Proteomics and Mass Spectrometry Course plan
|
|
|
- Agnes Nelson
- 6 years ago
- Views:
Transcription
1 Proteomics and Mass Spectrometry 2013 The team Stephen Barnes, PhD Matt Renfrow, PhD Helen Kim, PhD , MCLM , MCLM , MCLM 460A Jim Mobley, PhD Jeevan Prasain, PhD Peter E. Prevelige, PhD , THT , MCLM , BBRB 416 Course plan Meet Mondays/Wednesdays/Fridays in MCLM 401 from 9-10:30 am (Jan 7-Mar 22) Graduate Students taking this course are required to attend each session (unless there is advance communication with the instructor) Evaluations will be made from exams and in-class presentations Where possible, class notes will be available on the UAB proteomics website (go to - click on Class) 1
2 Recommended general texts Suggested text - Introduction to Proteomics by Daniel C. Liebler, 2002 Also see The Expanding Role of Mass Spectrometry in Biotechnology by Gary Siuzdak (a 2003 edition of the 1996 first edition) Mass spectrometry data analysis in proteomics, (ed., Mathiesson, R) in Methods in Molecular Biology, vol 367. Protein Mass Spectrometry, Volume 52 (Comprehensive Analytical Chemistry) (Julian Whitelegge (Editor) See also Suggested readings Kenyon G, et al. Defining the mandate of proteomics in the postgenomics era: workshop report. Mol Cell Proteomics, 1: (2002) Righetti P. et al. Prefractionation techniques in proteome analysis: the mining tools of the third millennium. Electrophoresis, 26: (2005) Anderson NL. The roles of multiple proteomic platforms in a pipeline for new diagnostics. Mol Cell Proteomics, 4: (2005) Venkatesan et al. An empirical framework for binary interactome mapping. Nat Methods, 6:83-90 (2009) PMID: Yan W et al. Evolution of organelle-associated protein profiling. J Proteomics, 72:4-11 (2009) PMID: Pan S, et al. Mass Spectrometry Based Targeted Protein Quantification: Methods and Applications. J Proteome Res, 8: (2009) PMID: Compton PD et al. On the Scalability and Requirements of Whole Protein Mass Spectrometry. Anal Chem, 83: (2011) PMID:
3 BMG/PHR section 1 Jan 7, Mon Barnes Analyzing biomolecules. The impact of omics on biomedical research Jan 9, Wed H. Kim Simplifying the proteome - techniques of protein purification Jan 11, Fri H. Kim Protein separation by electrophoresis and other 2D-methods Jan 14, Mon M. Renfrow The Mass Spectrum: What does it show you? MS 1, MS 2, MS n Jan 16, Wed M. Renfrow Mass Spectrometry: Detecting and moving ions in the gas phase. Instrumentation, Mass Analyzers, Ionization Jan 18, Fri M. Renfrow Biological Mass Spectrometry: Ionization, Calculating Mass, Charge Jan 21, Mon Martin Luther King Holiday Jan 23, Wed M. Renfrow Peptide/protein identification by MS, Search algorithms, false positives. Jan 25, Fri S. Barnes Qualitative burrowing of the proteome identifying PTMs Jan 28, Mon S. Barnes Quantitative burrowing of the proteome Labeling, label-free and absolute quantification Jan 30, Wed C. Crasto Web tools and the proteome/metabolome; Expasy, KEGG, NCBI, others Feb 1, Fri S. Barnes MRMPath; MRMutation; MRMass Space Feb 4, Mon J. Prasain Lipidomics and other small molecules by LC-MS Feb 6, Wed M. Renfrow Exam (Possible lecture catch up and questions) BMG/PHR section 2 Feb 8, Fri J. Prasain Metabolomics LC-MS, GC-MS and NMR Feb 11, Mon J. Prasain Quantitative analysis/method validation in metabolomics Feb 13, Wed S. Barnes Enzymology, metabolism and mass spectrometry Feb 15, Fri Student presentations Feb 18, Mon M. Renfrow Analysis of protein-protein interactions by affinity purification and mass spectrometry Feb 20, Wed M. Renfrow Applications of FT-ICR-MS Feb 22, Fri J. Novak/Renfrow Mass spectrometry in glycomics research Application to IgA nephropathy Feb 25, Mon E. Shonsey Application of MS in Forensics Feb 27, Wed S. Barnes Applications of MS to tissue imaging the lens and the metabolome 3
4 BMG/PHR section 3 Mar 1, Fri Student presentations Mar 4, Mon P. Prevelige Mass Spectrometry as a Tool for Studying Protein Structure Mar 6, Wed P. Prevelige Study of macromolecular structures protein complexes Mar 8, Fri H. Kim Use of proteomics and MS methods in the study of the brain proteome and neurodegenerative diseases Mar 11, Mon H. Kim/S. Barnes Putting it all together by-passing pyruvate kinase Mar 13, Wed S. Barnes Isotopes in mass spectrometry Mar 15, Fri S. Barnes Applying mass spectrometry to Free Radical Biology Mar 22, Fri Final report due Course learning objectives Introduction to the concepts and practice of systems biology where it involves mass spectrometry Sample ionization and mass spectrometers Mass spectrometry and its principal methods protein and peptide ID; peptide and metabolite ion fragmentation; stable isotope labeling; quantification 4
5 Course learning objectives Informatics, statistics and quality control in mass spectrometry Importance of prefractionation in proteomics - 2DE, LC and arrays Applying mass spectrometry to protein modifications, function, structure and biological location, and to other biological molecules History of proteomics Essentially preceded genomics Human protein index conceived in the 1970s by Norman and Leigh Anderson The term proteomics coined by Marc Wilkins in 1994 Human proteomics initiative (HPI) began in 2000 in Switzerland - Human Proteome Organization (HUPO) had meetings in 2002 in Versailles; 2003 in Montreal; 2004 in Beijing; 2005 in Munich; 2006 in Long Beach; 2007 in Seoul; 2008 in Amsterdam; 2009 in Toronto; 2010 in Sydney; 2011 in Geneva; 2012 in Boston; 2013 to be in Yokohoma 5
6 What proteomics is, what it isn t Proteomics is not just a mass spectrum of a spot on a gel George Kenyon, 2002 National Academy of Sciences Symposium Proteomics is the identities, quantities, structures, and biochemical and cellular functions of all proteins in an organism, organ or organelle, and how these vary in space, time and physiological state. Collapse of the single target paradigm - the need for systems biology Old paradigm Diseases are due to single genes - by knocking out the gene, or designing specific inhibitors to its protein, disease can be cured But the gene KO mouse didn t notice the loss of the gene New paradigm We have to understand gene and protein networks - proteins don t act alone - effective systems have built in redundancy 6
7 Classical NIH R01 Research styles A specific target and meaningful substrates Emphasis on mechanism Hypothesis-driven Linearizes locally multi-dimensional space Example Using an X-ray crystal structure of a protein to determine if a specific compound can fit into a binding pocket - from this a disease can be cured - this approach ignores whether the compound can get to the necessary biological site, whether it remains chemically intact, and where else it goes From substrates to targets to systems - a changing paradigm Classical approach - one substrate/one target Mid 1980s - use of a pure reagent to isolate DNAs from cdna libraries (multiple targets) Early 1990s - use of a reagent library (multiple ligands) to perfect interaction with a specific target effects of specific reagents on cell systems using DNA microarrays (500+ genes change, not just one) integration of transcriptomics, proteomics, peptidomics, metabolomics (everything changes, just like in ecology) 2010 NextGen and RNASeq analyses introduced (the canonical sequence is a myth and new transcriptome products) 7
8 Direction of NIH Research Metabolomics 1950s-60s emphasis on determining metabolic pathways 20+ Nobel prizes 1950s-early 1980s Identification and purification of proteins 1995-present Omics galore Transcriptomics (microarray, deep sequencing, RNA seq) Proteomics Sequencing of genes cdna libraries orthogonal research 2012 Human genome ENCODE project reveals the extent of DNA expression and roles for junk DNA, as well as intergenic proteins Sequencing of the human genome period of non-orthogonal research?junk DNA Sequencing of genomes of common experimental models 2006 First ENCODE project on 1% 2005 Tiling arrays reveal of the human genome reveals RNAs that there is far more coming from more than one gene expressed RNA than mrna Exploring information space - the Systems Biology approach Systems biology means measuring everything about a system at the same time For a long time, it was deemed as too complex for useful or purposeful investigation But are the tools available today? 8
