The Reelin Signaling Pathway: Some Recent Developments
|
|
|
- Archibald Gilmore
- 6 years ago
- Views:
Transcription
1 The Reelin Signaling Pathway: Some Recent Developments Yves Jossin, Isabelle Bar 1, Nina Ignatova, Fadel Tissir, Catherine Lambert de Rouvroit 1 and André M. Goffinet Developmental Genetics Unit, UCL GEDE 7382, 73 Avenue E. Mounier, B1200 Brussels and 1 Neurobiology Unit 2853, FUNDP Medical School, 61 Rue de Bruxelles, B5000 Namur, Belgium The Reelin signaling pathway plays a key role in the architectonic development of the central nervous system. Extracellular Reelin binds to receptors of the lipoprotein receptor family and induces tyrosine phosphorylation of the adaptor Dab1. In this paper, we discuss three recent developments. First, we show that the central part of Reelin is involved in receptor binding and signal activation as reflected in Dab1 phosphorylation. Second, we examine the genomic organization, alternative splicing and promoter use of the Dab1 gene, which hint at a particularly complex regulation. Third, we present preliminary studies by in situ hybridization that demonstrate regulated expression of Reelin receptors and Dab1 by radial precursors in the ventricular zone. Introduction The mammalian cerebral cortex develops by following a finely tuned sequence of events that regulate cell proliferation, neuronal migration and architectonic pattern formation, differentiation, synaptogenesis and myelination. Neuronal cell precursor proliferation takes place in germinative zones located along the cerebral ventricles, from which neurons migrate over variable distances to settle in the cortical plate. Like other developmental events, neuronal migration and architectonic pattern formation are under genetic control. Among the control mechanisms that have been identified, the Reelin signaling pathway plays a key role in cortical development in man, mouse and presumably all mammals. Reelin is a large (>400 kda) extracellular glycoprotein that is secreted by several neurons, particularly, in the embryonic cortex, by Cajal Retzius cells. Defective Reelin is the cause of the reeler brain malformation in mice (Lambert de Rouvroit and Goffinet, 1998; Rice and Curran, 2001) and of a peculiar type of lissencephaly in man (Hong et al., 2000). In reeler mice, neurons are generated in ventricular zones (VZ) and migrate initially as in normal animals. However, unlike normal cells, reeler neurons form defective architectonic patterns. Whereas normal cortical neurons form a dense, radially and laminarly organized cortical plate (CP) in which maturation proceeds from inside to outside, reeler neurons form a loose CP in which neurons are oriented obliquely and the gradient of maturation is almost inverted. The organization of the Reelin protein is as follows (D Arcangelo et al., 1995). The sequence begins with a signal peptide of residues, followed by a region with similarity to F-spondin (amino acids ). A unique region between amino acids 191 and 500 is followed by a succession of eight repeats (1 8) of amino acids (1, residues ; 2, ; 3, ; 4, ; 5, ; 6, ; 7, ; 8, ). Each Reelin repeat contains an EGF motif at its center, which divides the repeats into two sub-repeats, A and B, that show weak similarity to each other. The protein terminates with a basic stretch of 33 amino acids ( ). Furthermore, Reelin is cleaved in vivo at two sites approximately located between repeats 2 and 3 and between repeats 6 and 7, as schematized in Figure 1 (Lambert de Rouvroit et al., 1999). Reelin is not expressed in CP cells, but may act via the extracellular milieu, instructing them to assume their position and orientation. This view is supported by studies of three other genes, Disabled-1 (Dab1), VLDLR and ApoER2, mutations of which generate a reeler-like phenotype. The response of CP neurons to Reelin requires expression of at least one of two receptors, the Very Low Density Lipoprotein Receptor (VLDLR) and the Apolipoprotein-E Receptor Type 2 (ApoER2). Reelin does not bind to the closely related Low Density Lipoprotein Receptor (LDLR) nor to more distantly related members of the family such as LRP (Hiesberger et al., 1999; Trommsdorff et al., 1999). Whereas mutations of either VLDLR or ApoER2 generate subtle neurological phenotypes, mice deficient in both genes have a reeler-like phenotype, demonstrating a redundancy. VLDLR and ApoER2 are expressed in Reelin target neurons. Both have short cytoplasmic tails that contain an NPxY motif to bind Dab1, an intracellular adapter protein expressed in cells that respond to the Reelin signal. Dab1 exists as several predicted isoforms, as explained below. The Dab1 N-terminal region contains a PI-PTB domain to interact with non-phosphorylated tyrosine residues in the NPxY sequence (Howell et al., 1997b, 1999b, 2000). Dab1 contains five tyrosine residues and phosphorylation of these residue, particularly Y198 and Y220 (Keshvara et al., 2001), in response to the binding of Reelin to Figure 1. Proteolytic processing of Reelin. Lanes on the left are immunoblots of PAGE gels loaded with embryonic brain extracts immunoprecipitated (IP) with antibody G10 directed against the N-terminal region of Reelin (left lane) and with antibody 17 directed against the C-terminal region (right lane). Blots were revealed (WB) with G10 (left lane) and with a mix of antibodies directed against the C-terminal region (right lane) (de Bergeyck et al., 1998), respectively. The result shows that the Reelin protein (>400 kda) is cleaved at two main sites (arrowheads), generating fragments of 300, 220, 180 and 100 kda, as indicated in the schema. The CR50 epitope is approximately located as indicated. Oxford University Press All rights reserved. Cerebral Cortex Jun 2003;13: ; /03/$4.00
2 lipoprotein receptors is necessary for Dab1 function. Most but not all features of the Dab1 / mutant phenotype can be rescued by replacing the Dab1 gene by a partial Dab1 cdna encoding the PTB domain and the region with the five tyrosine residues (Herrick and Cooper, 2002). The rest of the Dab1 protein serves some unidentified function and this may be related to the presence of consensus S/T phosphorylation sites, some of which can be phosphorylated by the Cdk5/p35 kinase in a Reelinindependent manner (Keshvara et al., 2002). In the present paper, we focus on some recent results from our laboratories that concern: (i) the interaction between Reelin and its receptors and its relation to Dab1 phosphorylation; (ii) the presence and putative functional implications of alternative splicing and promoter utilization events in the Dab1 gene; (iii) mrna expression data that point to a possible role of Reelin signaling in cortical ventricular zones. Reelin Binding to Lipoprotein Receptors and Dab1 Phosphorylation Previous work showed that Reelin binding to the VLDLR and ApoER2 receptors does not require the N-terminal region of Reelin from the N-terminus up to the end of the second repeat (Hiesberger et al., 1999). However, the CR50 antibody, directed against an epitope in the N-terminal region, interferes with binding of full-length Reelin (D Arcangelo et al., 1999), possibly through a steric mechanism. Other studies show that Reelin has the capacity to aggregate and that this