Supplementary Information
|
|
|
- Angelica Barker
- 5 years ago
- Views:
Transcription
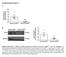
1 Supplementary Information Supplementary Figure 1: Identification of new regulators of MuSC by a proteome-based shrna screen. (a) FACS plots of GFP + and GFP - cells from Pax7 ICN -Z/EG (upper panel) and Pax7 ZsGreen (lower panel) reporter mice. (b) Immunofluorescence staining of myogenic factors in freshly isolated, quiescent (QSC) and 3 day cultured activated (ASC) satellite cells from Pax7 ICN -Z/EG mice. (c) Expansion and differentiation of FACS purified MuSC from Pax7 ICN - Z/EG mice in vitro. Scale bar 100 µm. (d) Upper panel: Images of a 384-well plate used for HTS. Lower panel: representative images of candidate target mrnas that promoted (Nf1, Cd180 and Fndc3b), or inhibited expansion of MuSC (Pax7, Prmt5 and Mustn1) or had no effects (Atg7) after shrna-mediated knockdown compared to control vectors (plko.1). Scale bar 50 µm. 1
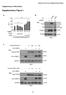
2 Supplementary Figure 2 2
3 Supplementary Figure 2: Prmt5 is dispensable for short-term maintenance of MuSC and skeletal muscle integrity. (a) Schematic outline of the Prmt5 gene locus with the floxed exon 7 that can be deleted by Cre recombinase-mediated recombination. Red arrowheads indicate positions of loxp sites. (b) Schematic outline of the TAM administration regimen. (c) RT-qPCR analysis of Prmt5 in isolated quiescent satellite cells of Ctrl (n=3) and Prmt5 sko (n=4) mice. Error bars represent standard deviations of the mean (t-test: **p < 0.01). (d) Representative images of control and Prmt5 sko mice after TAM administration for 21 days. (e) Bodyweight of control and Prmt5 sko mice after administration of TAM for 3 weeks. (n=3, each). Error bars represent standard deviations of the mean (t-test: ns p>0.5, n=3). (f) H&E staining of TA and Soleus muscles after TAM treatment of control and Prmt5 sko mice (n=3, each). Scale bar 20 µm. (g, h) Numbers of Pax7 expressing cells (Pax7 + ) on TA cryosections (g) or freshly isolated FDB myofibers (h) from control and Prmt5 sko mice 3 weeks after TAM treatment obtained by immunofluorescence staining (n=3, each). The numbers of Pax7 + cells per 10 mm 2 section area (g) or 100 myofibers (h) are displayed. Error bars represent standard deviations of the mean (t-test: ns p>0.5). (i) Schematic outline of the TAM regimen and CTX injection used for analysis of short-term (7 days) and long-term (4 months) muscle regeneration in control and Prmt5 sko littermates (n=3, each). Muscle regeneration was analyzed at different time points as indicated. (j, k) H&E (j) and Masson s Trichrome staining (k) of injured or non-injured TA muscles 7 days or 4 months after CTX injection showing impaired muscle regeneration and fibrosis of injured muscle in Prmt5 sko mice compared to control littermates (n=3, each). Scale bar 20 µm. 3
4 Supplementary Figure 3: RNA-seq and GO-analysis of up- and down-regulated genes in Prmt5 deficient MuSC. (a) Heat map of selected up- and down-regulated genes after inactivation of Prmt5 in MuSC (n=2, each). (b) GO term analysis of biological processes that are affected by the loss of Prmt5 in MuSCs (n=2, each). 4
5 Supplementary Figure 4: (a, b, c) Uncropped blots for Figure 6. (d) Uncropped gels for Figure 7. 5