9 Systems Biology To understand biology at the system level, we must examine the structure and dynamics of cellular and organismal function, rather than the characteristics of isolated parts of a cell or organism. Properties of systems, such as robustness, emerge as central issues, and understanding these properties may have an impact on the future of medicine. However, many breakthroughs in experimental devices, advanced software, and analytical methods are required before the achievements of systems biology can live up to their much-touted potential. Kitano, 2002 The Biological Data of the Future Destructive Qualitative Uni-dimensional Low temporal resolution Low data density Variable standards Non cumulative Non-destructive Quantitative Multi-dimensional and spatially resolved High Temporal resolution High data density Stricter standards Cumulative Current nature of data Elias Zerhouni, FASEB
10 Techniques in Systems Biology DNA microarrays to describe and quantify the transcriptosome Being replaced by NextGen sequencing and RNASeq Large scale and small scale proteomics Protein arrays Protein structure Metabolomics Integrated computational models Papers on systems biology Deo RC, MacRae CA. The zebrafish: scalable in vivo modeling for systems biology. WIRES Systems Biol 2011;3: Gardy JL et al. Enabling a systems biology approach to immunology: focus on innate immunity. Trends in Immunol 2011;30: Kriete A et al. Computational systems biology of aging. WIRES Systems Biol 2011;3: Shapira SD, Hacohen N. Systems biology approaches to dissect mammalian innate immunity. Current Opin Immunol 2011;23: Jorgenson JM, Haddow, PC. Visualization in simulation tools: requirements and a tool specification to support the teaching of dynamic biological processes. J Bioinform Comp Biol 2011;9: Gerdtzen ZP. Modeling Metabolic Networks for Mammalian Cell Systems: General Considerations, Modeling Strategies, and Available Tools. Adv Biochem Engin/Biotechnol DOI: /10_2011_120 10
11 High dimensionality of microarray or proteomics data means you must understand statistics While reproducible data can be obtained, the large numbers of parameters (individual genes or proteins) require large changes in expression before a change can be regarded as significant Use of the Bonferroni correction: A conservative correction number of observed genes Vulnerability of a system To really understand biological systems, you have to appreciate their dynamic state Read about control theory Realize that systems are subject to rhythms Subject them to fourier transform analysis to detect their resonance (requires far more data than we can currently collect) A small signal at the right frequency can disrupt the system Analogies the small boy in the bath and the screech of chalk on a chalk board 11
12 Hazards of interpreting transcriptomics (proteomic) data Expression patterns are the place where environmental variables and genetic variation come together. Environmental variables will affect gene expression levels. Don t we need to be very careful to understand the environmental inputs that might have an impact on that expression? Perhaps an over-thecounter herbal supplement might cause an expression pattern that looks like that of a very aggressive tumor. Abridged from Karen Kline, 2002 Why study the proteome when we can study DNA and RNA? NextGen and RNASeq analysis allows one to examine the mrna levels of thousands and thousands of genes However, the correlation between gene expression and protein levels is often poor, although that may be an issue of the timing of sampling Is this a new finding? No, before the age of molecular biology, it was well known 12
13 Apparent structured relationship between gene expression and protein content Ideker et al., Science 292: 929 (2001) Differing correlations between RNA expression of a gene and its associated protein amount in 23 different cell lines a) glucocorticoid-induced transcript 1 protein b) G1/S-specific cyclin-e2 c) jun-d, a transcription factor d) neurotransmitter transporter NTT73 From Gry et al. BMC Genomics 10:365 (2009) 13
14 Housekeeping genes and proteins are probably related This is the relationship between mrna (red) and protein (blue) levels expression of a house-keeping gene/protein, i.e., one that has to be expressed at all times Even with the small perturbation, the amounts of mrna and protein are well correlated to each other Time (hr) (Barnes & Allison, 2004) Sampling time affects interpretation of correlation between mrna and protein expression for important proteins Determining the relationship between mrna (red) and protein (blue) levels depends totally on when you measure them - for the figure opposite, the ratio at 2.5 hr is 10:1, whereas at 7.5 hr it s 1:100 better to measure the ratio over time and integrate the area under the curve mrna or protein concn Time (hr) (Barnes & Allison, 2004) 14
15 Figuring the right answer - the disadvantage of a static set of data Which ball in this picture is the real one? You have ten choices Take home lessons in analyzing proteins with proteomics methods The fewer proteins in the proteome you analyze, the better the chances of detecting the ones that matter. Genomics data can complement proteomics data. Understanding the biological properties of the proteins of interest can enhance proteomics analysis. Intrinsic properties of proteins form the basis of invaluable prefractionation prior to proteomics analysis. Quality control is an issue that becomes increasingly important with large datasets and measurement of small changes 15
16 Predicting the proteome Bioinformatics is the basis of high throughput proteome analysis using mass spectrometry. Protein sequences can be computationally predicted from the genome sequence However, bioinformatics is not able to predict with accuracy the sites or chemistry of posttranslational modifications - these need to be defined chemically (using mass spectrometry) Proteins in individuals will have different sequences there are 161 known natural mutations of the LDL receptor and 1361 mutations of human p53 Predicting the proteome Predicting the proteome has elements of a circular argument protein sequences were initially determined chemically and were correlated with the early gene sequences. It then became easier to sequence a protein from its mrna (captured from a cdna library). This could be checked (to a degree) by comparison to peptide sequences. Now we have the human genome. So, is it valid to predict the genes (and hence the proteome) from the sequence of the genome? We re doing this in current research. But as we ll see, the mass spectrometer is the ultimate test of this hypothesis - why? because of its mass accuracy 16
17 Pro immonium ion y What does this tell you? y [M+H] b b 2 -CO Mass/Charge, Da Protein structure Determined by folding - folding rules not yet defined - cannot predict structure de novo X-ray crystallography has been used to produce elegant structural information NMR and H-D exchange combined with mass spec enable the in-solution structure to be determined (see Peter Prevelige s lectures on March 6/8) 17