aggregation and the capacity to induce Dab1 phosphorylation are both inhibited by antibody CR50 (Utsunomiya-Tate et al., 2000; Kubo et al., 2002). Together, these data indicate that the N-terminus may contribute to, but is not sufficient for, Reelin signaling. In order to better understand the interaction of Reelin with VLDLR and ApoER2, we studied binding of different partial Reelin recombinant proteins to extracellular regions of receptors, fused to an Fc immunoglobulin fragment (provided by J. Herz). The reelin cdna construct pcrl (provided by T. Curran) was used to express Reelin and as a template to derive different parts of the reelin cdna, by nuclease restriction or PCR amplification. In the terminology of the Reelin constructs, R is used for repeat, N for N-terminus and Del for deletion of a given region. The constructs containing R3 R8, R3 R6, R3 R5, R3 R4, R4 R5, R5 R6, R7 R8, R4 and R6 were cloned in the psectag2b vector (Invitrogen), which encodes the polypeptide in phase with a signal peptide and a Myc epitope. Constructs N R6, DelR3 R5A, N R5A and N R2 were obtained from pcrl by nuclease restriction followed by ligation. Partial and full-length recombinant Reelin were produced by transient transfection of HEK293 cells and fresh supernatant was used without further purification. LDLR, VLDLR and ApoER2 receptor constructs were produced in the same manner. Receptor binding was estimated by co-immunoprecipitation. For estimation of Dab1 tyrosine phosphorylation, primary cortical neuronal cultures were incubated with full-length or recombinant proteins, the Dab1 protein was immunoprecipitated using a rabbit polyclonal antibody directed against the C-terminal peptide of Dab1 and the precipitates were analyzed on duplicate PAGE gels for estimation of Dab1 protein content using a monoclonal antibody against the PTB region of Dab1 and, for estimation of Dab1 tyrosine phosphorylation, using a commercial anti-phosphotyrosine antibody (clone 4G10; Upstate) (Howell et al., 1999a). Binding to Lipoprotein Receptors Binding to LDLR was very low or negative for all secreted proteins (Fig. 2A). In accord with published evidence, construct Figure 2. Reelin receptor interactions. (A) Binding of Reelin to lipoprotein receptors in vitro. The Reelin and R3 R6 cdnas were transfected into 293T cells. The supernatant was incubated with recombinant LDLR Fc, ApoER2 Fc and VLDLR Fc fusion proteins bound to protein A agarose beads. The complexes were immunoblotted and revealed with antibody G10 (Reelin) or anti-myc antibody (R3 R6). Both Reelin and R3 R6 bind to VLDLR and ApoER2, but not to LDLR. IP, immunoprecipitation with G10 or anti-myc, to demonstrate that the corresponding protein is well secreted. (B) Stimulation of Dab1 tyrosine phosphorylation by the Reelin protein. Mouse embryonic neurons (E16) were cultured for 3 days and treated with full-length Reelin, with the R3 R6 construct and with a supernatant of mock-transfected cells ( ). Dab1 was immunoprecipitated with a polyclonal anti-dab1 antibody and blots were revealed with another anti-dab1 antibody (upper panel) and with an anti-phosphotyrosine antibody (αpy). Unlike the control supernatant, both Reelin and R3 R6 are able to generate tyrosine phosphorylation of Dab1. N R2, composed of the N-terminal spondin similarity region, the unique segment and repeats 1 and 2, did not bind lipoprotein receptors. Even construct N R5A, which includes the first four reelin repeats and part of repeat 5, did not bind significantly. This confirmed that the N-terminal moiety of Reelin is not directly involved in receptor binding (Hiesberger et al., 1999). Similarly, all secreted constructs that contain one, two or three Reelin repeats failed to bind detectably to VLDLR and ApoER2. In contrast, constructs R3 R6 and R3 R8 and N R6 bound to VLDLR and ApoER2 similarly to full-length Reelin (Fig. 2A). Such an increase in binding capacity with increased repeat number may indicate cooperativity between repeats. Repeat number is not the sole factor involved, however. For example, construct DelR3 R5A, in which repeats 3, 4 and the N-terminal part of repeat 5 are deleted, did not bind detectably to either receptor, even though it contains five Reelin repeats. Altogether, these experiments suggest that at least four repeats are necessary to detect significant binding of Reelin to VLDLR and ApoER2. Furthermore, repeats contained in the central region of Reelin are particularly important for receptor binding. Dab1 Phosphorylation As a first approach to assess whether receptor binding is sufficient to trigger the Reelin signal or whether additional features are required, the ability of the different Reelin constructs to induce tyrosine phosphorylation of the Dab1 628 The Reelin Signaling Pathway Jossin et al.
3 adapter in neuronal cells was studied. Full-length Reelin and constructs R3 R6, R3 R8 and N R6 were equally able to induce Dab1 phosphorylation, whereas all other Reelin polypeptide constructs were inactive (Fig. 2B). The central part of Reelin defined in these experiments is large in comparison with the extracellular domains of VLDLR and ApoER2, which leaves ample room for interaction with several proteins in addition to lipoprotein receptors. However, the correlation between binding to receptors and stimulation of Dab1 tyrosine phosphorylation indicates that the central region of Reelin is sufficient to initiate the Dab1-dependent part of the signal. This central region of Reelin corresponds approximately to the fragment generated by cleavage; whether this is functionally relevant remains to be demonstrated. These observations are worth discussing in relation to published aspects of Reelin signaling. The protocadherin CNR1 and integrin α3β1 have been proposed as Reelin co-receptors (Senzaki et al., 1999; Dulabon et al., 2000). Whereas the part of Reelin implicated in integrin binding was not defined, binding of CNR1 to the N-terminal region of Reelin was considered critical. Our observations suggest that binding of Reelin to CNR is not necessary for its Dab1-dependent function, but may serve some other independent role that remains to be defined further. Another point is the recent observations that: (i) the N-terminal region of Reelin mediates Reelin aggregation; (ii) homodimerization of Reelin and Dab1 phosphorylation are both inhibited by the function blocking antibody CR50; (iii) Reelin devoid of its N-terminal region fails to trigger Dab1 phosphorylation, even though it is still able to bind to the VLDLR and ApoER2 receptors (Utsunomiya-Tate et al., 2000; Kubo et al., 2002). These findings suggested that Reelin homodimerization is necessary for activation of the signal as ref lected by Dab1 phosphorylation. For reasons that we do not understand, our observations that constructs containing Reelin R3 R8 and R3 R6 are able to induce Dab1 phosphorylation are at variance with that evidence. Our results, however, do not necessarily argue against a functional role of Reelin aggregation in vivo. Aggregation could increase the effective local concentration of Reelin, for example by anchoring the protein to the extracellular matrix. Due to the use of transfected cell supernatants and neuronal cultures, our binding and phosphorylation