6 Genotyping Primer Sequence (5'-->3') Pax7 CreERT2 Forward ACTAGGCTCCACTCTGTCCTTC Reverse GCAGATGTAGGGACATTCCAGTG ZsGreen Forward CTGCATGTACCACGAGTCCA Reverse GTCAGCTGCCACTTCTGGTT Rosa26 YFP RosaFA AAAGTCGCTCTGAGTTGTTAT RosaRF Rosa-SpliAC GGAGCGGGAGAAATGGATATG CATCAAGGAAACCCTGGACTACTG Mdx mdx_forward GCGCGAAACTCATCAAATATGCGTGTTAGTGT mdx WT Reverse mdx MT Reverse GATACGCTGCTTTAATGCCTTTAGTCACTCAGATAGTTG AAGCCATTTTG CGGCCTGTCACTCAGATAGTTGAAGCCATTTTA p21 p21 exon 144 GAACTTTGACTTCGTCACGG p21 genou p21 PGK neo3 ACAACACCTCCTGGTCAGAGG GAAGAACGAGATCAGCAG Pax7 ICN-Cre ck188 GCTCTGGATACACCTGAGTCT ck256 ba97 ck172 TCGGCCTTCTTCTAGGTTCTGCTC GATCTGGACGAAGAGCATCA GGATAGTGAAACAGGGGCAA Prmt5 Loxp3 Forward CCAGAACTTCCTCTGGTTTCTGG LoxP3 Reverse GAAAGCTGTGTGTCCTCACAC Supplementary Table 1: List of primers used for genotyping of different mouse strains. 6
7 Antibody Antigen Application Working Concentration Manufacturer Anti-Pax7 mouse Pax7 IF 0.5 µg/ml DSHB Anti-Myf5 rabbit Myf5 IF 0.2 µg/ml Santa Cruz Anti-MyoD rabbit MyoD IF 1 µg/ml Santa Cruz Anti-MyoD1 mouse MyoD IF 0.5 µg/ml LSBio Anti-Myogenin rabbit Myogenin IF 0.2 µg/ml Santa Cruz Anti-Myogenin mouse Myogenin IF 1:20 dilution DSHB MF20 Myosin heavy chain IF 1:20 dilution DSHB Anti-Gapdh Gapdh WB 1:2000 dilution Cell signaling Anti-Prmt5 Prmt5 WB,ChIP 0.3 µg/ml, 1 µg/100 µl Active Motif Anti-p53 p53 WB,ChIP 1:1000, 1:100 dilution Cell signaling Anti-H3R8me2s H3R8me2s ChIP 1 ug/100 µl Novus Anti-Histone H3 Histone H3 ChIP 1 ug/100 µl Abcam Rabbit IgG ChIP 1 ug/100 µl Diagenode Mouse IgG ChIP 1 ug/100 µl Diagenode Anti-CD11b PE-Cy7 CD11b FACS 2 µg/ml ebioscience Anti-CD45 PE CD45 FACS 2 µg/ml ebioscience Anti-CD31 PE PECAM1 FACS 2 µg/ml ebioscience Anti-CXCR4 APC CXCR4 FACS 2 µg/ml ebioscience Anti-CD34 A450 CD34 FACS 2 µg/ml ebioscience Anti-YFP YFP IF 0.2µg/ml Evrogen Supplementary Table 2: List of antibodies. Antibodies were used for immunofluorescence (IF), western blotting (WB), chromatin immunoprecipitation (ChIP), and FACS analysis (FACS). 7
8 ChIP-qPCR Primer forward (5-3 ) Primer reverse (5-3 ) p21 enhancer CACAGGGAAGAGAGCTCCAG AGCCAGGGCTACACAGAGAA p21 TSS TCCACAGCGATATCCAGACA GGACACACCTGTGACTCTGG p21 p53 binding site CAAGCCCTTCCCAGACTTCC TCTAGAGATCGCTGCCCAGA p21 CpG island CCTGTTTCGCGGTAGCCA ACAATGAGTCACCTCCTCGC Supplementary Table 3: List of primers used for ChIP-qPCR experiments. 8
9 RT-qPCR Primer forward(5-3 ) Primer reverse(5-3 ) Prmt5 Exon5-7 AGAATGCCCCGACTACACAC AGCCTCTGCTGCACCTTAGA p21 CGGTGTCAGAGTCTAGGGGA ATTGGAGTCAGGCGCAGATC p53 TGCTGTGCAATTAAAGGCTGT CGTGTTCTCCGAGATACTTGGT Pax7 GCTACCAGTACAGCCAGTATG GTCACTAAGCATGGGTAGATG Myf5 CCACCTCCAACTGCTCTGAC GCTTCAGGGCTTCTTTTCCT MyoD GAATGGCTACGACACCGCCTACTAC CCTACGGTGGTGCGCCCTCTGC Myogenin TTGCTCAGCTCCCTCAACCA TGGGCTGGGTGTTAGTCTTA m36b4 AGATTCGGGATATGCTGTTGGC TCGGGTCCTAGACCAGTGTTC Mdm4 AGTCAGGTGCGGCCAAAA CCCAAAAGATCTCCACCACA Mdm4 splicing TGTGGTGGAGATCTTTTGGG TCAGTTCTTTTTCTGGGATTGG Supplementary Table 4: List of primers used for RT-qPCR analysis. 9
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and
 Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Supplementary Figure 1: Analysis of monocyte subsets and lineage relationships. (a) Gating strategy for definition of MDP and cmop populations in BM
 Supplementary Figure 1: Analysis of monocyte subsets and lineage relationships. (a) Gating strategy for definition of MDP and cmop populations in BM of Cx3cr1 GFP/+ mice related to Fig. 1a. MDP was defined
Supplementary Figure 1: Analysis of monocyte subsets and lineage relationships. (a) Gating strategy for definition of MDP and cmop populations in BM of Cx3cr1 GFP/+ mice related to Fig. 1a. MDP was defined
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature875 a promoter firefly luciferase CNS b Supplementary Figure 1. Dual luciferase assays on enhancer activity of CNS1, 2, and 3. a. promoter sequence was inserted upstream of firefly luciferase
