol-0 arr7 ol-0 arr7 ARR7 ATIN7 D s of mrna/ng total RNA opie 0. ol-0 (x0 ) arr7 ARR7 Supplemental Fig.. Genotyping and
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplemental Data. Pillitteri et al. (2011) /tpc
 A B C D Supplemental Figure 1. Images of representative 5-dag seedlings used in this study.(a) WT; (B) spch; (C) scrm-d; (D) scrm-d. mute. Images were taken under the same magnification. Scale bar, 1 mm.
A B C D Supplemental Figure 1. Images of representative 5-dag seedlings used in this study.(a) WT; (B) spch; (C) scrm-d; (D) scrm-d. mute. Images were taken under the same magnification. Scale bar, 1 mm.
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast. Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice
 A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for
 Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Supplemental Data. Zhou et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. Confirmation of mutant mapping results. (A) Complementation assay with stably transformed genomic fragments (ComN-N) (2 kb upstream of TSS and 1.5 kb downstream of TES) and CaMV
Supplemental Figure 1. Confirmation of mutant mapping results. (A) Complementation assay with stably transformed genomic fragments (ComN-N) (2 kb upstream of TSS and 1.5 kb downstream of TES) and CaMV
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues.
 Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Supplementary information
 Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Nature Genetics: doi: /ng Supplementary Figure 1. ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets.
 Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplemental Data. Benstein et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Information
 Supplemental Information Supplemental Figure 1. The Heterologous Yeast System for Screening of Arabidopsis JA Transporters. (A) Exogenous JA inhibited yeast cell growth. Yeast cells were diluted to various
Supplemental Information Supplemental Figure 1. The Heterologous Yeast System for Screening of Arabidopsis JA Transporters. (A) Exogenous JA inhibited yeast cell growth. Yeast cells were diluted to various
A Repressor Complex Governs the Integration of
 Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Supplemental data. Zhao et al. (2009). The Wuschel-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice.
 Supplemental data. Zhao et al. (2009). The Wuschel-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. A B Supplemental Figure 1. Expression of WOX11p-GUS WOX11-GFP
Supplemental data. Zhao et al. (2009). The Wuschel-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. A B Supplemental Figure 1. Expression of WOX11p-GUS WOX11-GFP
Sperm cells are passive cargo of the pollen tube in plant fertilization
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
Supplemental Data. Zhang et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. uvs90 gene cloning The T-DNA insertion in uvs90 was identified using thermal asymmetric interlaced (TAIL)-PCR. Three rounds of amplification were performed; the second (2 nd ) and
Supplemental Figure 1. uvs90 gene cloning The T-DNA insertion in uvs90 was identified using thermal asymmetric interlaced (TAIL)-PCR. Three rounds of amplification were performed; the second (2 nd ) and
Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis
 Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis seedlings expressing ERF109pro-GUS. The GUS staining
Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis seedlings expressing ERF109pro-GUS. The GUS staining
Supplementary Information. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato
 Supplementary Information The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato Uri Krieger 1, Zachary B. Lippman 2 *, and Dani Zamir 1 * 1. The Hebrew University of Jerusalem Faculty
Supplementary Information The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato Uri Krieger 1, Zachary B. Lippman 2 *, and Dani Zamir 1 * 1. The Hebrew University of Jerusalem Faculty
Supplemental Data. Dai et al. (2013). Plant Cell /tpc Absolute FyPP3. Absolute
 A FyPP1 Absolute B FyPP3 Absolute Dry seeds Imbibed 24 hours Dry seeds Imbibed 24 hours C ABI5 Absolute Dry seeds Imbibed 24 hours Supplemental Figure 1. Expression of FyPP1, FyPP3 and ABI5 during seed
A FyPP1 Absolute B FyPP3 Absolute Dry seeds Imbibed 24 hours Dry seeds Imbibed 24 hours C ABI5 Absolute Dry seeds Imbibed 24 hours Supplemental Figure 1. Expression of FyPP1, FyPP3 and ABI5 during seed
Supplementary Materials: 1. Supplementary Figures S1-S9. 2. Supplementary Tables S1-S2
 Supplementary Materials: 1. Supplementary Figures S1-S9 2. Supplementary Tables S1-S2 S1 1. Supplementary Figures Fig. S1. Genotypes of tcp20 mutants. Fig. S2. Root phenotypes of tcp20 mutants. Fig. S3.
Supplementary Materials: 1. Supplementary Figures S1-S9 2. Supplementary Tables S1-S2 S1 1. Supplementary Figures Fig. S1. Genotypes of tcp20 mutants. Fig. S2. Root phenotypes of tcp20 mutants. Fig. S3.