assays could not be quantified using pure components and provide at best an estimate of the relative affinity of the different ligands. Aggregation of full-length Reelin may increase its affinity to an extent that we could not measure. Alternative Forms of Dab1 As Dab1 is a key component of the Reelin signaling pathway, a detailed study of the genomic organization and transcriptional regulation of the Dab1 gene was carried out in man and mouse. Here, we would like to emphasize some elements that concern the regulation of Dab1 expression and may be relevant to its function. Alternative First Exons and Organization of the 5 -Region (Figs 3 and 4) Comparison of the different Dab1 sequences revealed extensive variation in the 5 -untranslated regions (5 -UTR), as schematized in Figure 3. In mouse embryonic brain RNA, four different products named 1A, 1B, 1C and 1D were obtained. UTR 1B does not correspond to a single exon but is composed of 10 exons, 1B1 1B10. The Dab1 5 -UTR spreads over 850 kb of genomic DNA, pointing to a vast complexity. The human DAB1 gene has an almost identical organization, further suggesting that this evolutionarily conserved, complex organization may be functionally important. In order to assess whether the different 5 exons have different expression patterns, RT PCR reactions were carried out on mouse brain cdna at different stages from E11 to adult. As shown in Figure 4A, exons 1A and 1D are expressed at all stages tested. The complex UTR 1B is barely detectable at embryonic stages E11 and E12, whereas two main bands are amplified in RNA isolated from brain at E15 and later, including adult. We tested the expression of the alternative first exons in P19 cells, which differentiate into neurons in the presence of retinoic acid (R A) (Fig. 4B). Exons 1A and 1D are detected similarly in undifferentiated and differentiated P19 cells. In contrast, the expression of UTR 1B is complex. Multiple bands are amplified in undifferentiated cells and up to 4 days after RA induction. When neural induction is complete, only two bands are visible. This developmental regulation was confirmed in vivo: a complex pattern of multiple bands is amplified from mouse E11 RNA and becomes restricted to two main amplicons at P0 and in adult (Fig. 4C). These two amplicons contain fragments 1B1, 1B2 and 1B4, with alternative inclusion of 1B8. The more complex amplicons correspond to different combinations of exons 1B1 1B10 and contain variable numbers of ATG codons, suggesting that some upstream open reading frames (uorf) are excluded from segment 1B in parallel with neuronal differentiation. The ability of uorf to down-regulate translation has been documented, and our observations suggest that this phenomenon may play a role in the regulation of Dab1 expression. Internal Alternative Splicing Events (Fig. 5) Four different Dab1 cdna forms were initially described in the mouse (Howell et al., 1997a), as schematized in Figure 5A,B. The main form encodes a protein of 555 residues and is named Dab Another, Dab1-217, is due to recognition of an alternative polyadenylation signal in intron 7 and is predicted to encode a 217 amino acid polypeptide. A third form, Dab1-271, is due to insertion of an additional exon between exons 9 and 10 and results in the production of a 271 residue protein. Finally, another segment, named 555*, is included in some Dab1 cdna forms between exons 9 and 10. We showed that fragment 555* corresponds to two small exons of 51 and 48 bp separated by an intron of 91 bp and that both exons are consistently co-amplified. In undifferentiated P19 cells, the Dab1 cdna included fragment 555*. However, when differentiation of P19 cells was induced with R A, a proportion of Dab1 cdna without fragment 555* appeared at day 2 and increased progressively to become the major form at day 9 (Fig. 5C). In early embryonic mouse brain (E11 and E12), the Dab1 isoform with fragment 555* was predominant, but RNA from later developmental stages, E12 and later, and from primary neuronal cultures did not contain this fragment (Fig. 5D). In non-neural tissues such as liver and kidney, the Dab1 mrna contained fragment 555*. A similar pattern was found in chick, with inclusion of a small 57 nt exon in RNA from E6 embryo, but exclusion of that small exon from brain RNA at E20 (Fig. 4E). In situ hybridization with a cdna probe covering fragment 555* and adjacent segments confirmed expression in ventricular zones (Fig. 5F). Altogether, these observations suggest strongly that the exclusion of exon 555* occurs in parallel with neural differentiation. In summary, Dab1 mrna is expressed at high level in post-migratory neurons, but also at a more modest level in ventricular zones. Two main first exons and thus two main promoters are used for Dab1 translation in the brain. Dab1 Cerebral Cortex Jun 2003, V 13 N 6 629
4 Figure 3. Genomic organization of the mouse Dab1 gene. The main coding region is composed of 14 exons (ORF), plus exons 271 and 555*, and covers 300 kb of genomic DNA. The four alternative 5 -UTRs (UTR 1B, composed of 10 exons, and UTR 1D, 1A and 1C) are spread over 800 kb. Conserved 5 -UTR sequences in human and mouse are shown as filled boxes. Note the complexity of UTR region 1B, with 10 fragments (1B1 1B10). Tyrosine residues important for Dab1 function are indicated as Y and a number. Intron and exon sizes are indicated at the bottom. ATG, initiation codon; STOP, termination codon. Figure 4. Expression of different 5 -UTRs of Dab1. (A) RT PCR analysis of UTR 1A, 1B and 1D expression and the G3PDH reference gene. Lanes 1 8, mouse brain E11, E12, E14, E15, E18, P0, P5, P10 and adult. (B) RT PCR analysis of exons 1A, 1B, 1D during P19 cell differentiation induced by RA from day 0 to 9 and in control P0 brain using primers in exon 2 and in alternative first exons. HPRT, reference gene. (C) Illustration of the complexity of UTR 1B. Comparison of undifferentiated ( RA) and differentiated (+RA) P19 cells and of developing (E11) versus mature (Ad) mouse brain. Note the simplification of the UTR 1B amplification pattern in parallel with neural maturation. mrna in VZ includes the small exon 555*, whereas Dab1 mrna in post-migratory neurons does not. Expression of VLDLR and ApoER2 in the Developing Telencephalon The presence of some Dab1 mrna expression in the telencephalic VZ prompted us to study expression of Reelin receptors in more detail, using in situ hybridization. Preliminary results in early embryonic brain revealed expression of ApoER2 and VLDLR mrna in the VZ in addition to Dab1. AtE12,the preplate stage (Fig. 6), profuse reelin mrna expression was found in CR cells that are abundant all around the telencephalic vesicles, as well as in neurons in the basal forebrain. Expression of VLDLR and ApoER2 is also evident in precursor cells in the VZ in almost all sectors of the cortex as well as in post-mitotic, differentiating neurons. Dab1 expression is also concentrated in the ventricular zones, but is restricted to the cortical VZ and is not found, for example, in the VZ of the ganglionic eminences. Somewhat unexpectedly, the expression canvas of Dab1 and lipoprotein receptors are not identical. The expression pattern 630 The Reelin Signaling Pathway Jossin et al.