18 Protein informatics The predicted sequences of the proteins encoded by genes in sequenced genomes are available in many publicly available databases (subject to the limitations mentioned earlier) The mass of the protein is less useful (for bottom up, but not top-down analyses) than the masses of its fragment ions - as we ll see later, the masses of tryptic peptides can be used to identify a protein in a matter of seconds So, what do we do with all these data? Management of the data generated by DNA microarrays, NextGen Sequencing, RNASeq and proteomics/protein arrays High dimensional analysis Beyond the capabilities of individual investigators Urgent need for visualization tools The importance of new statistical methods for analysis of high dimensional systems 18
19 PROTIG and Videocast There is an NIH-based proteomics interest group (PROTIG) Proteomics and mass spec talks are available for viewing (using Real Player) Log on at Podcasts are also available 19
Proteomics and Protein Mass Spectrometry Course plan
 Proteomics and Protein Mass Spectrometry 2006 Stephen Barnes, PhD Helen Kim, PhD 4-7117, MCLM 452 4-3880, MCLM 460A sbarnes@uab.edu helenkim@uab.edu Matt Renfrow, PhD 6-4681, MCLM 570 Renfrow@uab.edu Course
Proteomics and Protein Mass Spectrometry 2006 Stephen Barnes, PhD Helen Kim, PhD 4-7117, MCLM 452 4-3880, MCLM 460A sbarnes@uab.edu helenkim@uab.edu Matt Renfrow, PhD 6-4681, MCLM 570 Renfrow@uab.edu Course
Metabolomics in Systems Biology
 Metabolomics in Systems Biology Basil J. Nikolau W.M. Keck Metabolomics Research Laboratory Iowa State University February 7, 2008 Outline What is metabolomics? Why is metabolomics important? What are
Metabolomics in Systems Biology Basil J. Nikolau W.M. Keck Metabolomics Research Laboratory Iowa State University February 7, 2008 Outline What is metabolomics? Why is metabolomics important? What are
Lecture 23: Metabolomics Technology
 MBioS 478/578 Bioinformatics Mark Lange Lecture 23: Metabolomics Technology Definitions and Background Technologies Nuclear Magnetic Resonance Mass Spectrometry General Introduction Fourier-Transform Mass
MBioS 478/578 Bioinformatics Mark Lange Lecture 23: Metabolomics Technology Definitions and Background Technologies Nuclear Magnetic Resonance Mass Spectrometry General Introduction Fourier-Transform Mass
Exam MOL3007 Functional Genomics
 Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Tuesday May 29 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Tuesday May 29 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
The Five Key Elements of a Successful Metabolomics Study
 The Five Key Elements of a Successful Metabolomics Study Metabolomics: Completing the Biological Picture Metabolomics is offering new insights into systems biology, empowering biomarker discovery, and
The Five Key Elements of a Successful Metabolomics Study Metabolomics: Completing the Biological Picture Metabolomics is offering new insights into systems biology, empowering biomarker discovery, and
MOLECULAR BIOLOGY OF EUKARYOTES 2016 SYLLABUS
 03-442 Lectures: MWF 9:30-10:20 a.m. Doherty Hall 2105 03-742 Advanced Discussion Section: Time and place to be announced Probably Mon 4-6 p.m. or 6-8p.m.? Once we establish who is taking the advanced
03-442 Lectures: MWF 9:30-10:20 a.m. Doherty Hall 2105 03-742 Advanced Discussion Section: Time and place to be announced Probably Mon 4-6 p.m. or 6-8p.m.? Once we establish who is taking the advanced
The Integrated Biomedical Sciences Graduate Program
 The Integrated Biomedical Sciences Graduate Program at the university of notre dame Cutting-edge biomedical research and training that transcends traditional departmental and disciplinary boundaries to
The Integrated Biomedical Sciences Graduate Program at the university of notre dame Cutting-edge biomedical research and training that transcends traditional departmental and disciplinary boundaries to
Advances in analytical biochemistry and systems biology: Proteomics
 Advances in analytical biochemistry and systems biology: Proteomics Brett Boghigian Department of Chemical & Biological Engineering Tufts University July 29, 2005 Proteomics The basics History Current
Advances in analytical biochemistry and systems biology: Proteomics Brett Boghigian Department of Chemical & Biological Engineering Tufts University July 29, 2005 Proteomics The basics History Current
Kinetics Review. Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets)
 Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Proteomics And Cancer Biomarker Discovery. Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar. Overview. Cancer.
 Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications
![MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications](/thumbs/74/71368870.jpg) MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
High Profile Publishing in Molecular Biology. Hélène Hodak, Marina Ostankovitch,
 High Profile Publishing in Molecular Biology Hélène Hodak, h.hodak@elsevier.com Marina Ostankovitch, m.ostankovitch@elsevier.com The field of Molecular Biology, inception to current trends Preparing a
High Profile Publishing in Molecular Biology Hélène Hodak, h.hodak@elsevier.com Marina Ostankovitch, m.ostankovitch@elsevier.com The field of Molecular Biology, inception to current trends Preparing a
Technician: Dionne Lutz, BS: Biology & MsED Office: Kanbar Center, Room 704, 41 Cooper Sq. (212) (office)
 BIO101: Molecular and Cellular Biology (WITH LABS!) Meeting Mondays, 6-9pm, in room 101 or in Kanbar Center on select dates (see schedule). (3 credits) Instructor: Oliver Medvedik, Ph.D Office: Room 206,
BIO101: Molecular and Cellular Biology (WITH LABS!) Meeting Mondays, 6-9pm, in room 101 or in Kanbar Center on select dates (see schedule). (3 credits) Instructor: Oliver Medvedik, Ph.D Office: Room 206,
Gene Regulation Solutions. Microarrays and Next-Generation Sequencing
 Gene Regulation Solutions Microarrays and Next-Generation Sequencing Gene Regulation Solutions The Microarrays Advantage Microarrays Lead the Industry in: Comprehensive Content SurePrint G3 Human Gene
Gene Regulation Solutions Microarrays and Next-Generation Sequencing Gene Regulation Solutions The Microarrays Advantage Microarrays Lead the Industry in: Comprehensive Content SurePrint G3 Human Gene
BIOCHEMISTRY 551: BIOCHEMICAL METHODS SYLLABUS
 BIOCHEMISTRY 551: BIOCHEMICAL METHODS SYLLABUS Course Description: Biochemistry 551 is an integrated lecture, lab and seminar course that covers biochemistry-centered theory and techniques. The course
BIOCHEMISTRY 551: BIOCHEMICAL METHODS SYLLABUS Course Description: Biochemistry 551 is an integrated lecture, lab and seminar course that covers biochemistry-centered theory and techniques. The course
Proteomics. Proteomics is the study of all proteins within organism. Challenges
 Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we
Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we
BIOLOGY Dr.Locke Lecture# 27 An Introduction to Polymerase Chain Reaction (PCR)
 BIOLOGY 207 - Dr.Locke Lecture# 27 An Introduction to Polymerase Chain Reaction (PCR) Required readings and problems: Reading: Open Genetics, Chapter 8.1 Problems: Chapter 8 Optional Griffiths (2008) 9
BIOLOGY 207 - Dr.Locke Lecture# 27 An Introduction to Polymerase Chain Reaction (PCR) Required readings and problems: Reading: Open Genetics, Chapter 8.1 Problems: Chapter 8 Optional Griffiths (2008) 9
2D gel Western blotting using antibodies against ubiquitin, SUMO and acetyl PTM
 2D gel Western blotting using antibodies against ubiquitin, SUMO and acetyl PTM Nancy Kendrick, Jon Johansen & Matt Hoelter, Kendrick Labs Inc www.kendricklabs.com Talk Outline Significance Method description
2D gel Western blotting using antibodies against ubiquitin, SUMO and acetyl PTM Nancy Kendrick, Jon Johansen & Matt Hoelter, Kendrick Labs Inc www.kendricklabs.com Talk Outline Significance Method description
Molecular and Cell Biology (MCB)
 Molecular and Cell Biology (MCB) Head of Department: Professor Michael Lynes Department Office: Room 104, Biology/Physics Building For major requirements, see the College of Liberal Arts and Sciences section
Molecular and Cell Biology (MCB) Head of Department: Professor Michael Lynes Department Office: Room 104, Biology/Physics Building For major requirements, see the College of Liberal Arts and Sciences section
