STAT3 signaling controls satellite cell expansion and skeletal muscle repair
|
|
|
- Leslie Leonard
- 5 years ago
- Views:
Transcription
1 SUPPLEMENTARY INFORMATION STAT3 signaling controls satellite cell expansion and skeletal muscle repair Matthew Timothy Tierney 1 *, Tufan Aydogdu 2,3 *, David Sala 2, Barbora Malecova 2, Sole Gatto 2, Pier Lorenzo Puri 2,4, Lucia Latella 4,5 and Alessandra Sacco 2 1 Graduate School of Biomedical Sciences, Sanford-Burnham Medical Research Institute, North Torrey Pines Road, La Jolla, CA Development, Aging and Regeneration Program (DARe), Sanford-Burnham Medical Research Institute, North Torrey Pines Rd, La Jolla, CA 92037, USA 3 Life Sciences Solutions, Thermo Fisher Scientific, 5781 Van Allen Way, Carlsbad, CA 92008, USA 4 Epigenetics and Regenerative Medicine, IRCCS Fondazione Santa Lucia, Via del Fosso di Fiorano 64, Rome, Italy 5 Institute of Translational Pharmacology, National Research Council of Italy, Via Fosso del Cavaliere 100, Rome, Italy * These authors equally contributed to this work. Contents: Supplementary Figures 1-8 Supplementary Figure Legends Supplementary Table 1 Supplementary Methods Supplementary References
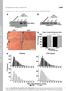
2 Supplementary Figure 1. STAT3 is transiently activated in SC during skeletal muscle regeneration. Representative images of uninjured and injured (notexin, NTX) tibialis anterior muscles of male, 2 mth C57BL/6 mice at 5 and 10 d after injury (green = Pax7, red = pstat3, white = Laminin, blue = Hoechst). Arrow indicates sublaminar Pax7 + pstat3 + SC, white arrowhead indicates a Pax7 pstat3 + SC. Scale bar, 20 µm. 2
3 Supplementary Figure 2. Stat3 promotes SC myogenic lineage progression. (a) Stat3 and Myod1 mrna levels in SC isolated from male, 2 mth C57BL/6 mice and infected with sh-stat3 or shcontrol and cultured in growth media for 72 h (n = 4). (b) Percentage of Pax7 + SC infected with sh- Stat3 or sh-control and cultured in growth media for 72 h (n = 2). (c) Representative images of TUNEL performed in SC infected with sh-stat3 or sh-control and cultured in growth media for 72 h (left). Quantification of the percentage of apoptotic TUNEL + nuclei upon sh-stat3 infection (n = 3, right). (d) Stat3 mrna levels in non-infected SC or infected with sh-stat3 or sh-control lentivirus were maintained in culture in growth media either in the absence or in the presence of recombinant Interleukin-6 (IL6) (100ng ml 1 ) for 96 h (n = 4). Data are represented as average ± s.e.m. Student s t test was used for all statistical analyses (***P < 0.001, **P < 0.01, *P < 0.05) except (d), where Oneway ANOVA with Tukey s post test was used. 3
4 Supplementary Figure 3. Stat3 regulates MyoD transcription. (a). Schematic representation of the Myod1-Luc reporter plasmid (top). Myod1 reporter plasmid transfected into 293 cells either alone or together with sh-stat3 (or sh-control) and Myod1 expression plasmids. Cells were cultured for 48 h in growth media after transfection before collection and analysis. Quantification of Firefly Luciferase activity normalized to Renilla luciferase (n = 7). (b) Chromatin ImmunoPrecipitation (ChIP) of H3K27Ac acetylation or IgG control in primary mouse myoblasts isolated from male, 2 mth C57BL/6 mice in the indicated regulatory regions: 590bp Myod1, +59bp Socs3, 1bp Oct4, +524bp Pdx1 and +1526bp IgH enhancer (n = 3). Graphs show H3K27Ac enrichment expressed as percentage of input DNA. (c) Quantification of Stat3 and Myod1 mrna levels in C2C12 myoblasts 96 h after infection with a lentivirus expressing sh-stat3 or sh-control (n = 3). (d) Western blotting of C2C12 cell extracts with the indicated antibodies after infection with a lentivirus expressing sh-stat3 or sh-control. Data 4