doi: 1.138/nature875 a promoter firefly luciferase CNS b Supplementary Figure 1. Dual luciferase assays on enhancer activity of CNS1, 2, and 3. a. promoter sequence was inserted upstream of firefly luciferase
SUPPLEMENTAL INFORMATION
 UTX/KDM6A demethylase activity is required for satellite cell-mediated muscle regeneration Hervé Faralli 1,2, Chaochen Wang 3, Kiran Nakka 1,2, Soji Sebastian 1,, Aissa Benyoucef 1,2, Lenan Zhuang 3, Alphonse
UTX/KDM6A demethylase activity is required for satellite cell-mediated muscle regeneration Hervé Faralli 1,2, Chaochen Wang 3, Kiran Nakka 1,2, Soji Sebastian 1,, Aissa Benyoucef 1,2, Lenan Zhuang 3, Alphonse
STAT3 signaling controls satellite cell expansion and skeletal muscle repair
 SUPPLEMENTARY INFORMATION STAT3 signaling controls satellite cell expansion and skeletal muscle repair Matthew Timothy Tierney 1 *, Tufan Aydogdu 2,3 *, David Sala 2, Barbora Malecova 2, Sole Gatto 2,
SUPPLEMENTARY INFORMATION STAT3 signaling controls satellite cell expansion and skeletal muscle repair Matthew Timothy Tierney 1 *, Tufan Aydogdu 2,3 *, David Sala 2, Barbora Malecova 2, Sole Gatto 2,
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in
 Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary Figure 1 Muscle dystrophic phenotype is absent at P6 in PKO mice. (a) Size
 Supplementary Figure 1 Muscle dystrophic phenotype is absent at P6 in PKO mice. (a) Size comparison of the P6 control and PKO mice. (b) H&E staining of hind-limb muscles from control and PKO mice at P6.
Supplementary Figure 1 Muscle dystrophic phenotype is absent at P6 in PKO mice. (a) Size comparison of the P6 control and PKO mice. (b) H&E staining of hind-limb muscles from control and PKO mice at P6.
Supplemental Table/Figure Legends
 MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
Multiple layers of B cell memory with different effector functions. Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos,
 Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Supplementary Figure S1: (A) Schematic representation of the Jarid2 Flox/Flox
 Supplementary Figure S1: (A) Schematic representation of the Flox/Flox locus and the strategy used to visualize the wt and the Flox/Flox mrna by PCR from cdna. An example of the genotyping strategy is
Supplementary Figure S1: (A) Schematic representation of the Flox/Flox locus and the strategy used to visualize the wt and the Flox/Flox mrna by PCR from cdna. An example of the genotyping strategy is
DRG Pituitary Cerebral Cortex
 Liver Spinal cord Pons Atg5 -/- Atg5 +/+ DRG Pituitary Cerebral Cortex WT KO Supplementary Figure S1 Ubiquitin-positive IBs accumulate in Atg5 -/- tissues. Atg5 -/- neonatal tissues were fixed and decalcified.
Liver Spinal cord Pons Atg5 -/- Atg5 +/+ DRG Pituitary Cerebral Cortex WT KO Supplementary Figure S1 Ubiquitin-positive IBs accumulate in Atg5 -/- tissues. Atg5 -/- neonatal tissues were fixed and decalcified.
Supplementary Material. Levels of S100B protein drive the reparative process in acute muscle injury and muscular dystrophy
 Supplementary Material Levels of protein drive the reparative process in acute muscle injury and muscular dystrophy Francesca Riuzzi 1,4 *, Sara Beccafico 1,4 *, Roberta Sagheddu 1,4, Sara Chiappalupi
Supplementary Material Levels of protein drive the reparative process in acute muscle injury and muscular dystrophy Francesca Riuzzi 1,4 *, Sara Beccafico 1,4 *, Roberta Sagheddu 1,4, Sara Chiappalupi
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector.
 Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Material. Disruption of spatiotemporal hypoxic signaling causes congenital heart disease
 Supplementary Material Disruption of spatiotemporal hypoxic signaling causes congenital heart disease Xuejun Yuan, Hui Qi, Xiang Li, Fan Wu, Jian Fang, Eva Bober, Gergana Dobreva, Yonggang Zhou, Thomas
Supplementary Material Disruption of spatiotemporal hypoxic signaling causes congenital heart disease Xuejun Yuan, Hui Qi, Xiang Li, Fan Wu, Jian Fang, Eva Bober, Gergana Dobreva, Yonggang Zhou, Thomas
The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells
 Supplementary Information The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells Bing Zhao 1,3, Zhen Qi 1,3, Yehua Li 1,3, Chongkai Wang 2,
Supplementary Information The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells Bing Zhao 1,3, Zhen Qi 1,3, Yehua Li 1,3, Chongkai Wang 2,
Supplemental Material
 Supplemental Material Figure S1.(A) Immuno-staining of freshly isolated Myf5-Cre:ROSA-YFP fiber (B) Satellite cell enumeration in WT and cko mice from resting hind limb muscles. Bulk hind limb muscles
Supplemental Material Figure S1.(A) Immuno-staining of freshly isolated Myf5-Cre:ROSA-YFP fiber (B) Satellite cell enumeration in WT and cko mice from resting hind limb muscles. Bulk hind limb muscles
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Nature Immunology: doi: /ni Supplementary Figure 1. Zranb1 gene targeting.
 Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure S1 Evi/Wls and Wnt3 are overexpressed in epithelial cells in colon cancers. (a) Validation of the specificity of the Wnt3
 Supplementary Figure S1 Evi/Wls and Wnt3 are overexpressed in epithelial cells in colon cancers. (a) Validation of the specificity of the Wnt3 antibody. HCT116 cells were reverse transfected with sicontrol
Supplementary Figure S1 Evi/Wls and Wnt3 are overexpressed in epithelial cells in colon cancers. (a) Validation of the specificity of the Wnt3 antibody. HCT116 cells were reverse transfected with sicontrol
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
H3K36me3 polyclonal antibody
 H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
(A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: WT; lower
 Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
supplementary information
 DOI: 10.1038/ncb2015 Figure S1 Confirmation of Sca-1 CD34 stain specificity using isotypematched control antibodies. Skeletal muscle preparations were stained and gated as described in the text (Hoechst
DOI: 10.1038/ncb2015 Figure S1 Confirmation of Sca-1 CD34 stain specificity using isotypematched control antibodies. Skeletal muscle preparations were stained and gated as described in the text (Hoechst
Supplementary Fig S1 Nutlin-3a treatment does not affect cell cycle progression in the absence
 Supplementary Figure Legends Supplementary Fig S1 Nutlin-3a treatment does not affect cell cycle progression in the absence of p53 or p21. HCT116 cells which were null for either p53 (A) or p21 (B) were
Supplementary Figure Legends Supplementary Fig S1 Nutlin-3a treatment does not affect cell cycle progression in the absence of p53 or p21. HCT116 cells which were null for either p53 (A) or p21 (B) were
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL MATERIALS AND METHODS Generation of TSPOΔ/Δ murine embryonic fibroblasts Embryos were harvested from 13.5-day pregnant TSPOfl/fl mice. After dissection to eviscerate and remove the
SUPPLEMENTAL MATERIAL MATERIALS AND METHODS Generation of TSPOΔ/Δ murine embryonic fibroblasts Embryos were harvested from 13.5-day pregnant TSPOfl/fl mice. After dissection to eviscerate and remove the
Supplementary Figure 1.
 Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Myofibroblasts in kidney fibrosis. LeBleu et al.