Tony Mire-Sluis Vice President, Corporate, Product and Device Quality Amgen Inc
 The Regulatory Implications of the ever increasing power of Mass Spectrometry and its role in the Analysis of Biotechnology Products Where do we draw the line? Tony Mire-Sluis Vice President, Corporate,
The Regulatory Implications of the ever increasing power of Mass Spectrometry and its role in the Analysis of Biotechnology Products Where do we draw the line? Tony Mire-Sluis Vice President, Corporate,
Introduction to Bioinformatics
 Introduction to Bioinformatics Alla L Lapidus, Ph.D. SPbSU St. Petersburg Term Bioinformatics Term Bioinformatics was invented by Paulien Hogeweg (Полина Хогевег) and Ben Hesper in 1970 as "the study of
Introduction to Bioinformatics Alla L Lapidus, Ph.D. SPbSU St. Petersburg Term Bioinformatics Term Bioinformatics was invented by Paulien Hogeweg (Полина Хогевег) and Ben Hesper in 1970 as "the study of
Gene expression connectivity mapping and its application to Cat-App
 Gene expression connectivity mapping and its application to Cat-App Shu-Dong Zhang Northern Ireland Centre for Stratified Medicine University of Ulster Outline TITLE OF THE PRESENTATION Gene expression
Gene expression connectivity mapping and its application to Cat-App Shu-Dong Zhang Northern Ireland Centre for Stratified Medicine University of Ulster Outline TITLE OF THE PRESENTATION Gene expression
AB SCIEX TripleTOF 5600 SYSTEM. High-resolution quant and qual
 AB SCIEX TripleTOF 5600 SYSTEM High-resolution quant and qual AB SCIEX TripleTOF 5600 SYSTEM AB SCIEX is a global leader in the development of life science analytical technologies that help answer complex
AB SCIEX TripleTOF 5600 SYSTEM High-resolution quant and qual AB SCIEX TripleTOF 5600 SYSTEM AB SCIEX is a global leader in the development of life science analytical technologies that help answer complex
Fundamentals of Bioinformatics: computation, biology, computational biology
 Fundamentals of Bioinformatics: computation, biology, computational biology Vasilis J. Promponas Bioinformatics Research Laboratory Department of Biological Sciences University of Cyprus A short self-introduction
Fundamentals of Bioinformatics: computation, biology, computational biology Vasilis J. Promponas Bioinformatics Research Laboratory Department of Biological Sciences University of Cyprus A short self-introduction
Proteomics: A Challenge for Technology and Information Science. What is proteomics?
 Proteomics: A Challenge for Technology and Information Science CBCB Seminar, November 21, 2005 Tim Griffin Dept. Biochemistry, Molecular Biology and Biophysics tgriffin@umn.edu What is proteomics? Proteomics
Proteomics: A Challenge for Technology and Information Science CBCB Seminar, November 21, 2005 Tim Griffin Dept. Biochemistry, Molecular Biology and Biophysics tgriffin@umn.edu What is proteomics? Proteomics
Towards unbiased biomarker discovery
 Towards unbiased biomarker discovery High-throughput molecular profiling technologies are routinely applied for biomarker discovery to make the drug discovery process more efficient and enable personalised
Towards unbiased biomarker discovery High-throughput molecular profiling technologies are routinely applied for biomarker discovery to make the drug discovery process more efficient and enable personalised
ATIP Avenir Program 2018 Young group leader
 ATIP Avenir Program 2018 Young group leader Important dates - October 17th (4 pm) 2017 : opening of the registrations online - November 23 th 2017: deadline for the online submission, the mailing of the
ATIP Avenir Program 2018 Young group leader Important dates - October 17th (4 pm) 2017 : opening of the registrations online - November 23 th 2017: deadline for the online submission, the mailing of the
Bioinformatics Analysis of Nano-based Omics Data
 Bioinformatics Analysis of Nano-based Omics Data Penny Nymark, Pekka Kohonen, Vesa Hongisto and Roland Grafström Hands-on Workshop on Nano Safety Assessment, 29 th September, 2016, National Technical University
Bioinformatics Analysis of Nano-based Omics Data Penny Nymark, Pekka Kohonen, Vesa Hongisto and Roland Grafström Hands-on Workshop on Nano Safety Assessment, 29 th September, 2016, National Technical University
Thermo Scientific Proteomics & Metabolomics Seminar Tour
 Proteomics & Seminar Tour Ignite your workflows with our with Cutting Edge LC-MS Technology Stop by to learn the latest advances in mass spectrometry workflows, technologies launched at 2012 ASMS and talk
Proteomics & Seminar Tour Ignite your workflows with our with Cutting Edge LC-MS Technology Stop by to learn the latest advances in mass spectrometry workflows, technologies launched at 2012 ASMS and talk
DEPARTMENT OF BIOCHEMISTRY AND MOLECULAR BIOLOGY
 Department of Biochemistry and Molecular Biology 1 DEPARTMENT OF BIOCHEMISTRY AND MOLECULAR BIOLOGY Office in Molecular and Radiological Biosciences Building, Room 111 (970) 491-5602 bmb.colostate.edu
Department of Biochemistry and Molecular Biology 1 DEPARTMENT OF BIOCHEMISTRY AND MOLECULAR BIOLOGY Office in Molecular and Radiological Biosciences Building, Room 111 (970) 491-5602 bmb.colostate.edu
Ontologies - Useful tools in Life Sciences and Forensics
 Ontologies - Useful tools in Life Sciences and Forensics How today's Life Science Technologies can shape the Crime Sciences of tomorrow 04.07.2015 Dirk Labudde Mittweida Mittweida 2 Watson vs Watson Dr.
Ontologies - Useful tools in Life Sciences and Forensics How today's Life Science Technologies can shape the Crime Sciences of tomorrow 04.07.2015 Dirk Labudde Mittweida Mittweida 2 Watson vs Watson Dr.
AGRO/ANSC/BIO/GENE/HORT 305 Fall, 2016 Overview of Genetics Lecture outline (Chpt 1, Genetics by Brooker) #1
 AGRO/ANSC/BIO/GENE/HORT 305 Fall, 2016 Overview of Genetics Lecture outline (Chpt 1, Genetics by Brooker) #1 - Genetics: Progress from Mendel to DNA: Gregor Mendel, in the mid 19 th century provided the
AGRO/ANSC/BIO/GENE/HORT 305 Fall, 2016 Overview of Genetics Lecture outline (Chpt 1, Genetics by Brooker) #1 - Genetics: Progress from Mendel to DNA: Gregor Mendel, in the mid 19 th century provided the
Lecture 23: Clinical and Biomedical Applications of Proteomics; Proteomics Industry
 Lecture 23: Clinical and Biomedical Applications of Proteomics; Proteomics Industry Clinical proteomics is the application of proteomic approach to the field of medicine. Proteome of an organism changes
Lecture 23: Clinical and Biomedical Applications of Proteomics; Proteomics Industry Clinical proteomics is the application of proteomic approach to the field of medicine. Proteome of an organism changes
BIOTECHNOLOGY. Course Syllabus. Section A: Engineering Mathematics. Subject Code: BT. Course Structure. Engineering Mathematics. General Biotechnology
 BIOTECHNOLOGY Subject Code: BT Course Structure Sections/Units Section A Section B Unit 1 Unit 2 Unit 3 Unit 4 Unit 5 Unit 6 Unit 7 Section C Section D Section E Topics Engineering Mathematics General
BIOTECHNOLOGY Subject Code: BT Course Structure Sections/Units Section A Section B Unit 1 Unit 2 Unit 3 Unit 4 Unit 5 Unit 6 Unit 7 Section C Section D Section E Topics Engineering Mathematics General
Network System Inference
 Network System Inference Francis J. Doyle III University of California, Santa Barbara Douglas Lauffenburger Massachusetts Institute of Technology WTEC Systems Biology Final Workshop March 11, 2005 What
Network System Inference Francis J. Doyle III University of California, Santa Barbara Douglas Lauffenburger Massachusetts Institute of Technology WTEC Systems Biology Final Workshop March 11, 2005 What
AP Biology Gene Expression/Biotechnology REVIEW
 AP Biology Gene Expression/Biotechnology REVIEW Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Gene expression can be a. regulated before transcription.