5 are represented as average ± s.e.m. Student s t test was used for all statistical analyses (***P < 0.001, **P < 0.01, *P < 0.05). 5
6 Supplementary Figure 4. Stat3 gene ablation does not break SC quiescence in healthy muscles. (a) Schematic representation of tamoxifen (tmx) treatment in male and female, 2-3 mth Pax7-CreER;STAT3 f/f and Pax7-CreER;STAT3 WT mice. EdU was administered daily for 5 d before harvesting. (b) Quantification of the percentage of Pax7 + SC isolated from uninjured skeletal muscles after 2 h in culture (n = 3). (c) Quantification of total SC number isolated from uninjured skeletal muscle of tmx-treated Pax7-CreER;STAT3 f/f and Pax7-CreER;STAT3 WT mice following culture in growth media for 96 h (n = 3). (d) Quantification of Pax7 + SC per mm 2 upon tmx treatment at either 2 or 3 mth of age (n = 3). (e) Representative images of uninjured skeletal muscle of tmx-treated Pax7- CreER;STAT3 f/f and Pax7-CreER;STAT3 WT mice (white = laminin). Scale bar, 100 µm (left). Quantification of average myofiber cross-sectional area (center) or their distribution (n = 3, right). 6
7 Data are represented as average ± s.e.m. Student s t test was used for all statistical analyses (***P < 0.001, **P < 0.01, *P < 0.05). 7
8 Supplementary Figure 5. Stat3 inhibitor treatment transiently increases the number of Pax7+ SC in the tissue, but does not affect mature myofibers. (a) Stat3, Myod1 and Socs3 mrna levels in SC isolated from male, 2 mth C57BL/6 mice and cultured for 96 h in the presence of vehicle (PBS) or 50 µm Stat3 inhibitor (n = 3). (b) Quantification of total SC number after culture in growth conditions for 96 h in the presence of vehicle (PBS) or Stat3 inhibitor (n = 10). (c) Schematic 8
9 representation of Stat3 inhibitor treatment and skeletal muscle injury in male, 2 mth wild-type mice for SC analysis at 5 d post-injury. (d) Representative images of Pax7 + SC in regenerating muscles, treated with Stat3 inhibitor or vehicle (PBS), 5 d post-injury (green = Pax7, white = Laminin, blue = nuclei). Scale bar, 20 µm (left). Quantification of Pax7 + SC per mm 2 (n = 3, right). (e) Schematic representation of Stat3 inhibitor treatment in uninjured muscles. (f) Representative images of hematoxylin and eosin staining of skeletal muscles 4 d after treatment. Scale bar, 50 µm (left). Quantification of average myofiber cross-sectional area (center) or their distribution (n = 3, right). Data are represented as average ± s.e.m. Student s t test was used for all statistical analyses (***P < 0.001, *P < 0.05). 9
10 Supplementary Figure 6. Stat3 transient inhibition enhances skeletal muscle tissue repair in aged muscles. (a) Quantification of myofiber cross sectional area distribution from male, 2 mth and 24 mth wild-type mice 5 d post-injury and treated with the Stat3 inhibitor or vehicle (PBS) (n = 3). (b) Representative images of hematoxylin and eosin staining of skeletal muscles from 2 mth and 24 mth wild-type mice 25 d post-injury, treated with the Stat3 inhibitor or vehicle (PBS). Scale bar, 50µm. (c) 10
11 Quantification of average myofiber cross-sectional area from male, 2 mth and 24 mth wild-type mice 25 d post-injury treated with the Stat3 inhibitor or vehicle (PBS) (n = 3). (d) Quantification of myofiber cross-sectional area distribution from male, 2 mth and 24 mth wild-type mice 25 d post-injury, treated with the Stat3 inhibitor or vehicle (PBS) (n = 3). Data are represented as average ± s.e.m. Student s t test was used for all statistical analyses (**P < 0.01, *P < 0.05). 11
