Supplemental Data. Wang et al. (2016). Plant Cell /tpc
|
|
|
- Emmeline Carter
- 5 years ago
- Views:
Transcription
1 Supplemental Figure 1. Time-Course Analysis of Storage Proteins during Endosperm Development of the Wild-type 9311 and the gpa4-1 Mutant. (A) SDS-PAGE analyses of seed storage proteins during wild-type and gpa4-1 endosperm development. Blue triangles indicate the 57-kDa proglutelins. Black and red hollow arrows denote the acidic and basic subunit, respectively. DAF, days after flowering. (B) Immunoblot analysis showing the delayed accumulation of glutelins, 26-kDa α-globulins, 13-kDa and 10-kDa prolamins in the gpa4-1 endosperm. 1
2 Supplemental Figure 2. RT-qPCR Assay of the Expression of Representative Genes Coding for Storage Proteins in 12 DAF Endosperm. All of the genes analyzed were down-regulated in the gpa4-1 mutant compared with wild type. DAF, days after flowering. Values are mean ± SD. n = 3. 2
3 Supplemental Figure 3. Immunoblot Analyses Showing Defective Export of Storage Proteins from the ER in gpa4-1. (A) Immunoblot analysis of GluA1, GluB1, α-globulin, 16-kD prolamin (Pro16), 13-kD prolamin-a (Pro13a), 13-kD prolamin-b (Pro13b), 10-kD prolamin (Pro10), and VPE1 showing their distribution in various differential centrifugation fractions using their specific antibodies. P2, pellet obtained following centrifugation at 2000g; P13, pellet obtained following centrifugation at 13,000g; P100, pellet obtained following centrifugation at 100,000g; S100, supernatant obtained following centrifugation at 100,000g. Blue triangles indicate the 57-kD proglutelins. Arrows indicate different forms of VPE1. pv, VPE1 precursor; iv, intermediate VPE1; mv, mature VPE1. (B) Immunoblot analysis using specific antibodies for chaperone proteins. The accumulation of Bip1, PDIL1-1, and Hsp90 proteins is increased in the gpa4 mutants compared with the corresponding wild types. (C) Expression analysis of ER stress response-related genes in 12 DAF endosperm. The transcript levels of these genes are highly elevated in the gpa4-1 mutant. DAF: days after flowering. Values are mean ± SD. n = 3. 3
4 Supplemental Figure 4. Rescue of the gpa4-2 Mutant Phenotype by the GFP-GOT1B Fusion Construct. (A) to (D) The fusion construct GFP-GOT1B under the control of GOT1B endogenous promoter rescues the grain appearance (A), the storage protein composition pattern (B), and the storage protein trafficking defects (C and D) of the gpa4-2 mutant. GL1 to GL3 denote the grains from three independent T1 transgenic lines. Stars represent the first type of novel structure with a glutelin core and a prolamin periphery. Bars = 2 μm in (C), and 5 μm in (D). 4
5 Supplemental Figure 5. Localization Pattern of GFP-GOT1B Fusion Protein. (A) GFP-GOT1B is localized in the punctate structures in the root cells of GFP-GOT1B transgenic lines. (B) and (C) Localization of GFP-GOT1B in N. benthamiana leaf epidermal cells. (B) GFP-GOT1B shows a typical punctate pattern in cells with lower expression. (C) Faint signals in ER networks are observed in cells with higher expression. Triangles indicate the punctate structures of GFP-GOT1B. Arrows indicate the faint signals of GFP-GOT1B in ER networks. Bars = 10 µm. 5