AP Biology Gene Expression/Biotechnology REVIEW Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Gene expression can be a. regulated before transcription.
Fragment-based screening of enzyme drug targets: Microfluidic mobility shift assay improves confidence in candidate selection
 Fragment-based screening of enzyme drug targets: Microfluidic mobility shift assay improves confidence in candidate selection Introduction Fragment-based lead discovery is establishing itself as valuable
Fragment-based screening of enzyme drug targets: Microfluidic mobility shift assay improves confidence in candidate selection Introduction Fragment-based lead discovery is establishing itself as valuable
BIOCHEMISTRY, BIOPHYSICS, AND MOLECULAR BIOLOGY (BBMB)
 Biochemistry, Biophysics, and Molecular Biology (BBMB) 1 BIOCHEMISTRY, BIOPHYSICS, AND MOLECULAR BIOLOGY (BBMB) Courses primarily for undergraduates: BBMB 101: Introduction to Biochemistry (1-0) Cr. 1.
Biochemistry, Biophysics, and Molecular Biology (BBMB) 1 BIOCHEMISTRY, BIOPHYSICS, AND MOLECULAR BIOLOGY (BBMB) Courses primarily for undergraduates: BBMB 101: Introduction to Biochemistry (1-0) Cr. 1.
Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs
 Technical Overview Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs Authors Stephen Madden, Crystal Cody, and Jungkap Park Agilent Technologies, Inc. Santa Clara, California,
Technical Overview Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs Authors Stephen Madden, Crystal Cody, and Jungkap Park Agilent Technologies, Inc. Santa Clara, California,
TECHNICAL NOTE Phosphorylation Site Specific Aptamers for Cancer Biomarker Peptide Enrichment and Detection in Mass Spectrometry Based Proteomics
 TECHNICAL NOTE Phosphorylation Site Specific Aptamers for Cancer Biomarker Peptide Enrichment and Detection in Mass Spectrometry Based Proteomics INTRODUCTION The development of renewable capture reagents
TECHNICAL NOTE Phosphorylation Site Specific Aptamers for Cancer Biomarker Peptide Enrichment and Detection in Mass Spectrometry Based Proteomics INTRODUCTION The development of renewable capture reagents
Functional Genomics Overview RORY STARK PRINCIPAL BIOINFORMATICS ANALYST CRUK CAMBRIDGE INSTITUTE 18 SEPTEMBER 2017
 Functional Genomics Overview RORY STARK PRINCIPAL BIOINFORMATICS ANALYST CRUK CAMBRIDGE INSTITUTE 18 SEPTEMBER 2017 Agenda What is Functional Genomics? RNA Transcription/Gene Expression Measuring Gene
Functional Genomics Overview RORY STARK PRINCIPAL BIOINFORMATICS ANALYST CRUK CAMBRIDGE INSTITUTE 18 SEPTEMBER 2017 Agenda What is Functional Genomics? RNA Transcription/Gene Expression Measuring Gene
Bioinformatics for Proteomics. Ann Loraine
 Bioinformatics for Proteomics Ann Loraine aloraine@uab.edu What is bioinformatics? The science of collecting, processing, organizing, storing, analyzing, and mining biological information, especially data
Bioinformatics for Proteomics Ann Loraine aloraine@uab.edu What is bioinformatics? The science of collecting, processing, organizing, storing, analyzing, and mining biological information, especially data
Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS
 Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS Michal Godula Ph.D. Thermo Fisher Scientific The world leader in serving science
Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS Michal Godula Ph.D. Thermo Fisher Scientific The world leader in serving science
LC/MS/MS Solutions for Biomarker Discovery QSTAR. Elite Hybrid LC/MS/MS System. More performance, more reliability, more answers
 LC/MS/MS Solutions for Biomarker Discovery QSTAR Elite Hybrid LC/MS/MS System More performance, more reliability, more answers More is better and the QSTAR Elite LC/MS/MS system has more to offer. More
LC/MS/MS Solutions for Biomarker Discovery QSTAR Elite Hybrid LC/MS/MS System More performance, more reliability, more answers More is better and the QSTAR Elite LC/MS/MS system has more to offer. More
Quantitation of mrna Using Real-Time Reverse Transcription PCR (RT-PCR)
 Quantitation of mrna Using Real-Time Reverse Transcription PCR (RT-PCR) Quantitative Real-Time RT-PCR Versus RT-PCR In Real-Time RT- PCR, DNA amplification monitored at each cycle but RT-PCR measures the
Quantitation of mrna Using Real-Time Reverse Transcription PCR (RT-PCR) Quantitative Real-Time RT-PCR Versus RT-PCR In Real-Time RT- PCR, DNA amplification monitored at each cycle but RT-PCR measures the
Identifying Signaling Pathways. BMI/CS 776 Spring 2016 Anthony Gitter
 Identifying Signaling Pathways BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2016 Anthony Gitter gitter@biostat.wisc.edu Goals for lecture Challenges of integrating high-throughput assays Connecting relevant
Identifying Signaling Pathways BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2016 Anthony Gitter gitter@biostat.wisc.edu Goals for lecture Challenges of integrating high-throughput assays Connecting relevant
Build Your Own Gene Expression Analysis Panels
 Build Your Own Gene Expression Analysis Panels George J. Quellhorst, Jr. Ph.D. Associate Director, R&D Agenda What Will We Discuss? Introduction Building Your Own Gene List Getting Started Increasing Coverage
Build Your Own Gene Expression Analysis Panels George J. Quellhorst, Jr. Ph.D. Associate Director, R&D Agenda What Will We Discuss? Introduction Building Your Own Gene List Getting Started Increasing Coverage
The Two-Hybrid System
 Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine The Two-Hybrid System Carolina Vollert & Peter Uetz Institut für Genetik Forschungszentrum Karlsruhe PO Box 3640 D-76021 Karlsruhe
Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine The Two-Hybrid System Carolina Vollert & Peter Uetz Institut für Genetik Forschungszentrum Karlsruhe PO Box 3640 D-76021 Karlsruhe
Protein Microarrays?
 Protein Microarrays? Protein Chemistry/Proteomics Unit and the Neuroscience Program,Biomedicum Helsinki E-Mail: marc.baumann@helsinki.fi (http://research.med.helsinki.fi/corefacilities/proteinchem) What
Protein Microarrays? Protein Chemistry/Proteomics Unit and the Neuroscience Program,Biomedicum Helsinki E-Mail: marc.baumann@helsinki.fi (http://research.med.helsinki.fi/corefacilities/proteinchem) What
Biology 142 Advanced Topics in Genetics and Molecular Biology Course Syllabus Spring 2006
 Biology 142 Advanced Topics in Genetics and Molecular Biology Course Syllabus Spring 2006 Faculty Information: Dr. Nitya Jacob, Office: Room 104, Pierce Hall; Phone: 770-784-8346 Office Hours: T 9:30-10:30
Biology 142 Advanced Topics in Genetics and Molecular Biology Course Syllabus Spring 2006 Faculty Information: Dr. Nitya Jacob, Office: Room 104, Pierce Hall; Phone: 770-784-8346 Office Hours: T 9:30-10:30
System Biology and Immune Response
 System Biology and Immune Response Fondation Mérieux Conference Centre Les Pensières Veyrier-du-Lac - France June 21-23, 2010 Steering Committee: Nicolas BURDIN Bruno GUY Michael KATZE Jacques LOUIS Ray
System Biology and Immune Response Fondation Mérieux Conference Centre Les Pensières Veyrier-du-Lac - France June 21-23, 2010 Steering Committee: Nicolas BURDIN Bruno GUY Michael KATZE Jacques LOUIS Ray
Biology 252 Nucleic Acid Methods
 Fall 2015 Biology 252 Nucleic Acid Methods COURSE OUTLINE Prerequisites: One semester of college biology (BIO 101 or BIO 173) and one semester of college English (ENG 111); completion of CHM 111is recommended.