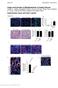 Origin and Function of Myofibroblasts in Kidney Fibrosis Valerie S. LeBleu, Gangadhar Taduri, Joyce O Connell, Yingqi Teng, Vesselina G. Cooke, Craig Woda, Hikaru Sugimoto and Raghu Kalluri Supplementary
Origin and Function of Myofibroblasts in Kidney Fibrosis Valerie S. LeBleu, Gangadhar Taduri, Joyce O Connell, Yingqi Teng, Vesselina G. Cooke, Craig Woda, Hikaru Sugimoto and Raghu Kalluri Supplementary
Supplemental material
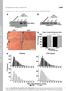 Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Supplemental Information. The TRAIL-Induced Cancer Secretome. Promotes a Tumor-Supportive Immune. Microenvironment via CCR2
 Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Supplementary Fig. 1
 a FL (1-2266) NL (1-1190) CL (1191-2266) HA-ICE1: - HA-ICE1: - - - FLAG-ICE2: + + + + FLAG-ELL: + + + + + + IP: anti-ha FLAG-ICE2 HA-ICE1-FL HA-ICE1-NL HA-ICE1-CL FLAG-ICE2 b IP: anti-ha FL (1-2266) NL
a FL (1-2266) NL (1-1190) CL (1191-2266) HA-ICE1: - HA-ICE1: - - - FLAG-ICE2: + + + + FLAG-ELL: + + + + + + IP: anti-ha FLAG-ICE2 HA-ICE1-FL HA-ICE1-NL HA-ICE1-CL FLAG-ICE2 b IP: anti-ha FL (1-2266) NL
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl
 Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Supplementary Table S1
 Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Control + SDS + 2BME. Control + SDS. Control
 Supplementary Figure 1 2BME:2-mercaptoethanol Control Control + SDS Control + SDS + 2BME Control Control + SDS Control + SDS + 2BME Control Control + SDS Control + SDS + 2BME Ephrin-B2 Oligomers Ephrin-B2
Supplementary Figure 1 2BME:2-mercaptoethanol Control Control + SDS Control + SDS + 2BME Control Control + SDS Control + SDS + 2BME Control Control + SDS Control + SDS + 2BME Ephrin-B2 Oligomers Ephrin-B2
Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin
 Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin locus. The EcoRI restriction site between exons M1 and M2
Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin locus. The EcoRI restriction site between exons M1 and M2
p53 is activated in response to disruption of the pre-mrna splicing machinery.
 p53 is activated in response to disruption of the pre-mrna splicing machinery. Nerea Allende-Vega, Saurabh Dayal, Usha Agarwala, Alison Sparks, Christophe Bourdon and Mark K. Saville. Jean- Supplementary
p53 is activated in response to disruption of the pre-mrna splicing machinery. Nerea Allende-Vega, Saurabh Dayal, Usha Agarwala, Alison Sparks, Christophe Bourdon and Mark K. Saville. Jean- Supplementary
Supplementary Information. Title BRD3 and BRD4 BET Bromodomain Proteins Differentially Regulate Skeletal Myogenesis