5 Figure 5. Alternative exon 555* is excluded from neurons. (A) Genomic localization of alternative exons 271 and 555* in the mouse Dab1 gene. (B) The various forms of the mouse Dab1 protein produced by alternative splicing. Form 217 results from the utilization of an alternative polyadenylation signal in intron 7. Three alternative exons are present between exons 9 and 10. Exon 271 contains a STOP codon. Form 555* is composed of two small exons of 51 (555*1) and 48 (555*2) bp. (C) Alternative splicing of exon 555* during P19 cell differentiation induced by RA from day 0 to 9 and P0 brain. Two products, with and without exon 555*, are amplified. During neuronal maturation, the larger product containing exon 555* is progressively replaced by a smaller amplicon lacking exon 555*. (D) Alternative exon 555* is expressed during early mouse brain development, but not in E19, P0 or adult brain. The Dab1 form containing the two exons 555* is expressed in the kidney. (E) An alternative exon corresponding to 555* is also included in chick at early embryonic stages (E6) and in embryonic eye but not in more mature brain (E20). (F) Analysis of the expression of alternative form 555* by in situ hybridization. The signal corresponding to 555* plus some surrounding sequence (c, d) is compared with that of a PTB probe (a, b) at E15. Both signals are detected in the cortex (simple arrow), cerebellum (arrowhead) and tectum (double arrow), but expression of the alternative exons 555* is relatively higher in the VZ at E15. Direct photography of autoradiographic film. Bar 1 mm. Cerebral Cortex Jun 2003, V 13 N 6 631
6 Figure 6. Comparative expression of Reelin, Dab1 and lipoprotein receptor VLDLR at E12. Selected frontal sections at two levels in the embryonic mouse brain at E12. The Reelin probe (Reln) reveals expression in Cajal Retzius cells at the level of the hippocampal anlage and future piriform cortex. Dab1 (Dab1-PTB) and the Reelin receptor (VLDLR) are expressed in the ventricular zone (VZ), in addition to expression in some post-mitotic neuronal fields; expression of ApoER2 was comparable to that of VLDLR (not shown). Although Dab1 and Reelin receptors are both expressed in the VZ, there are regional differences suggesting complementarity, possibly indicating some reciprocal feedback in mrna regulation. In situ hybridization with 33 P-labeled probes, dark field photography of sections after dipping in LM1 emulsion (Amersham). (Upper and lower panels) Frontal sections at rostral and posterior levels of the embryonic telencephalon. Hip, hippocampal anlage; NC, neocortical anlage; Pir, future piriform cortex; V, ventricle; EG, ganglionic eminence. Bar 100 µm. of Dab1 seems better correlated with the pattern of response to Reelin (as indicated by anomalies in reeler mutant mice) than the expression of VLDLR and ApoER2, which is more widespread. This result emphasizes the critical role of the Dab1 adaptor as the main switch in the Reelin signaling pathway. As a corollary, it raises the question of the function of VLDLR and ApoER2 in the sectors of the VZ that are Dab1-negative. In early post-natal brain, ApoER2 mrna expression remained high in all strata of the CP as well as in the VZ, whereas VLDLR mrna expression became restricted to the upper cortical plate where late generated neurons settle, but was no longer present in the VZ (not shown). The co-expression of lipoprotein receptors and Dab1 in the VZ suggests that Reelin may fulfill a novel, hitherto unknown, function at this level, for example in relation to neuron and glial cell precursor proliferation or to initiation of cell migration (Meyer et al., 2002). The late VZ contains progenitor cells destined for the olfactory bulb, and Reelin may play a role in migration of olfactory granule cells, as recently suggested (Hack et al., 2002). As far as we know, there is no expression of Reelin in the vicinity of the mammalian VZ. Some reelin mrna is present in subventricular zones in non-mammalian species (Bernier et al., 2000; Tissir et al., unpublished results), but not in mammals. A possibility could be that Reelin secreted in the external telencephalic field by CR cells may act locally and signal to the radial extensions from precursor cells in the VZ. Another possibility is that Reelin or Reelin fragments are able to diffuse from the MZ and reach cell bodies in the VZ. Further studies are needed, particularly to better define the expression and localization of the Dab1, ApoER2 and VLDLR proteins in radial precursor cells. Conclusions In conclusion, the recent data summarized above suggest that central Reelin repeats bind to VLDLR and ApoER2 receptors and this results in the organization of a supramolecular complex and in signal activation, ref lected by Dab1 phosphorylation. Other components of this complex remain unknown but are likely to include tyrosine kinase(s). However, no mice defective in tyrosine kinase genes have a reeler-like phenotype, and the role of Fyn and related kinases in Dab1 phosphorylation remains open to question. Another possibility is that unidentified components of the Reelin receptor complex might not be Reelin co-receptors in that they do not bind Reelin directly, but could be recruited to the complex via cis interaction with lipoprotein receptors. The central role of Dab1 in transduction of the Reelin signal is further emphasized by the complexity of its genomic organization and transcriptional regulation. Alternatively, spliced forms of Dab1 appear specifically expressed in precursor cells in the VZ, which also express VLDLR and ApoER2, pointing to a new role of the Reelin signal in precursor proliferation and/or rostral migration in the olfactory system. Several questions remain, particularly concerning the final action of Reelin on target cells and the identification of the downstream effectors of the Reelin signal. Notes We wish to thank T. Curran for the gift of the reelin pcrl plasmid, J. Herz for the gift of the lipoprotein receptor constructs and C. Dernoncourt for technical assistance. This work was supported by grants from FRIA (to Y.J.) and by grants FRFC and ARC /02, the Fondation Médicale Reine Elisabeth and by EU contract CONCORDE (QLG3-CT ). Address correspondence to André M. Goffinet, Developmental Genetics Unit, UCL GEDE 7382, 73 Avenue E. Mounier, B1200 Brussels, Belgium. andre.goffinet@gede.ucl.ac.be. References Bernier B, Bar I, D Arcangelo G, Curran T, Goffinet AM (2000) Reelin mrna expression during embryonic brain development in the chick. J Comp Neurol 422: D Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T (1995) A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 374: D Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, Curran T (1999) Reelin is a ligand for lipoprotein receptors. Neuron 24: de Bergeyck V, Naerhuyzen B, Goffinet AM, Lambert de Rouvroit C (1998) A panel of monoclonal antibodies against reelin, the extracellular matrix protein defective in reeler mutant mice. J Neurosci Methods 82: Dulabon L, Olson EC, Taglienti MG, Eisenhuth S, McGrath B, Walsh CA, Kreidberg JA, Anton ES (2000) Reelin binds alpha3beta1 integrin and inhibits neuronal migration. Neuron 27: Hack I, Loulier K, Carroll P, Cremer H (2002) A role for reelin as a detachment signal in adult neurogenesis. FENS Abstr 1:506. Herrick TM, Cooper JA (2002) A hypomorphic allele of dab1 reveals regional differences in reelin-dab1 signaling during brain development. Development 129: Hiesberger T, Trommsdorff M, Howell BW, Goffinet A, Mumby MC, Cooper JA, Herz J (1999) Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron 24: Hong SE, Shugart Y Y, Huang DT, Shahwan SA, Grant PE, Hourihane JO, Martin ND, Walsh CA (2000) Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nature Genet 26: Howell BW, Gertler FB, Cooper JA (1997a) Mouse disabled (mdab1): a Src binding protein implicated in neuronal development. EMBO J 16: Howell BW, Hawkes R, Soriano P, Cooper JA (1997b) Neuronal position in 632 The Reelin Signaling Pathway Jossin et al.