Poly-A signals. amirna. pentr/d-topo for Gateway cloning
 popon inducible system A Poly-A signals CaMV 35S minimal promoter TMV omega translational enhancer HYG KAN RR 35S 35S LhGR GUS Ω pop6 Ω Gene construct B - Dex + Dex CACC prs300 (mir319a) CACC amirna pentr/d-topo
popon inducible system A Poly-A signals CaMV 35S minimal promoter TMV omega translational enhancer HYG KAN RR 35S 35S LhGR GUS Ω pop6 Ω Gene construct B - Dex + Dex CACC prs300 (mir319a) CACC amirna pentr/d-topo
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplemental Data. Wang et al. (2017). Plant Cell /tpc NIP1;2 NIP7;1
 Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
Supplemental Figure 1. Immunoblot analysis of wild-type and mutant forms of JAZ3
 Supplemental Data. Chung and Howe (2009). A Critical Role for the TIFY Motif in Repression of Jasmonate Signaling by a Stabilized Splice Variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis.
Supplemental Data. Chung and Howe (2009). A Critical Role for the TIFY Motif in Repression of Jasmonate Signaling by a Stabilized Splice Variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis.
Supplemental Figure 1. VLN5 retains conserved residues at both type 1 and type 2 Ca 2+ -binding
 Supplemental Figure 1. VLN5 retains conserved residues at both type 1 and type 2 Ca 2+ -binding sites in the G1 domain. Multiple sequence alignment was performed with DNAMAN6.0.40. Secondary structural
Supplemental Figure 1. VLN5 retains conserved residues at both type 1 and type 2 Ca 2+ -binding sites in the G1 domain. Multiple sequence alignment was performed with DNAMAN6.0.40. Secondary structural
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
AD BD TOC1. Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between
 AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
What are the Properties of the Protein EMB2777?
 Of EMB2777 and F14J22.20 - exons & introns by Hanbee O What are the Properties of the Protein EMB2777?! Embryo defective / seed lethal transcriptional factor o Sas10 / U3 ribonucleoprotein (Utp) family
Of EMB2777 and F14J22.20 - exons & introns by Hanbee O What are the Properties of the Protein EMB2777?! Embryo defective / seed lethal transcriptional factor o Sas10 / U3 ribonucleoprotein (Utp) family
Supplemental Data. Seo et al. (2014). Plant Cell /tpc
 Supplemental Figure 1. Protein alignment of ABD1 from other model organisms. The alignment was performed with H. sapiens DCAF8, M. musculus DCAF8 and O. sativa Os10g0544500. The WD40 domains are underlined.
Supplemental Figure 1. Protein alignment of ABD1 from other model organisms. The alignment was performed with H. sapiens DCAF8, M. musculus DCAF8 and O. sativa Os10g0544500. The WD40 domains are underlined.
Supplementary Data Supplementary Figures
 Supplementary Data Supplementary Figures Supplementary Figure 1. Pi04314 is expressed during infection, each GFP-Pi04314 fusion is stable and myr GFP-Pi04314 is removed from the nucleus while NLS GFP-Pi04314
Supplementary Data Supplementary Figures Supplementary Figure 1. Pi04314 is expressed during infection, each GFP-Pi04314 fusion is stable and myr GFP-Pi04314 is removed from the nucleus while NLS GFP-Pi04314
Supplemental Figure S1. Nucleotide and deduced amino acid sequences of pepper CaHSP70a (Capsicum annuum heat shock protein 70a) cdna.