 Supplementary Information Title BRD3 and BRD4 BET Bromodomain Proteins Differentially Regulate Skeletal Myogenesis Authors Thomas C. Roberts 1,2, Usue Etxaniz 1, Alessandra Dall Agnese 1, Shwu-Yuan Wu
Supplementary Information Title BRD3 and BRD4 BET Bromodomain Proteins Differentially Regulate Skeletal Myogenesis Authors Thomas C. Roberts 1,2, Usue Etxaniz 1, Alessandra Dall Agnese 1, Shwu-Yuan Wu
Supplementary Information
 Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
0.9 5 H M L E R -C tr l in T w is t1 C M
 a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Chromatin immunoprecipitation (ChIP) ES and FACS-sorted GFP+/Flk1+ cells were fixed in 1% formaldehyde and sonicated until fragments of an average
 Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted
Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
a KYSE270-CON KYSE270-Id1
 a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
a Lamtor1 (gene) b Lamtor1 (mrna) c WT Lamtor1 Lamtor1 flox Lamtor2 A.U. p = LysM-Cre Lamtor3 Lamtor4 Lamtor5 BMDMs: Φ WT Φ KO β-actin WT BMDMs
 a Lamtor (gene) b Lamtor (mrna) c BMDMs: Φ WT Φ KO Lamtor flox 8 bp LysM-Cre 93 bp..5 p =.4 WT BMDMs: Φ WT Φ KO Lamtor (protein) BMDMs: Φ WT Φ KO Lamtor 8 kda Lamtor 4 kda Lamtor3 4 kda Lamtor4 kda Lamtor5.5
a Lamtor (gene) b Lamtor (mrna) c BMDMs: Φ WT Φ KO Lamtor flox 8 bp LysM-Cre 93 bp..5 p =.4 WT BMDMs: Φ WT Φ KO Lamtor (protein) BMDMs: Φ WT Φ KO Lamtor 8 kda Lamtor 4 kda Lamtor3 4 kda Lamtor4 kda Lamtor5.5
Supplementary Information
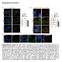 Supplementary Information Supplementary Figure S1 (a) P-cRAF colocalizes with LC3 puncta. Immunofluorescence (IF) depicting colocalization of P-cRAF (green) and LC3 puncta (red) in NIH/3T3 cells treated
Supplementary Information Supplementary Figure S1 (a) P-cRAF colocalizes with LC3 puncta. Immunofluorescence (IF) depicting colocalization of P-cRAF (green) and LC3 puncta (red) in NIH/3T3 cells treated
Supplementary Figure 1
 Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
A population of Nestin expressing progenitors in the cerebellum exhibits increased tumorigenicity
 A population of Nestin expressing progenitors in the cerebellum exhibits increased tumorigenicity Peng Li 1,2, Fang Du 1, Larra W. Yuelling 1, Tiffany Lin 3, Renata E. Muradimova 1, Rossella Tricarico
A population of Nestin expressing progenitors in the cerebellum exhibits increased tumorigenicity Peng Li 1,2, Fang Du 1, Larra W. Yuelling 1, Tiffany Lin 3, Renata E. Muradimova 1, Rossella Tricarico
Anti-ERK1/2, anti-p-erk1/2, anti-p38 anti-p-msk1 (Thr581) and anti-p-msk2
 Supplementary methods Antibodies Anti-ERK1/2, anti-p-erk1/2, anti-p38 anti-p-msk1 (Thr581) and anti-p-msk2 polyclonal were from Cell Signalling and the anti-p-histone H3(Ser1) antibody was from Upstate.