6 Supplemental Figure 6. Both the N- and C-termini Are Essential for the Proper Localization of GOT1B. (A) Scheme of GOT1B N- and C- terminus mutation constructs for expression in the leaf epidermal cells of N. benthamiana. TMD, transmembrane domain. (B) to (G) Typical subcellular localization patterns of the various GFP-fused GOT1B mutant forms co-expressed with a known ER marker (mrfp-hdel) in N. benthamiana leaf cells. Bars = 10 μm in (B to G). GFP-GOT1B * indicates different mutant isoforms of GFP-GOT1B as shown in (A). 6
7 Supplemental Figure 7. Interaction between GOT1B and Sec23. (A) Y2H assay showing that GOT1B specifically interacts with the COPII component Sec23c. (B) GOT1B interacts with Sec23b and Sec23c in the split-ubiquitin based Y2H assay. (C) BiFC assay showing that GOT1B can interact with both Sec23b and Sec23c in leaf epidermal cells of N. benthamiana. The Golgi-localized membrane proteins COG3 and COG8 (a pair of interacting proteins, Tan et al., 2016) were used as the negative control. Bars = 10 μm. (D) No detectable Sec23c protein was immunoprecipitated by anti-sar1b antibodies in the gpa4-1 mutant. (E) RT-qPCR assay showing that Sec23c has the highest expression in 12 DAF endosperm of the wild type. (F) Y2H assay showing that Got1p and human Got1 (hgot1a and hgot1b) not only interact with their corresponding Sec23, but also cross interact with rice Sec23c. 7
8 Supplemental Figure 8. Immunoblot Analyses Showing the Specificity of Antibodies. (A) to (E) Immunoblot analyses showing the specificity of antibodies for GOT1B (A), Sar1b (B), Sar1c (C), Sec23c (D), and Sec12b (E). (A) GOT1B protein can be detected only in IP sample precipitated by anti-got1b antibodies. Total extracts were loaded for immunoblot assays in (B) to (E). T, total extract; W, wash-off solution; IP, immunoprecipitation. Arrows indicate the corresponding protein bands. 8
9 Supplemental Figure 9. Rescue of the gpa4-2 Mutant Phenotypes by Yeast GOT1. (A) and (B) Yeast Got1 rescues the grain appearance (A), and the storage protein composition pattern (B) of the gpa4-2 mutant. YL1 to YL3 denote the grains from three independent T1 transgenic lines. 9
10 Supplemental Figure 10. The Seeds of Transgenic Rice Lines Expressing AtSar1b-GFP Show No Difference from Their Corresponding Recipient Plants. (A) and (B) The grain appearance (A) and the storage protein composition pattern (B) of Kinmaze and gpa4-2 transformed with AtSar1b-GFP. 1 to 4 are transgenic lines using Kinmaze (1 and 2) or gpa4-2 (3 and 4) as the recipients, respectively. 10
11 Supplemental Table 1. Comparison of Important Agronomic Traits between Wild Type and gpa4 Mutants Characteristic 9311 gpa4-1 Kinmaze gpa4-2 Plant height (cm) ± ± 4.7 ** 95.7 ± ±1.6 Panicle length (cm) ± ± ± ± 1.20 Tiller number 6.58 ± ± ± ± ,000-grain weight (g) ± ± 0.03 ** ± ± 0.13 ** Protein content (mg/grain) 2.88 ± ± 0.02 ** 2.05 ± ± 0.13 ** Amylose content (mg/grain) 4.56 ± ± 0.05 ** 3.02 ± ± 0.10 ** The asterisks indicate statistical significance between the gpa4 mutants and the corresponding wild type, as determined by a Student s t-test (**P < 0.01). 11
12 Supplemental Table 2. Segregation of Mutant Phenotype in Reciprocal Crosses between the Wild Type and the gpa4-1 Mutant. Cross Normal 57H/Floury X 2 3:1 gpa4-1/ /gpa