12 Supplementary Figure 7. Stat3 inhibition enhances skeletal muscle tissue repair in dystrophic muscles. Quantification of myofiber cross-sectional area distribution from male, 2 mth mx/mtr G2 mice at 28 d, treated with the Stat3 inhibitor or vehicle (PBS) (n = 3). Data are represented as average ± s.e.m. Student s t test was used for all statistical analyses (*P < 0.05). 12
13 Supplementary Figure 8. The effects of STAT3 inhibitor treatment is conserved in human muscle cells. (a) Representative immunofluorescence images of pstat3 in human myoblasts isolated from the quadriceps of patients yr of age and treated with the Stat3 inhibitor or vehicle for 72 h (blue = pstat3, white = nuclei). (b) Representative immunofluorescence images of TUNEL in human myoblasts treated with the Stat3 inhibitor or vehicle for 72 h (red = TUNEL, white = nuclei). (c) Representative immunofluorescence images of differentiated cultures of human myoblasts treated with the Stat3 inhibitor or vehicle in both growth and differentiation media (GM, then DM), or only in growth media and then switched to differentiation in the absence of Stat3 inhibitor (GM only) (red = myosin heavy chain, green = EdU, white = nuclei). Scale bar, 200 µm (left). Quantification of differentiation index in the three experimental conditions (n = 4). Data are represented as average±s.e.m. Student s t test was used for all statistical analyses. Student s t test was used for all statistical analyses (P > 0.05). 13
14 Supplementary Table 1. Primer sequences for RNA qpcr analysis and ChIP experiments. Myod1 fwd ggctacgacaccgcctacta rev cgactctggtggtgcatctg Socs3 fwd tgcaggagagcggattctac rev tgacgctcaacgtgaagaag Stat3 fwd tgaaggtggtggagaacctc rev gctgctgcatcttctgtctg Gapdh fwd tgcaccaccaactgcttag rev ggatgcagggatgatgttc Myod1 590bp ChIP fwd acagcgctggggttctaagc rev agcaagcctggcacaaatga Socs3 +59bp ChIP fwd cgcgcacagcctttcagtg rev tttacccggccagtacgcc Oct4 +1bp ChIP fwd agaggtgaaaccgtcccta rev gtgagaaggcgaagtctgaa Pdx1 +524bp ChIP fwd ctggctctttctccctttctc rev cgggtttcagaggaacttgt IgH +1526bp ChIP fwd aaccacagctacaagtttacc rev aaccagaacacctgcagcagc 14
Supplementary Information. Title BRD3 and BRD4 BET Bromodomain Proteins Differentially Regulate Skeletal Myogenesis
 Supplementary Information Title BRD3 and BRD4 BET Bromodomain Proteins Differentially Regulate Skeletal Myogenesis Authors Thomas C. Roberts 1,2, Usue Etxaniz 1, Alessandra Dall Agnese 1, Shwu-Yuan Wu
Supplementary Information Title BRD3 and BRD4 BET Bromodomain Proteins Differentially Regulate Skeletal Myogenesis Authors Thomas C. Roberts 1,2, Usue Etxaniz 1, Alessandra Dall Agnese 1, Shwu-Yuan Wu
Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and
 Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
Supplementary Information
 Supplementary Information Supplementary Figure 1: Identification of new regulators of MuSC by a proteome-based shrna screen. (a) FACS plots of GFP + and GFP - cells from Pax7 ICN -Z/EG (upper panel) and
Supplementary Information Supplementary Figure 1: Identification of new regulators of MuSC by a proteome-based shrna screen. (a) FACS plots of GFP + and GFP - cells from Pax7 ICN -Z/EG (upper panel) and
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in
 Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Supplementary Figure 1 Muscle dystrophic phenotype is absent at P6 in PKO mice. (a) Size
 Supplementary Figure 1 Muscle dystrophic phenotype is absent at P6 in PKO mice. (a) Size comparison of the P6 control and PKO mice. (b) H&E staining of hind-limb muscles from control and PKO mice at P6.
Supplementary Figure 1 Muscle dystrophic phenotype is absent at P6 in PKO mice. (a) Size comparison of the P6 control and PKO mice. (b) H&E staining of hind-limb muscles from control and PKO mice at P6.