 Supplemental Figure S1. Nucleotide and deduced amino acid sequences of pepper (Capsicum annuum heat shock protein 70a) cdna. Translation initiation codon is shown in bold typeface; termination codon is
Supplemental Figure S1. Nucleotide and deduced amino acid sequences of pepper (Capsicum annuum heat shock protein 70a) cdna. Translation initiation codon is shown in bold typeface; termination codon is
Supplementary Materials for
 www.sciencemag.org/cgi/content/full/science.1245533/dc1 Supplementary Materials for A Mechanism for Reorientation of Cortical Microtubule Arrays Driven by Microtubule Severing Jelmer J. Lindeboom, Masayoshi
www.sciencemag.org/cgi/content/full/science.1245533/dc1 Supplementary Materials for A Mechanism for Reorientation of Cortical Microtubule Arrays Driven by Microtubule Severing Jelmer J. Lindeboom, Masayoshi
Fig. S1. Semi-quantitative RT-PCR analysis of KOBITO1, ERECTA, ERL2 and actin transcripts in 15-day-old seedlings. Expression of KOBITO1 is not
 Fig. S1. Semi-quantitative RT-PCR analysis of KOBITO1, ERECTA, ERL2 and actin transcripts in 15-day-old seedlings. Expression of KOBITO1 is not changed in either or erl1 erl2 mutants. The mutation does
Fig. S1. Semi-quantitative RT-PCR analysis of KOBITO1, ERECTA, ERL2 and actin transcripts in 15-day-old seedlings. Expression of KOBITO1 is not changed in either or erl1 erl2 mutants. The mutation does
Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and
 Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and Tin TFBS motifs and located in the flanking or intronic
Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and Tin TFBS motifs and located in the flanking or intronic
Supplemental Data. Bai et al. Plant Cell. (2012) /tpc A
 A B Unconserved Conserved AIF4 AIF2 AIF3 AIF1 UPB1 IBH1 Supplemental Figure 1. IBH1, UPB1 and AIFs belong to the same HLH family. (A), Part of a phylogenetic tree constructed using conserved domains shows
A B Unconserved Conserved AIF4 AIF2 AIF3 AIF1 UPB1 IBH1 Supplemental Figure 1. IBH1, UPB1 and AIFs belong to the same HLH family. (A), Part of a phylogenetic tree constructed using conserved domains shows
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09861 & &' -(' ()*+ ')(+,,(','-*+,&,,+ ',+' ' 23,45/0*6787*9:./09 ;78?4?@*+A786?B- &' )*+*(,-* -(' ()*+ ')(+,,(','-*+,&,,+ ',+'./)*+*(,-*..)*+*(,-*./)*+*(,-*.0)*+*(,-*..)*+*(,-*
doi:10.1038/nature09861 & &' -(' ()*+ ')(+,,(','-*+,&,,+ ',+' ' 23,45/0*6787*9:./09 ;78?4?@*+A786?B- &' )*+*(,-* -(' ()*+ ')(+,,(','-*+,&,,+ ',+'./)*+*(,-*..)*+*(,-*./)*+*(,-*.0)*+*(,-*..)*+*(,-*
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/5/244/ra72/dc1 Supplementary Materials for An Interaction Between BZR1 and DELLAs Mediates Direct Signaling Crosstalk Between Brassinosteroids and Gibberellins
www.sciencesignaling.org/cgi/content/full/5/244/ra72/dc1 Supplementary Materials for An Interaction Between BZR1 and DELLAs Mediates Direct Signaling Crosstalk Between Brassinosteroids and Gibberellins
0.5% Sucrose. Supplemental Data. Chao et al. (2011). Plant Cell /tpc Col-0 tsc10a-2. Root length (mm)
 A 0% 0.5% 1% Col-0 tsc10a-2 B Root length (mm) 35 30 25 20 15 10 5 0 0% 0.50% 1% 2% 4% Sucrose concentration 2% 4% Sucrose Col-0 tsc10a-2 Supplemental Figure 1. The effect of sucrose on root growth of
A 0% 0.5% 1% Col-0 tsc10a-2 B Root length (mm) 35 30 25 20 15 10 5 0 0% 0.50% 1% 2% 4% Sucrose concentration 2% 4% Sucrose Col-0 tsc10a-2 Supplemental Figure 1. The effect of sucrose on root growth of
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
Supplemental Figure 1. Rosette Leaf Morphology of Single, Double and Triple Mutants of eid3, phya-201 and phyb-5. Photographs of Ler wild type,
 Supplemental Figure 1. Rosette Leaf Morphology of Single, Double and Triple Mutants of eid3, phya-201 and phyb-5. Photographs of Ler wild type, phya-201, phyb-5, phya-201 phyb-5, eid3, phya-201 eid3, phyb-5
Supplemental Figure 1. Rosette Leaf Morphology of Single, Double and Triple Mutants of eid3, phya-201 and phyb-5. Photographs of Ler wild type, phya-201, phyb-5, phya-201 phyb-5, eid3, phya-201 eid3, phyb-5
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Experimental Tools and Resources Available in Arabidopsis. Manish Raizada, University of Guelph, Canada
 Experimental Tools and Resources Available in Arabidopsis Manish Raizada, University of Guelph, Canada Community website: The Arabidopsis Information Resource (TAIR) at http://www.arabidopsis.org Can order
Experimental Tools and Resources Available in Arabidopsis Manish Raizada, University of Guelph, Canada Community website: The Arabidopsis Information Resource (TAIR) at http://www.arabidopsis.org Can order
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Erhard et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. c1-hbr allele structure. Diagram of the c1-hbr allele found in stocks segregating 1:1 for rpd1-1 and rpd1-2 homozygous mutants showing the presence of a 363 base pair (bp) Heartbreaker