7 the developing brain is regulated by mouse disabled-1. Nature 389: Howell BW, Herrick TM, Cooper JA (1999a) Reelin-induced tryosine phosphorylation of disabled 1 during neuronal positioning. Genes Dev 13: Howell BW, Lanier LM, Frank R, Gertler FB, Cooper JA (1999b) The disabled 1 phosphotyrosine-binding domain binds to the internalization signals of transmembrane glycoproteins and to phospholipids. Mol Cell Biol 19: Howell BW, Herrick TM, Hildebrand JD, Zhang Y, Cooper JA (2000) Dab1 tyrosine phosphorylation sites relay positional signals during mouse brain development. Curr Biol 10: Keshvara L, Benhayon D, Magdaleno S, Curran T (2001) Identification of reelin-induced sites of tyrosyl phosphorylation on disabled 1. J Biol Chem 276: Keshvara L, Magdaleno S, Benhayon D, Curran T (2002) Cyclin-dependent kinase 5 phosphorylates disabled 1 independently of Reelin signaling. J Neurosci 22: Kubo K, Mikoshiba K, Nakajima K (2002) Secreted Reelin molecules form homodimers. Neurosci Res 43: Lambert de Rouvroit C, Goffinet AM (1998) The reeler mouse as a model of brain development. Adv Anat Embryol Cell Biol 150: Lambert de Rouvroit C, de Bergeyck V, Cortvrindt C, Bar I, Eeckhout Y, Goffinet AM (1999) Reelin, the extracellular matrix protein deficient in reeler mutant mice, is processed by a metalloproteinase. Exp Neurol 156: Meyer G, Lambert de Rouvroit C, Goffinet A, Wahle P (2002) DAB1 mrna and protein expression in developing human cortex. FENS Abstr 1:506. Rice DS, Curran T (2001) Role of the reelin signaling pathway in central nervous system development. Annu Rev Neurosci 24: Senzaki K, Ogawa M, Yagi T (1999) Proteins of the CNR family are multiple receptors for Reelin. Cell 99: Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J (1999) Reeler/ Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell 97: Utsunomiya-Tate N, Kubo K, Tate S, Kainosho M, Katayama E, Nakajima K, Mikoshiba K (2000) Reelin molecules assemble together to form a large protein complex, which is inhibited by the function-blocking CR-50 antibody. Proc Natl Acad Sci USA 97: Cerebral Cortex Jun 2003, V 13 N 6 633
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
Alzheimer s Disease. Joachim Herz. STARS January 11, 2010
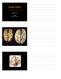 Alzheimer s Disease Joachim Herz STARS January 11, 2010 1 The Hallmarks of Alzheimer Disease Amyloid Plaques Neurofibrillary Tangles From: Medical Library of Utah 1906 First description of Alzheimer s
Alzheimer s Disease Joachim Herz STARS January 11, 2010 1 The Hallmarks of Alzheimer Disease Amyloid Plaques Neurofibrillary Tangles From: Medical Library of Utah 1906 First description of Alzheimer s
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10163 Supplementary Table 1 Efficiency of vector construction. Process wells recovered efficiency (%) Recombineering* 480 461 96 Intermediate plasmids 461 381 83 Recombineering efficiency
doi:10.1038/nature10163 Supplementary Table 1 Efficiency of vector construction. Process wells recovered efficiency (%) Recombineering* 480 461 96 Intermediate plasmids 461 381 83 Recombineering efficiency
Advances in Anatomy Embryology and Cell Biology
 Advances in Anatomy Embryology and Cell Biology Vol. 150 Editors F. Beck, Melbourne D. Brown, Charlestown B. Christ, Freiburg W. -Kriz, Heidelberg E. Marani, Leiden R. Putz, Munchen Y. Sano, Kyoto T. H.
Advances in Anatomy Embryology and Cell Biology Vol. 150 Editors F. Beck, Melbourne D. Brown, Charlestown B. Christ, Freiburg W. -Kriz, Heidelberg E. Marani, Leiden R. Putz, Munchen Y. Sano, Kyoto T. H.
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
GENETICS - CLUTCH CH.15 GENOMES AND GENOMICS.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF GENOMICS Genomics is the study of genomes in their entirety Bioinformatics is the analysis of the information content of genomes - Genes, regulatory sequences,
!! www.clutchprep.com CONCEPT: OVERVIEW OF GENOMICS Genomics is the study of genomes in their entirety Bioinformatics is the analysis of the information content of genomes - Genes, regulatory sequences,
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
7.012 Problem Set 5. Question 1
 Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Problem Set 4
 7.016- Problem Set 4 Question 1 Arginine biosynthesis is an example of multi-step biochemical pathway where each step is catalyzed by a specific enzyme (E1, E2 and E3) as is outlined below. E1 E2 E3 A
7.016- Problem Set 4 Question 1 Arginine biosynthesis is an example of multi-step biochemical pathway where each step is catalyzed by a specific enzyme (E1, E2 and E3) as is outlined below. E1 E2 E3 A
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
3. Translation. 2. Transcription. 1. Replication. and functioning through their expression in. Genes are units perpetuating themselves
 Central Dogma Genes are units perpetuating themselves and functioning through their expression in the form of proteins 1 DNA RNA Protein 2 3 1. Replication 2. Transcription 3. Translation Spring 2002 21
Central Dogma Genes are units perpetuating themselves and functioning through their expression in the form of proteins 1 DNA RNA Protein 2 3 1. Replication 2. Transcription 3. Translation Spring 2002 21
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Chapter 18: Regulation of Gene Expression. 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer
 Chapter 18: Regulation of Gene Expression 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer Gene Regulation Gene regulation refers to all aspects of controlling
Chapter 18: Regulation of Gene Expression 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer Gene Regulation Gene regulation refers to all aspects of controlling
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Additional Practice Problems for Reading Period
 BS 50 Genetics and Genomics Reading Period Additional Practice Problems for Reading Period Question 1. In patients with a particular type of leukemia, their leukemic B lymphocytes display a translocation
BS 50 Genetics and Genomics Reading Period Additional Practice Problems for Reading Period Question 1. In patients with a particular type of leukemia, their leukemic B lymphocytes display a translocation
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
C) Based on your knowledge of transcription, what factors do you expect to ultimately identify? (1 point)
 Question 1 Unlike bacterial RNA polymerase, eukaryotic RNA polymerases need additional assistance in finding a promoter and transcription start site. You wish to reconstitute accurate transcription by
Question 1 Unlike bacterial RNA polymerase, eukaryotic RNA polymerases need additional assistance in finding a promoter and transcription start site. You wish to reconstitute accurate transcription by
Bi 8 Lecture 5. Ellen Rothenberg 19 January 2016
 Bi 8 Lecture 5 MORE ON HOW WE KNOW WHAT WE KNOW and intro to the protein code Ellen Rothenberg 19 January 2016 SIZE AND PURIFICATION BY SYNTHESIS: BASIS OF EARLY SEQUENCING complex mixture of aborted DNA