Fall 2015 Biology 252 Nucleic Acid Methods COURSE OUTLINE Prerequisites: One semester of college biology (BIO 101 or BIO 173) and one semester of college English (ENG 111); completion of CHM 111is recommended.
Leonardo Mariño-Ramírez, PhD NCBI / NLM / NIH. BIOL 7210 A Computational Genomics 2/18/2015
 Leonardo Mariño-Ramírez, PhD NCBI / NLM / NIH BIOL 7210 A Computational Genomics 2/18/2015 The $1,000 genome is here! http://www.illumina.com/systems/hiseq-x-sequencing-system.ilmn Bioinformatics bottleneck
Leonardo Mariño-Ramírez, PhD NCBI / NLM / NIH BIOL 7210 A Computational Genomics 2/18/2015 The $1,000 genome is here! http://www.illumina.com/systems/hiseq-x-sequencing-system.ilmn Bioinformatics bottleneck
The Nobel Prize in Chemistry 2006
 I n f o r m a t i o n f o r t h e p u b l i c The Nobel Prize in Chemistry 2006 This year s Nobel Prize in Chemistry is awarded to Roger D. Kornberg for his fundamental studies concerning how the information
I n f o r m a t i o n f o r t h e p u b l i c The Nobel Prize in Chemistry 2006 This year s Nobel Prize in Chemistry is awarded to Roger D. Kornberg for his fundamental studies concerning how the information
Genetics Lecture 21 Recombinant DNA
 Genetics Lecture 21 Recombinant DNA Recombinant DNA In 1971, a paper published by Kathleen Danna and Daniel Nathans marked the beginning of the recombinant DNA era. The paper described the isolation of
Genetics Lecture 21 Recombinant DNA Recombinant DNA In 1971, a paper published by Kathleen Danna and Daniel Nathans marked the beginning of the recombinant DNA era. The paper described the isolation of
F 11/23 Happy Thanksgiving! 8 M 11/26 Gene identification in the genomic era Bamshad et al. Nature Reviews Genetics 12: , 2011
 3 rd Edition 4 th Edition Lecture Day Date Topic Reading Problems Reading Problems 1 M 11/5 Complementation testing reveals that genes are distinct entities Ch. 7 224-232 2 W 11/7 One gene makes one protein
3 rd Edition 4 th Edition Lecture Day Date Topic Reading Problems Reading Problems 1 M 11/5 Complementation testing reveals that genes are distinct entities Ch. 7 224-232 2 W 11/7 One gene makes one protein
Chem Lecture 1 Introduction to Biochemistry Part 3
 Chem 452 - Lecture 1 Introduction to Biochemistry Part 3 Question of the Day: This (last) week, big news was made in the field of genomic. It was not only reported in the journals Nature and Science, but
Chem 452 - Lecture 1 Introduction to Biochemistry Part 3 Question of the Day: This (last) week, big news was made in the field of genomic. It was not only reported in the journals Nature and Science, but
Biology (BIOL) Biology BIOL 1620
 (BIOL) BIOL 1010 General Biology, Summer Introduces major themes and concepts of biology including cell and molecular biology, genetics, diversity, evolution, and ecology. Provides students with necessary
(BIOL) BIOL 1010 General Biology, Summer Introduces major themes and concepts of biology including cell and molecular biology, genetics, diversity, evolution, and ecology. Provides students with necessary
Bayesian Networks as framework for data integration
 Bayesian Networks as framework for data integration Jun Zhu, Ph. D. Department of Genomics and Genetic Sciences Icahn Institute of Genomics and Multiscale Biology Icahn Medical School at Mount Sinai New
Bayesian Networks as framework for data integration Jun Zhu, Ph. D. Department of Genomics and Genetic Sciences Icahn Institute of Genomics and Multiscale Biology Icahn Medical School at Mount Sinai New
Outline. Analysis of Microarray Data. Most important design question. General experimental issues
 Outline Analysis of Microarray Data Lecture 1: Experimental Design and Data Normalization Introduction to microarrays Experimental design Data normalization Other data transformation Exercises George Bell,
Outline Analysis of Microarray Data Lecture 1: Experimental Design and Data Normalization Introduction to microarrays Experimental design Data normalization Other data transformation Exercises George Bell,
Technical Review. Real time PCR
 Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
LTQ Orbitrap XL Hybrid FT Mass Spectrometer Unrivaled Performance and Flexibility
 m a s s s p e c t r o m e t r y LTQ Orbitrap XL Hybrid FT Mass Spectrometer Unrivaled Performance and Flexibility Part of Thermo Fisher Scientific LTQ Orbitrap XL OFFERING OUTSTANDING MASS ACCURACY, RESOLVING
m a s s s p e c t r o m e t r y LTQ Orbitrap XL Hybrid FT Mass Spectrometer Unrivaled Performance and Flexibility Part of Thermo Fisher Scientific LTQ Orbitrap XL OFFERING OUTSTANDING MASS ACCURACY, RESOLVING
Introduction to Microarray Data Analysis and Gene Networks. Alvis Brazma European Bioinformatics Institute
 Introduction to Microarray Data Analysis and Gene Networks Alvis Brazma European Bioinformatics Institute A brief outline of this course What is gene expression, why it s important Microarrays and how
Introduction to Microarray Data Analysis and Gene Networks Alvis Brazma European Bioinformatics Institute A brief outline of this course What is gene expression, why it s important Microarrays and how
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
RNA-Sequencing analysis
 RNA-Sequencing analysis Markus Kreuz 25. 04. 2012 Institut für Medizinische Informatik, Statistik und Epidemiologie Content: Biological background Overview transcriptomics RNA-Seq RNA-Seq technology Challenges
RNA-Sequencing analysis Markus Kreuz 25. 04. 2012 Institut für Medizinische Informatik, Statistik und Epidemiologie Content: Biological background Overview transcriptomics RNA-Seq RNA-Seq technology Challenges
Metabolomics: Sifting Through Complex Samples
 Metabolomics: Sifting Through Complex Samples Thursday, March 14, 2013 Print Jeffrey M. Perkel Look between the covers of any biochemistry textbook and you ll likely find an illustration outlining the
Metabolomics: Sifting Through Complex Samples Thursday, March 14, 2013 Print Jeffrey M. Perkel Look between the covers of any biochemistry textbook and you ll likely find an illustration outlining the
Overview of Health Informatics. ITI BMI-Dept
 Overview of Health Informatics ITI BMI-Dept Fellowship Week 5 Overview of Health Informatics ITI, BMI-Dept Day 10 7/5/2010 2 Agenda 1-Bioinformatics Definitions 2-System Biology 3-Bioinformatics vs Computational
Overview of Health Informatics ITI BMI-Dept Fellowship Week 5 Overview of Health Informatics ITI, BMI-Dept Day 10 7/5/2010 2 Agenda 1-Bioinformatics Definitions 2-System Biology 3-Bioinformatics vs Computational
Synthetic Biology. Sustainable Energy. Therapeutics Industrial Enzymes. Agriculture. Accelerating Discoveries, Expanding Possibilities. Design.