Supplemental Figure 1. c1-hbr allele structure. Diagram of the c1-hbr allele found in stocks segregating 1:1 for rpd1-1 and rpd1-2 homozygous mutants showing the presence of a 363 base pair (bp) Heartbreaker
Map-Based Cloning of Qualitative Plant Genes
 Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
Supplemental Data. Jing et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Characterization of epp1 Mutants. (A) Cotyledon angles of 5-d-old Col wild-type (gray bars) and epp1-1 (black bars) seedlings under red (R), far-red (FR) and blue (BL) light conditions,
Supplemental Figure 1. Characterization of epp1 Mutants. (A) Cotyledon angles of 5-d-old Col wild-type (gray bars) and epp1-1 (black bars) seedlings under red (R), far-red (FR) and blue (BL) light conditions,
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase
 A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase tag insertion at the 3 end using CRISPR-Cas9 genome editing technique Sambuddha Basu 1, 3, Levi Adams 1, 3, Subhrangshu
A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase tag insertion at the 3 end using CRISPR-Cas9 genome editing technique Sambuddha Basu 1, 3, Levi Adams 1, 3, Subhrangshu
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
Supplemental Data. Li et al. (2015). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1: Characterization of asr3 T-DNA knockout lines and complementation transgenic lines. (A) The scheme of At2G33550 (ASR3) with gray boxes indicating exons and dash
Supplemental Data Supplemental Figure 1: Characterization of asr3 T-DNA knockout lines and complementation transgenic lines. (A) The scheme of At2G33550 (ASR3) with gray boxes indicating exons and dash
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Supplemental Figure 1
 Supplemental Figure 1 A gta2-1 gta2-2 1kb AT4G08350 B Col-0 gta2-1 LP+RP+LBb1.3 C Col-0 gta2-2 LP+RP+LB1 D Col-0 gta2-2 gta2-1 GTA2 TUBLIN E Col0 gta2-1 gta2-2 Supplemental Figure 1. Phenotypic analysis
Supplemental Figure 1 A gta2-1 gta2-2 1kb AT4G08350 B Col-0 gta2-1 LP+RP+LBb1.3 C Col-0 gta2-2 LP+RP+LB1 D Col-0 gta2-2 gta2-1 GTA2 TUBLIN E Col0 gta2-1 gta2-2 Supplemental Figure 1. Phenotypic analysis
Supplementary Material
 Supplementary Material The Cerato-Platanin protein Epl-1 from Trichoderma harzianum is involved in mycoparasitism, plant resistance induction and self cell wall protection Eriston Vieira Gomes 1, Mariana
Supplementary Material The Cerato-Platanin protein Epl-1 from Trichoderma harzianum is involved in mycoparasitism, plant resistance induction and self cell wall protection Eriston Vieira Gomes 1, Mariana
Supplemental Data. Furlan et al. Plant Cell (2017) /tpc
 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure.
Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure.
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
SUPPLEMENTAL FILES. Supplemental Figure 1. Expression domains of Arabidopsis HAM orthologs in both shoot meristem and root tissues.
 SUPPLEMENTAL FILES SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Expression domains of Arabidopsis HAM orthologs in both shoot meristem and root tissues. RT-PCR amplification of AtHAM1, AtHAM2, AtHAM3,
SUPPLEMENTAL FILES SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Expression domains of Arabidopsis HAM orthologs in both shoot meristem and root tissues. RT-PCR amplification of AtHAM1, AtHAM2, AtHAM3,
Supplementary Information. Isl2b regulates anterior second heart field development in zebrafish
 Supplementary Information Isl2b regulates anterior second heart field development in zebrafish Hagen R. Witzel 1, Sirisha Cheedipudi 1, Rui Gao 1, Didier Y.R. Stainier 2 and Gergana D. Dobreva 1,3* 1 Origin
Supplementary Information Isl2b regulates anterior second heart field development in zebrafish Hagen R. Witzel 1, Sirisha Cheedipudi 1, Rui Gao 1, Didier Y.R. Stainier 2 and Gergana D. Dobreva 1,3* 1 Origin
(A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1
 SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
Supplementary Information
 Supplementary Information Supplementary Fig. 1. Seed dormancy and germination responses of RVE1 and PIF1. (a) Diagram of RVE1 and the T-DNA insertion of the rve1-2 mutant (SAIL_326_A01). Black boxes represent
Supplementary Information Supplementary Fig. 1. Seed dormancy and germination responses of RVE1 and PIF1. (a) Diagram of RVE1 and the T-DNA insertion of the rve1-2 mutant (SAIL_326_A01). Black boxes represent
Chinese Academy of Sciences, Datun Road, Chaoyang District, Beijing, 10101, China
 SUPPLEMENTARY INFORMATION Title: Bi-directional processing of pri-mirnas with branched terminal loops by Arabidopsis Dicer-like1 Hongliang Zhu 1,2,3, Yuyi Zhou 1,2,4, Claudia Castillo-González 1,2, Amber
SUPPLEMENTARY INFORMATION Title: Bi-directional processing of pri-mirnas with branched terminal loops by Arabidopsis Dicer-like1 Hongliang Zhu 1,2,3, Yuyi Zhou 1,2,4, Claudia Castillo-González 1,2, Amber
Supplemental Figure 1. Transcript profiles of Arabidopsis IPMS1 and IPMS2 in different tissues and developmental stages.