Bi 8 Lecture 5 MORE ON HOW WE KNOW WHAT WE KNOW and intro to the protein code Ellen Rothenberg 19 January 2016 SIZE AND PURIFICATION BY SYNTHESIS: BASIS OF EARLY SEQUENCING complex mixture of aborted DNA
Site directed mutagenesis, Insertional and Deletion Mutagenesis. Mitesh Shrestha
 Site directed mutagenesis, Insertional and Deletion Mutagenesis Mitesh Shrestha Mutagenesis Mutagenesis (the creation or formation of a mutation) can be used as a powerful genetic tool. By inducing mutations
Site directed mutagenesis, Insertional and Deletion Mutagenesis Mitesh Shrestha Mutagenesis Mutagenesis (the creation or formation of a mutation) can be used as a powerful genetic tool. By inducing mutations
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES
 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Generation of inducible BICD2 knock-out mice. A) The mouse BICD2 locus and gene targeting constructs. To generate an inducible Bicd2
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Generation of inducible BICD2 knock-out mice. A) The mouse BICD2 locus and gene targeting constructs. To generate an inducible Bicd2
UTR Reporter Vectors and Viruses
 UTR Reporter Vectors and Viruses 3 UTR, 5 UTR, Promoter Reporter (Version 1) Applied Biological Materials Inc. #1-3671 Viking Way Richmond, BC V6V 2J5 Canada Notice to Purchaser All abm products are for
UTR Reporter Vectors and Viruses 3 UTR, 5 UTR, Promoter Reporter (Version 1) Applied Biological Materials Inc. #1-3671 Viking Way Richmond, BC V6V 2J5 Canada Notice to Purchaser All abm products are for
Genome annotation & EST
 Genome annotation & EST What is genome annotation? The process of taking the raw DNA sequence produced by the genome sequence projects and adding the layers of analysis and interpretation necessary
Genome annotation & EST What is genome annotation? The process of taking the raw DNA sequence produced by the genome sequence projects and adding the layers of analysis and interpretation necessary
To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression
 Supplemental figures Supplemental Figure. 1. Silencing expression of Celsr3 by shrna. To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression plasmids for the shrna
Supplemental figures Supplemental Figure. 1. Silencing expression of Celsr3 by shrna. To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression plasmids for the shrna
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Control of Eukaryotic Gene Expression (Learning Objectives)
 Control of Eukaryotic Gene Expression (Learning Objectives) 1. Compare and contrast chromatin and chromosome: composition, proteins involved and level of packing. Explain the structure and function of
Control of Eukaryotic Gene Expression (Learning Objectives) 1. Compare and contrast chromatin and chromosome: composition, proteins involved and level of packing. Explain the structure and function of
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM Part I: Definitions. Homology: Reverse transcriptase. Allostery: cdna library
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX
 Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Test Bank for Molecular Cell Biology 7th Edition by Lodish
 Test Bank for Molecular Cell Biology 7th Edition by Lodish Link download full: http://testbankair.com/download/test-bank-formolecular-cell-biology-7th-edition-by-lodish/ Chapter 5 Molecular Genetic Techniques
Test Bank for Molecular Cell Biology 7th Edition by Lodish Link download full: http://testbankair.com/download/test-bank-formolecular-cell-biology-7th-edition-by-lodish/ Chapter 5 Molecular Genetic Techniques
Switch On Protein-Protein Interactions
 Switch On Protein-Protein Interactions Rapid and specific control of signal transduction pathways, protein activity, protein localization, transcription, and more Dimerizer protein 1 protein 2 Clontech
Switch On Protein-Protein Interactions Rapid and specific control of signal transduction pathways, protein activity, protein localization, transcription, and more Dimerizer protein 1 protein 2 Clontech
32 Gene regulation in Eukaryotes Lecture Outline 11/28/05. Gene Regulation in Prokaryotes and Eukarykotes
 3 Gene regulation in Eukaryotes Lecture Outline /8/05 Gene regulation in eukaryotes Chromatin remodeling More kinds of control elements Promoters, Enhancers, and Silencers Combinatorial control Cell-specific
3 Gene regulation in Eukaryotes Lecture Outline /8/05 Gene regulation in eukaryotes Chromatin remodeling More kinds of control elements Promoters, Enhancers, and Silencers Combinatorial control Cell-specific
Supplemental Materials and Methods. Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2,
 Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Reelin Binds 3 1 Integrin and Inhibits Neuronal Migration
 Neuron, Vol. 27, 33 44, July, 2000, Copyright 2000 by Cell Press Reelin Binds 3 1 Integrin and Inhibits Neuronal Migration Lori Dulabon,* Eric C. Olson, Mary G. Taglienti, Scott Eisenhuth,* Barbara McGrath,*
Neuron, Vol. 27, 33 44, July, 2000, Copyright 2000 by Cell Press Reelin Binds 3 1 Integrin and Inhibits Neuronal Migration Lori Dulabon,* Eric C. Olson, Mary G. Taglienti, Scott Eisenhuth,* Barbara McGrath,*
PrimePCR Assay Validation Report
 Gene Information Gene Name fibroblast growth factor receptor 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID FGFR1 Human The protein encoded by this gene
Gene Information Gene Name fibroblast growth factor receptor 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID FGFR1 Human The protein encoded by this gene
Reading Lecture 8: Lecture 9: Lecture 8. DNA Libraries. Definition Types Construction
 Lecture 8 Reading Lecture 8: 96-110 Lecture 9: 111-120 DNA Libraries Definition Types Construction 142 DNA Libraries A DNA library is a collection of clones of genomic fragments or cdnas from a certain
Lecture 8 Reading Lecture 8: 96-110 Lecture 9: 111-120 DNA Libraries Definition Types Construction 142 DNA Libraries A DNA library is a collection of clones of genomic fragments or cdnas from a certain
Table S1. Primers used in the study
 Table S1. Primers used in the study Primer name Application Sequence I1F16 Genotyping GGCAAGTGAGTGAGTGCCTA I1R11 Genotyping CCCACTCGTATTGACGCTCT V19 Genotyping GGGTCTCAAAGTCAGGGTCA D18Mit184-F Genotyping
Table S1. Primers used in the study Primer name Application Sequence I1F16 Genotyping GGCAAGTGAGTGAGTGCCTA I1R11 Genotyping CCCACTCGTATTGACGCTCT V19 Genotyping GGGTCTCAAAGTCAGGGTCA D18Mit184-F Genotyping
Introducing new DNA into the genome requires cloning the donor sequence, delivery of the cloned DNA into the cell, and integration into the genome.
 Key Terms Chapter 32: Genetic Engineering Cloning describes propagation of a DNA sequence by incorporating it into a hybrid construct that can be replicated in a host cell. A cloning vector is a plasmid
Key Terms Chapter 32: Genetic Engineering Cloning describes propagation of a DNA sequence by incorporating it into a hybrid construct that can be replicated in a host cell. A cloning vector is a plasmid
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr.
 MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
Gene Expression: Transcription
 Gene Expression: Transcription The majority of genes are expressed as the proteins they encode. The process occurs in two steps: Transcription = DNA RNA Translation = RNA protein Taken together, they make
Gene Expression: Transcription The majority of genes are expressed as the proteins they encode. The process occurs in two steps: Transcription = DNA RNA Translation = RNA protein Taken together, they make
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
TRANSGENIC ANIMALS. transient. stable. - Two methods to produce transgenic animals:
 Only for teaching purposes - not for reproduction or sale CELL TRANSFECTION transient stable TRANSGENIC ANIMALS - Two methods to produce transgenic animals: 1- DNA microinjection 2- embryonic stem cell-mediated
Only for teaching purposes - not for reproduction or sale CELL TRANSFECTION transient stable TRANSGENIC ANIMALS - Two methods to produce transgenic animals: 1- DNA microinjection 2- embryonic stem cell-mediated
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Figure S1 is related to Figure 1B, showing more details of outer segment of
 Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Chapter 14 Regulation of Transcription
 Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Bootcamp: Molecular Biology Techniques and Interpretation
 Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Year III Pharm.D Dr. V. Chitra
 Year III Pharm.D Dr. V. Chitra 1 Genome entire genetic material of an individual Transcriptome set of transcribed sequences Proteome set of proteins encoded by the genome 2 Only one strand of DNA serves
Year III Pharm.D Dr. V. Chitra 1 Genome entire genetic material of an individual Transcriptome set of transcribed sequences Proteome set of proteins encoded by the genome 2 Only one strand of DNA serves
Biol 321 Spring 2013 Quiz 4 25 pts NAME
 Biol 321 Spring 2013 Quiz 4 25 pts NAME 1. (3 pts.) a. What is the name of this compound? BE EXPLICIT deoxyribose 5 b. Number the carbons on this structure: 4 1 3 2 2. (4 pts.) Circle True or False. If
Biol 321 Spring 2013 Quiz 4 25 pts NAME 1. (3 pts.) a. What is the name of this compound? BE EXPLICIT deoxyribose 5 b. Number the carbons on this structure: 4 1 3 2 2. (4 pts.) Circle True or False. If
Problem Set 8. Answer Key
 MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
Document S1. Supplemental Experimental Procedures and Three Figures (see next page)
 Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Introduction to Genome Biology
 Introduction to Genome Biology Sandrine Dudoit, Wolfgang Huber, Robert Gentleman Bioconductor Short Course 2006 Copyright 2006, all rights reserved Outline Cells, chromosomes, and cell division DNA structure
Introduction to Genome Biology Sandrine Dudoit, Wolfgang Huber, Robert Gentleman Bioconductor Short Course 2006 Copyright 2006, all rights reserved Outline Cells, chromosomes, and cell division DNA structure
Supplementary Figure 1. Drawing of spinal cord open-book preparations and DiI tracing. Nature Neuroscience: doi: /nn.3893
 Supplementary Figure 1 Drawing of spinal cord open-book preparations and DiI tracing. Supplementary Figure 2 In ovo electroporation of dominant-negative PlexinA1 in commissural neurons induces midline
Supplementary Figure 1 Drawing of spinal cord open-book preparations and DiI tracing. Supplementary Figure 2 In ovo electroporation of dominant-negative PlexinA1 in commissural neurons induces midline
Cloning, chromosome localization and features of a novel human gene, MATH2
 Indian Academy of Sciences Cloning, chromosome localization and features of a novel human gene, MATH2 LINGCHEN GUO 1, MIN JIANG 1, YUSHU MA 1, HAIPENG CHENG 1, XIAOHUA NI 1, YANGSHENG JIN 2, YI XIE 1 *
Indian Academy of Sciences Cloning, chromosome localization and features of a novel human gene, MATH2 LINGCHEN GUO 1, MIN JIANG 1, YUSHU MA 1, HAIPENG CHENG 1, XIAOHUA NI 1, YANGSHENG JIN 2, YI XIE 1 *
A Modified Digestion-Circularization PCR (DC-PCR) Approach to Detect Hypermutation- Associated DNA Double-Strand Breaks
 A Modified Digestion-Circularization PCR (DC-PCR) Approach to Detect Hypermutation- Associated DNA Double-Strand Breaks SARAH K. DICKERSON AND F. NINA PAPAVASILIOU Laboratory of Lymphocyte Biology, The
A Modified Digestion-Circularization PCR (DC-PCR) Approach to Detect Hypermutation- Associated DNA Double-Strand Breaks SARAH K. DICKERSON AND F. NINA PAPAVASILIOU Laboratory of Lymphocyte Biology, The
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Chapter 10 (Part II) Gene Isolation and Manipulation
 Biology 234 J. G. Doheny Chapter 10 (Part II) Gene Isolation and Manipulation Practice Questions: Answer the following questions with one or two sentences. 1. What does PCR stand for? 2. What does the
Biology 234 J. G. Doheny Chapter 10 (Part II) Gene Isolation and Manipulation Practice Questions: Answer the following questions with one or two sentences. 1. What does PCR stand for? 2. What does the
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
7.06 Problem Set #3, 2006
 7.06 Problem Set #3, 2006 1. You are studying the EGF/Ras/MAPK pathway in cultured cells. When the pathway is activated, cells are signaled to proliferate. You generate various mutants described below.
7.06 Problem Set #3, 2006 1. You are studying the EGF/Ras/MAPK pathway in cultured cells. When the pathway is activated, cells are signaled to proliferate. You generate various mutants described below.
INVESTIGATION OF THE BINDING SPECIFICITY OF IGF-IR USING MONOCLONAL ANTIBODIES
 INVESTIGATION OF THE BINDING SPECIFICITY OF IGF-IR USING MONOCLONAL ANTIBODIES By Mehrnaz Keyhanfar, Pharm.D. A thesis submitted to the University of Adelaide, South Australia in fulfilment of the requirements
INVESTIGATION OF THE BINDING SPECIFICITY OF IGF-IR USING MONOCLONAL ANTIBODIES By Mehrnaz Keyhanfar, Pharm.D. A thesis submitted to the University of Adelaide, South Australia in fulfilment of the requirements
Supplement Figure S1:
 Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
GENETICS EXAM 3 FALL a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size.
 Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Supplementary Methods
 Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
CHAPTER 4 Cloning, expression, purification and preparation of site-directed mutants of NDUFS3 and NDUFS7
 CHAPTER 4 Cloning, expression, purification and preparation of site-directed mutants of NDUFS3 and NDUFS7 subunits of human mitochondrial Complex-I Q module N DUFS2, 3, 7 and 8 form the core subunits of
CHAPTER 4 Cloning, expression, purification and preparation of site-directed mutants of NDUFS3 and NDUFS7 subunits of human mitochondrial Complex-I Q module N DUFS2, 3, 7 and 8 form the core subunits of
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit
 Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1154040/dc1 Supporting Online Material for Selective Blockade of MicroRNA Processing by Lin-28 Srinivas R. Viswanathan, George Q. Daley,* Richard I. Gregory* *To whom
www.sciencemag.org/cgi/content/full/1154040/dc1 Supporting Online Material for Selective Blockade of MicroRNA Processing by Lin-28 Srinivas R. Viswanathan, George Q. Daley,* Richard I. Gregory* *To whom
Characterization and Functional Analysis of a Newly Identified Human MT5-MMP Transcript Variant Isolated from Multipotent NT2 Cells
 Virginia Commonwealth University VCU Scholars Compass Theses and Dissertations Graduate School 2006 Characterization and Functional Analysis of a Newly Identified Human MT5-MMP Transcript Variant Isolated
Virginia Commonwealth University VCU Scholars Compass Theses and Dissertations Graduate School 2006 Characterization and Functional Analysis of a Newly Identified Human MT5-MMP Transcript Variant Isolated
Lecture for Wednesday. Dr. Prince BIOL 1408
 Lecture for Wednesday Dr. Prince BIOL 1408 THE FLOW OF GENETIC INFORMATION FROM DNA TO RNA TO PROTEIN Copyright 2009 Pearson Education, Inc. Genes are expressed as proteins A gene is a segment of DNA that
Lecture for Wednesday Dr. Prince BIOL 1408 THE FLOW OF GENETIC INFORMATION FROM DNA TO RNA TO PROTEIN Copyright 2009 Pearson Education, Inc. Genes are expressed as proteins A gene is a segment of DNA that
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene. Andrew ElBardissi, The Pennsylvania State University
 Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MCB 102 University of California, Berkeley August 11 13, Problem Set 8
 MCB 102 University of California, Berkeley August 11 13, 2009 Isabelle Philipp Handout Problem Set 8 The answer key will be posted by Tuesday August 11. Try to solve the problem sets always first without
MCB 102 University of California, Berkeley August 11 13, 2009 Isabelle Philipp Handout Problem Set 8 The answer key will be posted by Tuesday August 11. Try to solve the problem sets always first without
Transcriptomics. Marta Puig Institut de Biotecnologia i Biomedicina Universitat Autònoma de Barcelona
 Transcriptomics Marta Puig Institut de Biotecnologia i Biomedicina Universitat Autònoma de Barcelona Central dogma of molecular biology Central dogma of molecular biology Genome Complete DNA content of
Transcriptomics Marta Puig Institut de Biotecnologia i Biomedicina Universitat Autònoma de Barcelona Central dogma of molecular biology Central dogma of molecular biology Genome Complete DNA content of
Chapter 5. Genetic Models. Organization and Expression of Immunoglobulin Genes 3. The two-gene model: Models to Explain Antibody Diversity
 Chapter 5 Organization and Expression of Immunoglobulin Genes 3 4 5 6 Genetic Models How to account for: ) Vast diversity of antibody specificities ) Presence of Variable regions at the amino end of Heavy
Chapter 5 Organization and Expression of Immunoglobulin Genes 3 4 5 6 Genetic Models How to account for: ) Vast diversity of antibody specificities ) Presence of Variable regions at the amino end of Heavy
The Two-Hybrid System
 Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine The Two-Hybrid System Carolina Vollert & Peter Uetz Institut für Genetik Forschungszentrum Karlsruhe PO Box 3640 D-76021 Karlsruhe
Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine The Two-Hybrid System Carolina Vollert & Peter Uetz Institut für Genetik Forschungszentrum Karlsruhe PO Box 3640 D-76021 Karlsruhe
Supplementary Information. A novel human endogenous retroviral protein inhibits cell-cell fusion. Supplementary Figures:
 Supplementary Information A novel human endogenous retroviral protein inhibits cell-cell fusion Jun Sugimoto, Makiko Sugimoto, Helene Bernstein, Yoshihiro Jinno and Danny J. Schust Supplementary Figures:
Supplementary Information A novel human endogenous retroviral protein inhibits cell-cell fusion Jun Sugimoto, Makiko Sugimoto, Helene Bernstein, Yoshihiro Jinno and Danny J. Schust Supplementary Figures:
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-18 CELL BIOLOGY BIO-5005B Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-18 CELL BIOLOGY BIO-5005B Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
Themes: RNA and RNA Processing. Messenger RNA (mrna) What is a gene? RNA is very versatile! RNA-RNA interactions are very important!
 Themes: RNA is very versatile! RNA and RNA Processing Chapter 14 RNA-RNA interactions are very important! Prokaryotes and Eukaryotes have many important differences. Messenger RNA (mrna) Carries genetic
Themes: RNA is very versatile! RNA and RNA Processing Chapter 14 RNA-RNA interactions are very important! Prokaryotes and Eukaryotes have many important differences. Messenger RNA (mrna) Carries genetic
PLNT2530 (2018) Unit 6b Sequence Libraries
 PLNT2530 (2018) Unit 6b Sequence Libraries Molecular Biotechnology (Ch 4) Analysis of Genes and Genomes (Ch 5) Unless otherwise cited or referenced, all content of this presenataion is licensed under the
PLNT2530 (2018) Unit 6b Sequence Libraries Molecular Biotechnology (Ch 4) Analysis of Genes and Genomes (Ch 5) Unless otherwise cited or referenced, all content of this presenataion is licensed under the
GENES AND CHROMOSOMES V. Lecture 7. Biology Department Concordia University. Dr. S. Azam BIOL 266/
 1 GENES AND CHROMOSOMES V Lecture 7 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University 2 CELL NUCLEUS AND THE CONTROL OF GENE EXPRESSION An Overview of Gene Regulation in Eukaryotes
1 GENES AND CHROMOSOMES V Lecture 7 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University 2 CELL NUCLEUS AND THE CONTROL OF GENE EXPRESSION An Overview of Gene Regulation in Eukaryotes
a previous report, Lee and colleagues showed that Oct4 and Sox2 bind to DXPas34 and Xite
 SUPPLEMENTARY INFORMATION doi:1.138/nature9496 a 1 Relative RNA levels D12 female ES cells 24h Control sirna 24h sirna Oct4 5 b,4 Oct4 Xist (wt) Xist (mut) Tsix 3' (wt) Tsix 5' (wt) Beads Oct4,2 c,2 Xist
SUPPLEMENTARY INFORMATION doi:1.138/nature9496 a 1 Relative RNA levels D12 female ES cells 24h Control sirna 24h sirna Oct4 5 b,4 Oct4 Xist (wt) Xist (mut) Tsix 3' (wt) Tsix 5' (wt) Beads Oct4,2 c,2 Xist
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Bi 8 Lecture 4. Ellen Rothenberg 14 January Reading: from Alberts Ch. 8
 Bi 8 Lecture 4 DNA approaches: How we know what we know Ellen Rothenberg 14 January 2016 Reading: from Alberts Ch. 8 Central concept: DNA or RNA polymer length as an identifying feature RNA has intrinsically
Bi 8 Lecture 4 DNA approaches: How we know what we know Ellen Rothenberg 14 January 2016 Reading: from Alberts Ch. 8 Central concept: DNA or RNA polymer length as an identifying feature RNA has intrinsically
Divergent roles of ApoER2 and Vldlr in the migration of cortical neurons
 RESEARCH ARTICLE 3883 Development 134, 3883-3891 (2007) doi:10.1242/dev.005447 Divergent roles of ApoER2 and Vldlr in the migration of cortical neurons Iris Hack 1, *, Sabine Hellwig 1,2, *, Dirk Junghans
RESEARCH ARTICLE 3883 Development 134, 3883-3891 (2007) doi:10.1242/dev.005447 Divergent roles of ApoER2 and Vldlr in the migration of cortical neurons Iris Hack 1, *, Sabine Hellwig 1,2, *, Dirk Junghans
GH RECEPTOR (GHR) is a transmembrane protein that
 0163-769X/98/$03.00/0 Endocrine Reviews 19(5): 559 582 Copyright 1998 by The Endocrine Society Printed in U.S.A. Alternative Processing of Growth Hormone Receptor Transcripts* ALEXANDER EDENS AND FRANK
0163-769X/98/$03.00/0 Endocrine Reviews 19(5): 559 582 Copyright 1998 by The Endocrine Society Printed in U.S.A. Alternative Processing of Growth Hormone Receptor Transcripts* ALEXANDER EDENS AND FRANK
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