Supplementary methods Antibodies Anti-ERK1/2, anti-p-erk1/2, anti-p38 anti-p-msk1 (Thr581) and anti-p-msk2 polyclonal were from Cell Signalling and the anti-p-histone H3(Ser1) antibody was from Upstate.
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supporting Online Material. Methods
 Supporting Online Material Methods Cell culture Cells were prepared from muscle tissue by enzymatic dissociation as previously described. Cells were plated on gelatin coated dishes in a 1:1mixture (v/v)
Supporting Online Material Methods Cell culture Cells were prepared from muscle tissue by enzymatic dissociation as previously described. Cells were plated on gelatin coated dishes in a 1:1mixture (v/v)
Yalin Emre, Magali Irla, Isabelle Dunand- Sauthier, Romain Ballet, Mehdi Meguenani, Stephane Jemelin, Christian Vesin, Walter Reith and Beat A.
 Supplementary information Thymic epithelial cell expansion through matricellular protein CYR61 boosts progenitor homing and T- cell output Yalin Emre, Magali Irla, Isabelle Dunand- Sauthier, Romain Ballet,
Supplementary information Thymic epithelial cell expansion through matricellular protein CYR61 boosts progenitor homing and T- cell output Yalin Emre, Magali Irla, Isabelle Dunand- Sauthier, Romain Ballet,
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles
 Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
Pei et al. Supplementary Figure S1
 Pei et al. Supplementary Figure S1 C H-CUL9: + + + + + Myc-ROC1: - - + + + U2OS/pcDN3 U2OS/H-CUL9 U2OS/ + H-CUL9 IP: -H -myc input -H -myc 1 2 3 4 5 H-CUL9 Myc-ROC1 -H -H -H -H H-CUL9: wt RR myc-roc1:
Pei et al. Supplementary Figure S1 C H-CUL9: + + + + + Myc-ROC1: - - + + + U2OS/pcDN3 U2OS/H-CUL9 U2OS/ + H-CUL9 IP: -H -myc input -H -myc 1 2 3 4 5 H-CUL9 Myc-ROC1 -H -H -H -H H-CUL9: wt RR myc-roc1:
Nanog-Luc. R-Luc. 40 HA-Klf Relative protein level of Klf USP21. Vector USP2 USP21 *** *** *** ***
 :Flag Relative protein level of Relative protein level of : Flag Vec 21LV 21SV 2 : HA HA- HA- HA- HA-Klf4 Relative mrna expression Relative protein level of Relative protein level of Relative protein level
:Flag Relative protein level of Relative protein level of : Flag Vec 21LV 21SV 2 : HA HA- HA- HA- HA-Klf4 Relative mrna expression Relative protein level of Relative protein level of Relative protein level
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Figure S1. Efficacy of STIM1 knockdown by electroporation of STIM1 sirnas. (a) Western blot of FDB muscles from 4 mice treated with either STIM1-specific sirna (KD)
SUPPLEMENTARY INFORMATION Supplementary Figure S1. Efficacy of STIM1 knockdown by electroporation of STIM1 sirnas. (a) Western blot of FDB muscles from 4 mice treated with either STIM1-specific sirna (KD)
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplemental Figure 1: Podocyte specific knockdown of Klf4 in Podocin-Cre Klf4 flox/flox mice was confirmed. (A) Primary glomerular epithelial cells
 Supplemental Figure 1: Podocyte specific knockdown of Klf4 in Podocin-Cre Klf4 flox/flox mice was confirmed. (A) Primary glomerular epithelial cells were isolated from 10-week old Podocin-Cre Klf4 flox/flox
Supplemental Figure 1: Podocyte specific knockdown of Klf4 in Podocin-Cre Klf4 flox/flox mice was confirmed. (A) Primary glomerular epithelial cells were isolated from 10-week old Podocin-Cre Klf4 flox/flox
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figures
 Supplementary Figures Fig. S1. Specificity of perilipin antibody in SAT compared to various organs. (A) Images of SAT at E18.5 stained with perilipin and CD31 antibody. Scale bars: 100 μm. SAT stained
Supplementary Figures Fig. S1. Specificity of perilipin antibody in SAT compared to various organs. (A) Images of SAT at E18.5 stained with perilipin and CD31 antibody. Scale bars: 100 μm. SAT stained
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1 Characterization of GSCs. a. Immunostaining of primary GSC spheres from GSC lines. Nestin (neural progenitor marker, red), TLX (green). Merged images of nestin,
Supplementary Figures Supplementary Fig. 1 Characterization of GSCs. a. Immunostaining of primary GSC spheres from GSC lines. Nestin (neural progenitor marker, red), TLX (green). Merged images of nestin,
Supplementary Fig. 1 Generation and genotyping of ThPOK / mice
 Supplementary Fig. 1 Generation and genotyping of ThPOK / mice (a). The strategy used to target the ThPOK locus is schematically depicted. A targeting construct was generated by inserting a loxp site (filled
Supplementary Fig. 1 Generation and genotyping of ThPOK / mice (a). The strategy used to target the ThPOK locus is schematically depicted. A targeting construct was generated by inserting a loxp site (filled
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplementary Figures and Figure legends
 Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
Description of Supplementary Files. File name: Supplementary Information Description: Supplementary figures and supplementary tables.
 Description of Supplementary Files File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Supplementary Data 1 Description: Differential expression
Description of Supplementary Files File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Supplementary Data 1 Description: Differential expression
neutrophils (CD11b+, the total Statistical multiple
 Supplementary Figure 1. (A) Wild type and Vim / mice were treated with (24mg/kg LPS); lungs were harvested at 48h and enzymaticallyy digested. CD45+ cells were excluded from the total cell population and
Supplementary Figure 1. (A) Wild type and Vim / mice were treated with (24mg/kg LPS); lungs were harvested at 48h and enzymaticallyy digested. CD45+ cells were excluded from the total cell population and
Supplementary Figure 1. Bone density was decreased in osteoclast-lineage cell specific Gna13 deficient mice. (a-c) PCR genotyping of mice by mouse
 Supplementary Figure 1. Bone density was decreased in osteoclast-lineage cell specific Gna13 deficient mice. (a-c) PCR genotyping of mice by mouse tail DNA. Primers were designed to detect Gna13-WT/f (~400bp/470bp)
Supplementary Figure 1. Bone density was decreased in osteoclast-lineage cell specific Gna13 deficient mice. (a-c) PCR genotyping of mice by mouse tail DNA. Primers were designed to detect Gna13-WT/f (~400bp/470bp)
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1.
 Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
amplify the conditional allele (2-lox) and recombined allele (1-lox) following tamoxifen
 Supplementary data Supplementary Table 1. Real-time PCR primer sequences Supplementary Fig. 1. (A) Genomic map of the Vhl (2-lox) conditional allele. Numbered boxes represent exon 1, which is targeted
Supplementary data Supplementary Table 1. Real-time PCR primer sequences Supplementary Fig. 1. (A) Genomic map of the Vhl (2-lox) conditional allele. Numbered boxes represent exon 1, which is targeted
Supplementary Information Supplementary Figure 1
 Bararia, Kwok et al, Supplementary Page 1 Supplementary Information Supplementary Figure 1 S1 Bararia, Kwok et al, Supplementary Page 2 Supplementary Figure 1 (cont.) S2 Bararia, Kwok et al, Supplementary
Bararia, Kwok et al, Supplementary Page 1 Supplementary Information Supplementary Figure 1 S1 Bararia, Kwok et al, Supplementary Page 2 Supplementary Figure 1 (cont.) S2 Bararia, Kwok et al, Supplementary
Supplementary Table 3d: PCR primers for genomic sequencing-based phasereconstruction
 Supplementary Table 3: Detailed lists of grnas, primers, donor constructs, antibodies, Gene Expression Omnibus (GEO) identifiers for Chip-seq and DHSs datasets and CRISPR/Cas9 genome edited cell lines
Supplementary Table 3: Detailed lists of grnas, primers, donor constructs, antibodies, Gene Expression Omnibus (GEO) identifiers for Chip-seq and DHSs datasets and CRISPR/Cas9 genome edited cell lines
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
SUPPLEMENTARY MATERIALS
 SUPPLEMENTARY MATERIALS Supplementary Table S1: List of primers used for ChIP analysis and Oligo-pull-down assay. Genes Sequence mppargc1a FW 5 -GCGAGGTTTCTGCTTAGTCA-3 (-2317) RV 5 -ACAATGACTAAGCAGAAACCTCG-3
SUPPLEMENTARY MATERIALS Supplementary Table S1: List of primers used for ChIP analysis and Oligo-pull-down assay. Genes Sequence mppargc1a FW 5 -GCGAGGTTTCTGCTTAGTCA-3 (-2317) RV 5 -ACAATGACTAAGCAGAAACCTCG-3
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell
 Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
The cell-cycle regulator c-myc is essential for the formation and maintenance of germinal centers
 SUPPLEMENTARY INFORMATION The cell-cycle regulator c-myc is essential for the formation and maintenance of germinal centers Dinis Pedro Calado 1,2, Yoshiteru Sasaki 3, Susana A Godinho 4, Alex Pellerin
SUPPLEMENTARY INFORMATION The cell-cycle regulator c-myc is essential for the formation and maintenance of germinal centers Dinis Pedro Calado 1,2, Yoshiteru Sasaki 3, Susana A Godinho 4, Alex Pellerin
Flowcytometry-based purity analysis of peritoneal macrophage culture.
 Liao et al., KLF4 regulates macrophage polarization Revision of Manuscript 45444-RG- Supplementary Figure Legends Figure S Flowcytometry-based purity analysis of peritoneal macrophage culture. Thioglycollate
Liao et al., KLF4 regulates macrophage polarization Revision of Manuscript 45444-RG- Supplementary Figure Legends Figure S Flowcytometry-based purity analysis of peritoneal macrophage culture. Thioglycollate
Mannen et al., http :// /cgi /content /full /jcb /DC1
 Supplemental material JCB Mannen et al., http ://www.jcb.org /cgi /content /full /jcb.201601024 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Characterization of SNB components. (A) SNB localization of Venus-tagged
Supplemental material JCB Mannen et al., http ://www.jcb.org /cgi /content /full /jcb.201601024 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Characterization of SNB components. (A) SNB localization of Venus-tagged
SUPPLEMENTARY DATA. Mice
 Mice All animals were handled in accordance with protocols approved by the Animal Use and Care Committees of Washington State University. Wild-type (WT) C57BL/6 mice, green fluorescent mice (EGFP (B6-Tg(CAG-EGFP)131Osb/LeySopJ,
Mice All animals were handled in accordance with protocols approved by the Animal Use and Care Committees of Washington State University. Wild-type (WT) C57BL/6 mice, green fluorescent mice (EGFP (B6-Tg(CAG-EGFP)131Osb/LeySopJ,
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
The trithorax protein partner menin acts in tandem with EZH2 to suppress C/EBPα and differentiation in MLL-AF9 leukemia
 SUPPLEMENTARY APPENDIX The trithorax protein partner menin acts in tandem with EZH2 to suppress C/EBPα and differentiation in MLL-AF9 leukemia Austin T. Thiel, Zijie Feng, Dhruv K. Pant, Lewis A. Chodosh,
SUPPLEMENTARY APPENDIX The trithorax protein partner menin acts in tandem with EZH2 to suppress C/EBPα and differentiation in MLL-AF9 leukemia Austin T. Thiel, Zijie Feng, Dhruv K. Pant, Lewis A. Chodosh,
Supplementary information. Supplementary Figures
 Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/4/e1602814/dc1 Supplementary Materials for CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice Yu Zhang, Chengzu Long, Hui
advances.sciencemag.org/cgi/content/full/3/4/e1602814/dc1 Supplementary Materials for CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice Yu Zhang, Chengzu Long, Hui
Cell extracts and western blotting RNA isolation and real-time PCR Chromatin immunoprecipitation (ChIP)
 Cell extracts and western blotting Cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed with lysis buffer. 1 Total cell extracts were separated by SDS-PAGE and transferred to nitrocellulose
Cell extracts and western blotting Cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed with lysis buffer. 1 Total cell extracts were separated by SDS-PAGE and transferred to nitrocellulose