 Transcript level (intensity) Supplemental Data. Xing et al. Plant Cell (2017) 10.1105/tpc.17.00186 1600 1400 1200 1000 800 600 400 200 0 IPMS1 IPMS2 Supplemental Figure 1. Transcript profiles of Arabidopsis
Transcript level (intensity) Supplemental Data. Xing et al. Plant Cell (2017) 10.1105/tpc.17.00186 1600 1400 1200 1000 800 600 400 200 0 IPMS1 IPMS2 Supplemental Figure 1. Transcript profiles of Arabidopsis
Supplementary Information. c d e
 Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
Supplemental Data. Wu and Xue (2010). Plant Cell /tpc
 Supplemental Data. Wu and Xue (21). Plant Cell 1.115/tpc.11.75564 A P1-S P1-A P2-S P2-A P3-S P3-A P4-S P4-A B Relative expression Relative expression C 5. 4.5 4. 3.5 3. 2.5 2. 1.5 1..5 5. 4.5 4. 3.5 3.
Supplemental Data. Wu and Xue (21). Plant Cell 1.115/tpc.11.75564 A P1-S P1-A P2-S P2-A P3-S P3-A P4-S P4-A B Relative expression Relative expression C 5. 4.5 4. 3.5 3. 2.5 2. 1.5 1..5 5. 4.5 4. 3.5 3.
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplemental Figure 1.
 Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Data. Osakabe et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Phylogenetic analysis of KUP in various species. The amino acid sequences of KUPs from green algae and land plants were identified with a BLAST search and aligned using ClustalW
Supplemental Figure 1. Phylogenetic analysis of KUP in various species. The amino acid sequences of KUPs from green algae and land plants were identified with a BLAST search and aligned using ClustalW
Fig. S1. TPL and TPL N176H protein interactions. (A) Semi-in vivo pull-down assays using recombinant GST N-TPL and GST N-TPL N176H fusions and
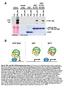 Fig. S1. TPL and TPL N176H protein interactions. (A) Semi-in vivo pull-down assays using recombinant GST N-TPL and GST N-TPL N176H fusions and transgenic Arabidopsis TPL-HA lysates. Immunoblotting of input
Fig. S1. TPL and TPL N176H protein interactions. (A) Semi-in vivo pull-down assays using recombinant GST N-TPL and GST N-TPL N176H fusions and transgenic Arabidopsis TPL-HA lysates. Immunoblotting of input
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Supplemental Figure 1. Conserved regions of the kinase domain of PEPR1, PEPR2, CLV1 and BRI1.
 Supplemental Figure 1. Conserved regions of the kinase domain of PEPR1, PEPR2, CLV1 and BRI1. The asterisks and colons indicate the important residues for ATP-binding pocket and substrate binding pocket,
Supplemental Figure 1. Conserved regions of the kinase domain of PEPR1, PEPR2, CLV1 and BRI1. The asterisks and colons indicate the important residues for ATP-binding pocket and substrate binding pocket,
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
that two types of cis-acting elements control the abaxial epidermis-specific
 FEBS Letters 583 (2009) 3711 3717 journal homepage: www.febsletters.org Two types of cis-acting elements control the abaxial epidermis-specific transcription of the MIR165a and MIR166a genes Xiaozhen Yao,
FEBS Letters 583 (2009) 3711 3717 journal homepage: www.febsletters.org Two types of cis-acting elements control the abaxial epidermis-specific transcription of the MIR165a and MIR166a genes Xiaozhen Yao,
Genome Sequence Assembly
 Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