Supplemental Table/Figure Legends
 MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
SUPPLEMENTAL INFORMATION
 UTX/KDM6A demethylase activity is required for satellite cell-mediated muscle regeneration Hervé Faralli 1,2, Chaochen Wang 3, Kiran Nakka 1,2, Soji Sebastian 1,, Aissa Benyoucef 1,2, Lenan Zhuang 3, Alphonse
UTX/KDM6A demethylase activity is required for satellite cell-mediated muscle regeneration Hervé Faralli 1,2, Chaochen Wang 3, Kiran Nakka 1,2, Soji Sebastian 1,, Aissa Benyoucef 1,2, Lenan Zhuang 3, Alphonse
Supplementary Information
 Supplementary Information Negative regulation of initial steps in skeletal myogenesis by mtor and other kinases Raphael A. Wilson 1*, Jing Liu 1*, Lin Xu 1, James Annis 2, Sara Helmig 1, Gregory Moore
Supplementary Information Negative regulation of initial steps in skeletal myogenesis by mtor and other kinases Raphael A. Wilson 1*, Jing Liu 1*, Lin Xu 1, James Annis 2, Sara Helmig 1, Gregory Moore
Supplementary Material. Levels of S100B protein drive the reparative process in acute muscle injury and muscular dystrophy
 Supplementary Material Levels of protein drive the reparative process in acute muscle injury and muscular dystrophy Francesca Riuzzi 1,4 *, Sara Beccafico 1,4 *, Roberta Sagheddu 1,4, Sara Chiappalupi
Supplementary Material Levels of protein drive the reparative process in acute muscle injury and muscular dystrophy Francesca Riuzzi 1,4 *, Sara Beccafico 1,4 *, Roberta Sagheddu 1,4, Sara Chiappalupi
Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern
 Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern blot. Northern blot analysis of mir-302b expression following infection with PAO1, PAK and Kp in (A) lung
Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern blot. Northern blot analysis of mir-302b expression following infection with PAO1, PAK and Kp in (A) lung
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
TITLE: Enhancement of Skeletal Muscle Repair by the Urokinase Type Plasminogen Activator System
 AD Award Number: W81XWH-05-1-0159 TITLE: Enhancement of Skeletal Muscle Repair by the Urokinase Type Plasminogen Activator System PRINCIPAL INVESTIGATOR: Timothy J. Koh, Ph.D. CONTRACTING ORGANIZATION:
AD Award Number: W81XWH-05-1-0159 TITLE: Enhancement of Skeletal Muscle Repair by the Urokinase Type Plasminogen Activator System PRINCIPAL INVESTIGATOR: Timothy J. Koh, Ph.D. CONTRACTING ORGANIZATION:
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplemental Figure 1 (Figure S1), related to Figure 1 Figure S1 provides evidence to demonstrate Nfatc1Cre is a mouse line that directed gene
 Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
SUPPLEMENTAL MATERIALS SIRTUIN 1 PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF2 ACTIVATION
 SUPPLEMENTAL MATERIALS SIRTUIN PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF ACTIVATION Haranatha R. Potteti*, Subbiah Rajasekaran*, Senthilkumar B. Rajamohan*, Chandramohan R. Tamatam,
SUPPLEMENTAL MATERIALS SIRTUIN PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF ACTIVATION Haranatha R. Potteti*, Subbiah Rajasekaran*, Senthilkumar B. Rajamohan*, Chandramohan R. Tamatam,
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Table S1
 Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Supplementary Figure S1. Expression of complement components in E4 chick eyes. (a) A
 Supplementary Figure S1. Expression of complement components in E4 chick eyes. (a) A schematic of the eye showing the ciliary margin (CM) of the anterior region, and the neuroepithelium (NE) and retinal
Supplementary Figure S1. Expression of complement components in E4 chick eyes. (a) A schematic of the eye showing the ciliary margin (CM) of the anterior region, and the neuroepithelium (NE) and retinal
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas.
 Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
SUPPLEMENTARY INFORMATION
 Elastase levels and activity are increased in dystrophic muscle and impair myoblast cell survival, proliferation and differentiation N. Arecco 1,2,3, C.J. Clarke 1,2, F.K. Jones 1,2, D. Simpson 1,4, D.
Elastase levels and activity are increased in dystrophic muscle and impair myoblast cell survival, proliferation and differentiation N. Arecco 1,2,3, C.J. Clarke 1,2, F.K. Jones 1,2, D. Simpson 1,4, D.
Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates
 Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates vascular calcification (VC). (a) Von Kossa staining shows that TSA potentiated the Pi-induced VC. Scale bar, 100
Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates vascular calcification (VC). (a) Von Kossa staining shows that TSA potentiated the Pi-induced VC. Scale bar, 100
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles
 Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector.
 Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary Information
 Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
The Acute Phase Protein Orosomucoid Regulates Food Intake and Energy Homeostasis via Leptin Receptor Signaling Pathway. Shanghai , China
 The Acute Phase Protein Orosomucoid Regulates Food Intake and Energy Homeostasis via Leptin Receptor Signaling Pathway Yang Sun, 1 Yili Yang, 2 Zhen Qin, 1 Jinya Cai, 3 Xiuming Guo, 1 Yun Tang, 3 Jingjing
The Acute Phase Protein Orosomucoid Regulates Food Intake and Energy Homeostasis via Leptin Receptor Signaling Pathway Yang Sun, 1 Yili Yang, 2 Zhen Qin, 1 Jinya Cai, 3 Xiuming Guo, 1 Yun Tang, 3 Jingjing
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
SUPPLEMENTARY INFORMATION
 ARTICLE NUMBER: 0 DOI: 0.0/NMICROBIOL.0. The host protein CLUH participates in the subnuclear transport of influenza virus ribonucleoprotein complexes Tomomi Ando,, Seiya Yamayoshi, Yuriko Tomita,, Shinji
ARTICLE NUMBER: 0 DOI: 0.0/NMICROBIOL.0. The host protein CLUH participates in the subnuclear transport of influenza virus ribonucleoprotein complexes Tomomi Ando,, Seiya Yamayoshi, Yuriko Tomita,, Shinji
Supplemental Material
 Supplemental Material Figure S1.(A) Immuno-staining of freshly isolated Myf5-Cre:ROSA-YFP fiber (B) Satellite cell enumeration in WT and cko mice from resting hind limb muscles. Bulk hind limb muscles
Supplemental Material Figure S1.(A) Immuno-staining of freshly isolated Myf5-Cre:ROSA-YFP fiber (B) Satellite cell enumeration in WT and cko mice from resting hind limb muscles. Bulk hind limb muscles
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/259/ra5/dc1 Supplementary Materials for Systems Biology Approach Identifies the Kinase Csnk1a1 as a Regulator of the DNA Damage Response in Embryonic Stem Cells
www.sciencesignaling.org/cgi/content/full/6/259/ra5/dc1 Supplementary Materials for Systems Biology Approach Identifies the Kinase Csnk1a1 as a Regulator of the DNA Damage Response in Embryonic Stem Cells
- Supplementary Information -
 TRIM32 dependent transcription in adult neural progenitor cells regulates neuronal differentiation. - Supplementary Information - Anna-Lena Hillje 1, 2#, Maria Angeliki S. Pavlou 1, 2#, Elisabeth Beckmann
TRIM32 dependent transcription in adult neural progenitor cells regulates neuronal differentiation. - Supplementary Information - Anna-Lena Hillje 1, 2#, Maria Angeliki S. Pavlou 1, 2#, Elisabeth Beckmann
Supplemental Figure 1: Podocyte specific knockdown of Klf4 in Podocin-Cre Klf4 flox/flox mice was confirmed. (A) Primary glomerular epithelial cells
 Supplemental Figure 1: Podocyte specific knockdown of Klf4 in Podocin-Cre Klf4 flox/flox mice was confirmed. (A) Primary glomerular epithelial cells were isolated from 10-week old Podocin-Cre Klf4 flox/flox
Supplemental Figure 1: Podocyte specific knockdown of Klf4 in Podocin-Cre Klf4 flox/flox mice was confirmed. (A) Primary glomerular epithelial cells were isolated from 10-week old Podocin-Cre Klf4 flox/flox
Figure S1. nuclear extracts. HeLa cell nuclear extract. Input IgG IP:ORC2 ORC2 ORC2. MCM4 origin. ORC2 occupancy
 A nuclear extracts B HeLa cell nuclear extract Figure S1 ORC2 (in kda) 21 132 7 ORC2 Input IgG IP:ORC2 32 ORC C D PRKDC ORC2 occupancy Directed against ORC2 C-terminus (sc-272) MCM origin 2 2 1-1 -1kb
A nuclear extracts B HeLa cell nuclear extract Figure S1 ORC2 (in kda) 21 132 7 ORC2 Input IgG IP:ORC2 32 ORC C D PRKDC ORC2 occupancy Directed against ORC2 C-terminus (sc-272) MCM origin 2 2 1-1 -1kb
Resveratrol inhibits epithelial-mesenchymal transition of retinal. pigment epithelium and development of proliferative vitreoretinopathy
 Resveratrol inhibits epithelial-mesenchymal transition of retinal pigment epithelium and development of proliferative vitreoretinopathy Keijiro Ishikawa, 1,2 Shikun He, 2, 3 Hiroto Terasaki, 1 Hossein
Resveratrol inhibits epithelial-mesenchymal transition of retinal pigment epithelium and development of proliferative vitreoretinopathy Keijiro Ishikawa, 1,2 Shikun He, 2, 3 Hiroto Terasaki, 1 Hossein
Supplementary Figure 1. Generation of B2M -/- ESCs. Nature Biotechnology: doi: /nbt.3860
 Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Nature Biotechnology: doi: /nbt Supplementary Figure 1. In vitro validation of OTC sgrnas and donor template.
 Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and
 SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
Supplementary Figure 1. GST pull-down analysis of the interaction of GST-cIAP1 (A, B), GSTcIAP1
 Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
a KYSE270-CON KYSE270-Id1
 a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
supplementary information
 Figure S1 ZEB1 full length mrna. (a) Analysis of the ZEB1 mrna using the UCSC genome browser (http://genome.ucsc.edu) revealed truncation of the annotated Refseq sequence (NM_030751). The probable terminus
Figure S1 ZEB1 full length mrna. (a) Analysis of the ZEB1 mrna using the UCSC genome browser (http://genome.ucsc.edu) revealed truncation of the annotated Refseq sequence (NM_030751). The probable terminus
MLN8237 induces proliferation arrest, expression of differentiation markers and
 Supplementary Figure Legends Supplementary Figure 1 827 induces proliferation arrest, expression of differentiation markers and polyploidization of a human erythroleukemia cell line with the activating
Supplementary Figure Legends Supplementary Figure 1 827 induces proliferation arrest, expression of differentiation markers and polyploidization of a human erythroleukemia cell line with the activating
Supplementary Figure 1. Nur77 and leptin-controlled obesity. (A) (B) (C)
 Supplementary Figure 1. Nur77 and leptin-controlled obesity. (A) Effect of leptin on body weight and food intake between WT and KO mice at the age of 12 weeks (n=7). Mice were i.c.v. injected with saline
Supplementary Figure 1. Nur77 and leptin-controlled obesity. (A) Effect of leptin on body weight and food intake between WT and KO mice at the age of 12 weeks (n=7). Mice were i.c.v. injected with saline
Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3'
 Supplementary Table 1: Real-time PCR primer sequences for ΔEGFR, wtegfr, IL-6, LIF and GAPDH. Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3' wtegfr
Supplementary Table 1: Real-time PCR primer sequences for ΔEGFR, wtegfr, IL-6, LIF and GAPDH. Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3' wtegfr
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators.
 Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/8/347/347ra94/d Supplementary Materials for n mirn-mediated therapy for S6 blocks IRES-driven translation of the N second cistron Yu Miyazaki, Xiaofei
www.sciencetranslationalmedicine.org/cgi/content/full/8/347/347ra94/d Supplementary Materials for n mirn-mediated therapy for S6 blocks IRES-driven translation of the N second cistron Yu Miyazaki, Xiaofei
TRIM31 is recruited to mitochondria after infection with SeV.
 Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Xiaoqing Zhang, Guo Zhang, Hai Zhang, Michael Karin, Hua Bai, and Dongsheng Cai
 Cell, Volume 135 Supplemental Data Hypothalamic IKKβ/NF-κB and ER Stress Link Overnutrition to Energy Imbalance and Obesity Xiaoqing Zhang, Guo Zhang, Hai Zhang, Michael Karin, Hua Bai, and Dongsheng Cai
Cell, Volume 135 Supplemental Data Hypothalamic IKKβ/NF-κB and ER Stress Link Overnutrition to Energy Imbalance and Obesity Xiaoqing Zhang, Guo Zhang, Hai Zhang, Michael Karin, Hua Bai, and Dongsheng Cai
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Data. Flvcr1a TCTAAGGCCCAGTAGGACCC GGCCTCAACTGCCTGGGAGC AGAGGGCAACCTCGGTGTCC
 Supplementary Data Supplementary Materials and Methods Measurement of reactive oxygen species accumulation in fresh intestinal rings Accumulation of reactive oxygen species in fresh intestinal rings was
Supplementary Data Supplementary Materials and Methods Measurement of reactive oxygen species accumulation in fresh intestinal rings Accumulation of reactive oxygen species in fresh intestinal rings was
Supplementary Figure 1 A
 Supplementary Figure A B M. mullata p53, 3 UTR Luciferase activity (%) mir-5b 8 Le et al. Supplementary Information NC-DP - + - - - - - NC-DP - - + - - - - NC-DP3 - - - + - - - 5b-DP - - - - + + + NC-AS
Supplementary Figure A B M. mullata p53, 3 UTR Luciferase activity (%) mir-5b 8 Le et al. Supplementary Information NC-DP - + - - - - - NC-DP - - + - - - - NC-DP3 - - - + - - - 5b-DP - - - - + + + NC-AS
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Mammosphere formation assay. Mammosphere culture was performed as previously described (13,
 Supplemental Text Materials and Methods Mammosphere formation assay. Mammosphere culture was performed as previously described (13, 17). For co-culture with fibroblasts and treatment with CM or CCL2, fibroblasts
Supplemental Text Materials and Methods Mammosphere formation assay. Mammosphere culture was performed as previously described (13, 17). For co-culture with fibroblasts and treatment with CM or CCL2, fibroblasts
Detection of Histone Modifications at Specific Gene Loci in Single Cells in Histological Sections
 Detection of Histone Modifications at Specific Gene Loci in Single Cells in Histological Sections Delphine Gomez; Laura L Shankman; Anh T Nguyen; Gary K Owens. Supplementary Figures 1-13 Supplementary
Detection of Histone Modifications at Specific Gene Loci in Single Cells in Histological Sections Delphine Gomez; Laura L Shankman; Anh T Nguyen; Gary K Owens. Supplementary Figures 1-13 Supplementary
Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death
 SUPPLEMENTAL DATA Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death Huajun Jin 1, Arthi Kanthasamy 1, Vellareddy Anantharam
SUPPLEMENTAL DATA Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death Huajun Jin 1, Arthi Kanthasamy 1, Vellareddy Anantharam
Wnt16 smact merge VK/AB
 A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Myofibroblasts in kidney fibrosis. LeBleu et al.