 Synthetic Biology Accelerating Discoveries, Expanding Possibilities Sustainable Energy Therapeutics Industrial Enzymes Agriculture Design Build Generate Solutions to Advance Synthetic Biology Research
Synthetic Biology Accelerating Discoveries, Expanding Possibilities Sustainable Energy Therapeutics Industrial Enzymes Agriculture Design Build Generate Solutions to Advance Synthetic Biology Research
Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis
 Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis SWATH Acquisition on the TripleTOF 5600+ System Samuel L. Bader, Robert L. Moritz Institute of Systems Biology,
Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis SWATH Acquisition on the TripleTOF 5600+ System Samuel L. Bader, Robert L. Moritz Institute of Systems Biology,
Microarray Technique. Some background. M. Nath
 Microarray Technique Some background M. Nath Outline Introduction Spotting Array Technique GeneChip Technique Data analysis Applications Conclusion Now Blind Guess? Functional Pathway Microarray Technique
Microarray Technique Some background M. Nath Outline Introduction Spotting Array Technique GeneChip Technique Data analysis Applications Conclusion Now Blind Guess? Functional Pathway Microarray Technique
Xevo G2-S QTof and TransOmics: A Multi-Omics System for the Differential LC/MS Analysis of Proteins, Metabolites, and Lipids
 Xevo G2-S QTof and TransOmics: A Multi-Omics System for the Differential LC/MS Analysis of Proteins, Metabolites, and Lipids Ian Edwards, Jayne Kirk, and Joanne Williams Waters Corporation, Manchester,
Xevo G2-S QTof and TransOmics: A Multi-Omics System for the Differential LC/MS Analysis of Proteins, Metabolites, and Lipids Ian Edwards, Jayne Kirk, and Joanne Williams Waters Corporation, Manchester,
EECS730: Introduction to Bioinformatics
 EECS730: Introduction to Bioinformatics Lecture 14: Microarray Some slides were adapted from Dr. Luke Huan (University of Kansas), Dr. Shaojie Zhang (University of Central Florida), and Dr. Dong Xu and
EECS730: Introduction to Bioinformatics Lecture 14: Microarray Some slides were adapted from Dr. Luke Huan (University of Kansas), Dr. Shaojie Zhang (University of Central Florida), and Dr. Dong Xu and
Introduction to Bioinformatics and Gene Expression Technologies
 Introduction to Bioinformatics and Gene Expression Technologies Utah State University Fall 2017 Statistical Bioinformatics (Biomedical Big Data) Notes 1 1 Vocabulary Gene: hereditary DNA sequence at a
Introduction to Bioinformatics and Gene Expression Technologies Utah State University Fall 2017 Statistical Bioinformatics (Biomedical Big Data) Notes 1 1 Vocabulary Gene: hereditary DNA sequence at a
Engineering Genetic Circuits
 Engineering Genetic Circuits I use the book and slides of Chris J. Myers Lecture 0: Preface Chris J. Myers (Lecture 0: Preface) Engineering Genetic Circuits 1 / 19 Samuel Florman Engineering is the art
Engineering Genetic Circuits I use the book and slides of Chris J. Myers Lecture 0: Preface Chris J. Myers (Lecture 0: Preface) Engineering Genetic Circuits 1 / 19 Samuel Florman Engineering is the art
Protein-Protein-Interaction Networks. Ulf Leser, Samira Jaeger
 Protein-Protein-Interaction Networks Ulf Leser, Samira Jaeger SHK Stelle frei Ab 1.9.2015, 2 Jahre, 41h/Monat Verbundprojekt MaptTorNet: Pankreatische endokrine Tumore Insb. statistische Aufbereitung und
Protein-Protein-Interaction Networks Ulf Leser, Samira Jaeger SHK Stelle frei Ab 1.9.2015, 2 Jahre, 41h/Monat Verbundprojekt MaptTorNet: Pankreatische endokrine Tumore Insb. statistische Aufbereitung und
Introduction, by Tortora, Funke and Case, 11th Ed. TENTATIVE LECTURE OUTLINE DATE TOPIC CHAPTER
 MICROBIOLOGY 220 Spring 2014 TTh Section Professor: Scott Rose Text: Microbiology; An Introduction, by Tortora, Funke and Case, 11th Ed. TENTATIVE LECTURE OUTLINE DATE TOPIC CHAPTER Jan. 23 Introduction
MICROBIOLOGY 220 Spring 2014 TTh Section Professor: Scott Rose Text: Microbiology; An Introduction, by Tortora, Funke and Case, 11th Ed. TENTATIVE LECTURE OUTLINE DATE TOPIC CHAPTER Jan. 23 Introduction
Introduction to BIOINFORMATICS
 Introduction to BIOINFORMATICS Antonella Lisa CABGen Centro di Analisi Bioinformatica per la Genomica Tel. 0382-546361 E-mail: lisa@igm.cnr.it http://www.igm.cnr.it/pagine-personali/lisa-antonella/ What
Introduction to BIOINFORMATICS Antonella Lisa CABGen Centro di Analisi Bioinformatica per la Genomica Tel. 0382-546361 E-mail: lisa@igm.cnr.it http://www.igm.cnr.it/pagine-personali/lisa-antonella/ What
Roche Molecular Biochemicals Technical Note No. LC 10/2000
 Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Bioinformatics of Transcriptional Regulation
 Bioinformatics of Transcriptional Regulation Carl Herrmann IPMB & DKFZ c.herrmann@dkfz.de Wechselwirkung von Maßnahmen und Auswirkungen Einflussmöglichkeiten in einem Dialog From genes to active compounds
Bioinformatics of Transcriptional Regulation Carl Herrmann IPMB & DKFZ c.herrmann@dkfz.de Wechselwirkung von Maßnahmen und Auswirkungen Einflussmöglichkeiten in einem Dialog From genes to active compounds
Antibody Analysis by ESI-TOF LC/MS
 Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS Antibody Analysis by ESI-TOF LC/MS Authors Lorenzo Chen, Merck & Company,
Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS Antibody Analysis by ESI-TOF LC/MS Authors Lorenzo Chen, Merck & Company,
Introduction to BioMEMS & Medical Microdevices DNA Microarrays and Lab-on-a-Chip Methods
 Introduction to BioMEMS & Medical Microdevices DNA Microarrays and Lab-on-a-Chip Methods Companion lecture to the textbook: Fundamentals of BioMEMS and Medical Microdevices, by Prof., http://saliterman.umn.edu/
Introduction to BioMEMS & Medical Microdevices DNA Microarrays and Lab-on-a-Chip Methods Companion lecture to the textbook: Fundamentals of BioMEMS and Medical Microdevices, by Prof., http://saliterman.umn.edu/
Feature Selection of Gene Expression Data for Cancer Classification: A Review
 Available online at www.sciencedirect.com ScienceDirect Procedia Computer Science 50 (2015 ) 52 57 2nd International Symposium on Big Data and Cloud Computing (ISBCC 15) Feature Selection of Gene Expression
Available online at www.sciencedirect.com ScienceDirect Procedia Computer Science 50 (2015 ) 52 57 2nd International Symposium on Big Data and Cloud Computing (ISBCC 15) Feature Selection of Gene Expression
less sensitive than RNA-seq but more robust analysis pipelines expensive but quantitiatve standard but typically not high throughput
 Chapter 11: Gene Expression The availability of an annotated genome sequence enables massively parallel analysis of gene expression. The expression of all genes in an organism can be measured in one experiment.
Chapter 11: Gene Expression The availability of an annotated genome sequence enables massively parallel analysis of gene expression. The expression of all genes in an organism can be measured in one experiment.
What we ll do today. Types of stem cells. Do engineered ips and ES cells have. What genes are special in stem cells?
 Do engineered ips and ES cells have similar molecular signatures? What we ll do today Research questions in stem cell biology Comparing expression and epigenetics in stem cells asuring gene expression
Do engineered ips and ES cells have similar molecular signatures? What we ll do today Research questions in stem cell biology Comparing expression and epigenetics in stem cells asuring gene expression
Current and emerging services in proteomics and metabolomics through the Center for Mass Spectrometry and Proteomics
 Current and emerging services in proteomics and metabolomics through the Center for Mass Spectrometry and Proteomics Pratik Jagtap, PhD Managing Director, CMSP z.umn.edu/cmsppresents Who are we? Center
Current and emerging services in proteomics and metabolomics through the Center for Mass Spectrometry and Proteomics Pratik Jagtap, PhD Managing Director, CMSP z.umn.edu/cmsppresents Who are we? Center
Blot: a spot or stain, especially of ink on paper.
 Blotting technique Blot: a spot or stain, especially of ink on paper. 2/27 In molecular biology and genetics, a blot is a method of transferring proteins, DNA or RNA, onto a carrier (for example, a nitrocellulose,pvdf
Blotting technique Blot: a spot or stain, especially of ink on paper. 2/27 In molecular biology and genetics, a blot is a method of transferring proteins, DNA or RNA, onto a carrier (for example, a nitrocellulose,pvdf
Call for Pilot Project Applications Chemical Biology for Infectious Diseases NIH Center of Biomedical Research Excellence University of Kansas
 Call for Pilot Project Applications Chemical Biology for Infectious Diseases NIH Center of Biomedical Research Excellence University of Kansas Applications due: Oct 3 rd, 2016 Anticipated Decision Date:
Call for Pilot Project Applications Chemical Biology for Infectious Diseases NIH Center of Biomedical Research Excellence University of Kansas Applications due: Oct 3 rd, 2016 Anticipated Decision Date:
Biochemistry 9/1/17. Course No: CHM4006; Credits: Day / Time: Wednesday & Friday: 13:30-15:00. Ro # H & 208 (IT/BT)
 You Do Not Need to write down the following infos because all the following slides and all lecture notes will be uploaded at the link: http://itbe.hanyang.ac.kr This/today s file will be uploaded next
You Do Not Need to write down the following infos because all the following slides and all lecture notes will be uploaded at the link: http://itbe.hanyang.ac.kr This/today s file will be uploaded next
Do engineered ips and ES cells have similar molecular signatures?
 Do engineered ips and ES cells have similar molecular signatures? Comparing expression and epigenetics in stem cells George Bell, Ph.D. Bioinformatics and Research Computing 2012 Spring Lecture Series
Do engineered ips and ES cells have similar molecular signatures? Comparing expression and epigenetics in stem cells George Bell, Ph.D. Bioinformatics and Research Computing 2012 Spring Lecture Series
Controlling DNA. Ethical guidelines for the use of DNA technology. Module Type: Discussion, literature review, and debate
 Ethical guidelines for the use of DNA technology Author: Tara Cornelisse, Ph.D. Candidate, Environmental Studies, University of California Santa Cruz. Field-tested with: 11 th -12 th grade students in
Ethical guidelines for the use of DNA technology Author: Tara Cornelisse, Ph.D. Candidate, Environmental Studies, University of California Santa Cruz. Field-tested with: 11 th -12 th grade students in
Protein Folding Problem I400: Introduction to Bioinformatics
 Protein Folding Problem I400: Introduction to Bioinformatics November 29, 2004 Protein biomolecule, macromolecule more than 50% of the dry weight of cells is proteins polymer of amino acids connected into
Protein Folding Problem I400: Introduction to Bioinformatics November 29, 2004 Protein biomolecule, macromolecule more than 50% of the dry weight of cells is proteins polymer of amino acids connected into
Suggest a technique that could be used to provide molecular evidence that all English Elm trees form a clone. ... [1]
![Suggest a technique that could be used to provide molecular evidence that all English Elm trees form a clone. ... [1] Suggest a technique that could be used to provide molecular evidence that all English Elm trees form a clone. ... [1]](/thumbs/77/74807071.jpg) 1 Molecular evidence E Ulmus procera, form a genetically isolated clone. English Elms developed from a variety of elm brought to Britain from Rome in the first century A.D. Although English Elm trees make
1 Molecular evidence E Ulmus procera, form a genetically isolated clone. English Elms developed from a variety of elm brought to Britain from Rome in the first century A.D. Although English Elm trees make
Recent Advancements in Analytical Methods of Drug Detection
 Recent Advancements in Analytical Methods of Drug Detection Mario Thevis Gene and Cell Doping Symposium Beijing, June 5/6, 2013 www.wada-ama.org Challenges at the time accepted by anti-doping authorities
Recent Advancements in Analytical Methods of Drug Detection Mario Thevis Gene and Cell Doping Symposium Beijing, June 5/6, 2013 www.wada-ama.org Challenges at the time accepted by anti-doping authorities
Next-Generation Sequencing Gene Expression Analysis Using Agilent GeneSpring GX
 Next-Generation Sequencing Gene Expression Analysis Using Agilent GeneSpring GX Technical Overview Introduction RNA Sequencing (RNA-Seq) is one of the most commonly used next-generation sequencing (NGS)
Next-Generation Sequencing Gene Expression Analysis Using Agilent GeneSpring GX Technical Overview Introduction RNA Sequencing (RNA-Seq) is one of the most commonly used next-generation sequencing (NGS)
Additional file 2. Figure 1: Receiver operating characteristic (ROC) curve using the top
 Additional file 2 Figure Legends: Figure 1: Receiver operating characteristic (ROC) curve using the top discriminatory features between HIV-infected (n=32) and HIV-uninfected (n=15) individuals. The top
Additional file 2 Figure Legends: Figure 1: Receiver operating characteristic (ROC) curve using the top discriminatory features between HIV-infected (n=32) and HIV-uninfected (n=15) individuals. The top
IPA Advanced Training Course
 IPA Advanced Training Course Academia Sinica 2015 Oct Gene( 陳冠文 ) Supervisor and IPA certified analyst 1 Review for Introductory Training course Searching Building a Pathway Editing a Pathway for Publication
IPA Advanced Training Course Academia Sinica 2015 Oct Gene( 陳冠文 ) Supervisor and IPA certified analyst 1 Review for Introductory Training course Searching Building a Pathway Editing a Pathway for Publication
Supplementary Figure 1. Utilization of publicly available antibodies in different applications.
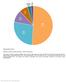 Supplementary Figure 1 Utilization of publicly available antibodies in different applications. The fraction of publicly available antibodies toward human protein targets is shown with data for the following
Supplementary Figure 1 Utilization of publicly available antibodies in different applications. The fraction of publicly available antibodies toward human protein targets is shown with data for the following
Designing Complex Omics Experiments
 Designing Complex Omics Experiments Xiangqin Cui Section on Statistical Genetics Department of Biostatistics University of Alabama at Birmingham 6/15/2015 Some slides are from previous lectures given by
Designing Complex Omics Experiments Xiangqin Cui Section on Statistical Genetics Department of Biostatistics University of Alabama at Birmingham 6/15/2015 Some slides are from previous lectures given by