 Origin and Function of Myofibroblasts in Kidney Fibrosis Valerie S. LeBleu, Gangadhar Taduri, Joyce O Connell, Yingqi Teng, Vesselina G. Cooke, Craig Woda, Hikaru Sugimoto and Raghu Kalluri Supplementary
Origin and Function of Myofibroblasts in Kidney Fibrosis Valerie S. LeBleu, Gangadhar Taduri, Joyce O Connell, Yingqi Teng, Vesselina G. Cooke, Craig Woda, Hikaru Sugimoto and Raghu Kalluri Supplementary
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Nature Medicine: doi: /nm.4169
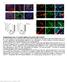 Supplementary Fig.1. EC-specific deletion of Ccm3 by Cdh5-CreERT2. a. mt/mg reporter mice were bred with Cdh5CreERT2 deleter mice followed by tamoxifen feeding from P1 to P3. mg expression was specifically
Supplementary Fig.1. EC-specific deletion of Ccm3 by Cdh5-CreERT2. a. mt/mg reporter mice were bred with Cdh5CreERT2 deleter mice followed by tamoxifen feeding from P1 to P3. mg expression was specifically
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Table 1 Supplementary Figures 1, 2, 3, 4, 5, 6, 7, 8 and legends Supplementary Video 1, 2 legends AON Sequence Position Length MW tcdna 5 -AACCTCGGCTTACCT-3 +2-13
SUPPLEMENTARY INFORMATION Supplementary Table 1 Supplementary Figures 1, 2, 3, 4, 5, 6, 7, 8 and legends Supplementary Video 1, 2 legends AON Sequence Position Length MW tcdna 5 -AACCTCGGCTTACCT-3 +2-13
Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex
 POSTER PRESENTATION Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex Hai-Chon Lee *, Je-In Youn, Kyungwha Lee, Hwanyul Yong, Seung-Yong
POSTER PRESENTATION Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex Hai-Chon Lee *, Je-In Youn, Kyungwha Lee, Hwanyul Yong, Seung-Yong
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplementary Figure 1.
 Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Transcriptional regulation of BRCA1 expression by a metabolic switch: Di, Fernandez, De Siervi, Longo, and Gardner. H3K4Me3
 ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Nanog-Luc. R-Luc. 40 HA-Klf Relative protein level of Klf USP21. Vector USP2 USP21 *** *** *** ***
 :Flag Relative protein level of Relative protein level of : Flag Vec 21LV 21SV 2 : HA HA- HA- HA- HA-Klf4 Relative mrna expression Relative protein level of Relative protein level of Relative protein level
:Flag Relative protein level of Relative protein level of : Flag Vec 21LV 21SV 2 : HA HA- HA- HA- HA-Klf4 Relative mrna expression Relative protein level of Relative protein level of Relative protein level
SUPPLEMENTARY INFORMATION
 The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
SUPPLEMENTAL EXPERIMENTAL PROCEDURES
 SUPPLEMENTAL EXPERIMENTAL PROCEDURES Luciferase Assays Cells were seeded on 24well plates and grown to 7% confluency. Cells were then transfected with ng of reporter constructs and 1 ng of the renilla
SUPPLEMENTAL EXPERIMENTAL PROCEDURES Luciferase Assays Cells were seeded on 24well plates and grown to 7% confluency. Cells were then transfected with ng of reporter constructs and 1 ng of the renilla
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated
 Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
Supplementary Figure 1. Bone density was decreased in osteoclast-lineage cell specific Gna13 deficient mice. (a-c) PCR genotyping of mice by mouse
 Supplementary Figure 1. Bone density was decreased in osteoclast-lineage cell specific Gna13 deficient mice. (a-c) PCR genotyping of mice by mouse tail DNA. Primers were designed to detect Gna13-WT/f (~400bp/470bp)
Supplementary Figure 1. Bone density was decreased in osteoclast-lineage cell specific Gna13 deficient mice. (a-c) PCR genotyping of mice by mouse tail DNA. Primers were designed to detect Gna13-WT/f (~400bp/470bp)
Kostandin V. Pajcini, Stephane Y. Corbel, Julien Sage, Jason H. Pomerantz, and Helen M. Blau
 Cell Stem Cell, Volume 7 Supplemental Information Transient Inactivation of Rb and ARF Yields Regenerative Cells from Postmitotic Mammalian Muscle Kostandin V. Pajcini, Stephane Y. Corbel, Julien Sage,
Cell Stem Cell, Volume 7 Supplemental Information Transient Inactivation of Rb and ARF Yields Regenerative Cells from Postmitotic Mammalian Muscle Kostandin V. Pajcini, Stephane Y. Corbel, Julien Sage,
Supplementary figures
 Relative intensity Relative intensity Relative intensity Supplementary figures a None Caffeine None Caffeine c None Caffeine 6 6 6 ISG 6 6 6 UBE1L 6 6 6 UBCH8 6 6 6 EFP 1 1 DOX (h) 1 1 CPT (h) 1 1 UV (h)
Relative intensity Relative intensity Relative intensity Supplementary figures a None Caffeine None Caffeine c None Caffeine 6 6 6 ISG 6 6 6 UBE1L 6 6 6 UBCH8 6 6 6 EFP 1 1 DOX (h) 1 1 CPT (h) 1 1 UV (h)
SUPPLEMENTARY MATERIALS
 SUPPLEMENTARY MATERIALS Supplementary Table S1: List of primers used for ChIP analysis and Oligo-pull-down assay. Genes Sequence mppargc1a FW 5 -GCGAGGTTTCTGCTTAGTCA-3 (-2317) RV 5 -ACAATGACTAAGCAGAAACCTCG-3
SUPPLEMENTARY MATERIALS Supplementary Table S1: List of primers used for ChIP analysis and Oligo-pull-down assay. Genes Sequence mppargc1a FW 5 -GCGAGGTTTCTGCTTAGTCA-3 (-2317) RV 5 -ACAATGACTAAGCAGAAACCTCG-3
b alternative classical none
 Supplementary Figure. 1: Related to Figure.1 a d e b alternative classical none NIK P-IkBa Total IkBa Tubulin P52 (Lighter) P52 (Darker) RelB (Lighter) RelB (Darker) HDAC1 Control-Sh RelB-Sh NF-kB2-Sh
Supplementary Figure. 1: Related to Figure.1 a d e b alternative classical none NIK P-IkBa Total IkBa Tubulin P52 (Lighter) P52 (Darker) RelB (Lighter) RelB (Darker) HDAC1 Control-Sh RelB-Sh NF-kB2-Sh
Supplementary information. Supplementary Figures
 Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
Genomic DNA fragments identified by ChIP-chip assay for MR [28] corresponding to two
![Genomic DNA fragments identified by ChIP-chip assay for MR [28] corresponding to two Genomic DNA fragments identified by ChIP-chip assay for MR [28] corresponding to two](/thumbs/89/98549522.jpg) Supplemental Materials and Methods Genomic DNA fragments identified by ChIP-chip assay for MR [28] corresponding to two upstream regions of the human Klf9 gene (-5139 to -5771 bp and -3875 to -4211 bp)
Supplemental Materials and Methods Genomic DNA fragments identified by ChIP-chip assay for MR [28] corresponding to two upstream regions of the human Klf9 gene (-5139 to -5771 bp and -3875 to -4211 bp)
8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and
 1 Supplemental information 2 3 Materials and Methods 4 Reagents and animals 5 8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and 6 Silencer Select Pre-designed sirna
1 Supplemental information 2 3 Materials and Methods 4 Reagents and animals 5 8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and 6 Silencer Select Pre-designed sirna