13 Supplemental Table 3. The Distribution of COPII-Related Proteins in the Endomembrane System P2 P13 P100 S100 Sar1b ± ± ± ± 1.72 gpa ± 0.61** ± 1.56** ± 1.21** ± 1.84 Sar1c ± ± ± ± 0.94 gpa ± 1.52** ± 0.68** ± ± 1.32 Sec23c ± ± ± ± 2.73 gpa ± ± ± ± 3.15 Sec12b ± ± ± ± 0.23 gpa ± ± ± ± 0.25 The asterisks indicate statistical significance between the wild type and the gpa4-1 mutant, as determined by a Student s t-test (**P < 0.01). 13
14 Supplemental Table 4. Comparison of Important Agronomic Traits between AtSar1b-GFP Transgenic Rice Lines and Their Corresponding Recipient Plants. Kinmaze AtSar1b-GFP /Kinmaze gpa4-2 AtSar1b-GFP /gpa4-2 Plant height (cm) 93.8 ± ± ± ±2.3 Panicle length (cm) ± ± ± ± 1.23 Tiller number 6.87 ± ± ± ± grain weight (g) ± ± ± ±
15 Supplemental Table 5. Primers Used in This Study. Usage Primer name Sequence G1-F 5' TTCCCACCACCTCCTCCG 3' G1-R 5' GCCACTCCTCCACGCACA 3' G3-F 5' CTGCCCCCTTGCCCCTTC 3' G3-R 5' CTCACCACGCCACCCTCG 3' G4-F 5' TGCTGCTCCGCTACACAAA 3' Primers for fine mapping G4-R 5' CCTCCTTCCCCTCCTCACT 3' G9-F 5' TAGCAGCTACCTCACCGC 3' G9-R 5' ACCAGGACCTCCCGACCC 3' G11-F 5' ACCTGCGAACCACCACGGAC 3' G11-R 5' GCGAGAGGCGAAGGAAATAG 3' G15-F 5' GTGCTGCTTTAGCCGTTTAC 3' G15-R 5' TCAGGGTGTGGGAATGGTAG 3' GluA1-F 5' TGATGGTGAAGTGCCAGTTGTTGC 3' GluA1-R 5' ACGCCTGTATGCTTGAGGGTTTCT 3' GluB2-F 5' TGTCCTTCGCCCTGGACAACTATT 3' GluB2-R 5' TGGTAAGGCGCGGAATACTGAGTT 3' GluC1-F 5' ATGTAAAGTTCAACCGCGGCGATG 3' GluC1-R 5' TCTCGTTGATCTGCCACTCAGCAT 3' GluD1-F 5' AAATGAGCAGTTTCGATGCGCTGG 3' GluD1-R 5' TGTTCGAGTATCGAGGCACCACAA 3' RT-qPCR primers for storage protein genes Pro10.1-F 5' GTTGCATGCAGCTACAAGGCATGA 3' Pro10.1-R 5' TCTGCATCGCCATCTTCACCATCT 3' Pro13a.2-F 5' TTCAGGCGATTGTGCAGCAACTAC 3' Pro13a.2-R 5' GGATGGCAAGTTTAAGGCCAGCAA 3' RT-qPCR primers for internal control Pro13b.2-F 5' GTATAGCATTGCGGCAAGCACCTT 3' Pro13b.2-R 5' ATGGCCTGGACAACGTTAATGTCC 3' Pro13b.18-F 5' ACATTGTTCAGGCCATAGTGCAGC 3' Pro13b.18-R 5' ACAGAGCTTGAGCTTGAGCCAGAT 3' Pro16.2-F 5' GCTCTCAATTTGCCCTCCATGTGT 3' Pro16.2-R 5' TGGTACACACTACCAAGAACCGCA 3' α-globulin-f 5' GATCAGTTCATCACCAACAAAACA 3' α-globulin-r 5' TCCAGAACGCACAAAATCAT 3' Ubiquitin-F 5' TCTTCCCCTTCAGGACGTGT 3' Ubiquitin-R 5' GGACCAAAGGTCACCACCAT 3' 15
16 RT-qPCR primers for GPA4 and its homologous genes GPA4 cdna cloning GPA4 gdna cloning Membrane protein verification Subcellular localization GPA4-F 5' TGTTCTGCTCTTCACTGGTGTGGT 3' GPA4-R 5' AGCCTAGCCCAATCTTCTTGAGGT 3' Os10g F 5' ATTGGGATAGGGCTGACAGGCTTT 3' Os10g R 5' GCCTGACAGGAAGAGAATATTGCCCA 3' Os02g F 5' TGCTTGGTTGGCAATCTATGTGGC 3' Os02g R 5' ACAGGCCATCGGACAAAGAGAAGA 3' GPA4-cDNA-F 5' GAAAACGCCGCTACGCTACCAC 3' GPA4-cDNA-R 5' GCTCAAGCATAAGCCGACAAGG 3' GPA4-gDNA-F 5' GCACCTAACTGTTTCACTTTCTG 3' GPA4-gDNA-R 5' CCTCTACTGCTACCTTGTCCC 3' 1305GOT1B-F 5' TCTAGAATGGTTTCCTTCGAGATGAA 3' 1305GOT1B-R 5' AGATCTGCTCAAGCATAAGCCGACAA 3' 1305OsTip3-1-F 5' TCTAGAATGAGCACGGCGGCGGCGA 3' 1305OsTip3-1-R 5' AGATCTCTAGTAGTCCTCCGGCGC 3' 1305GFP-GOT1B 5' TGGACGAGCTGTACAGATCTATGGTTTCCTTCGAGATG -F 3' 1305GFP-GOT1B 5' GGGCGGCCGCTTTAAGATCTCTACACTGGAACCCGTTT -R 3' 1305-GOT1B-GF P-F 5' TCTAGAATGGTTTCCTTCGAGATG 3' 1305-GOT1B-GF P-R 5' GGATCCCACTGGAACCCGTTTTCC 3' pgbt9-got1b-f 5' GAATTCATGGTTTCCTTCGAGATGA 3' pgbt9-got1b-r 5' GGATCCGCTCAAGCATAAGCCGACAA 3' pgadsec12b-f 5' GGAGGCCAGTGAATTCATGGCGCGGCGCGGCGGC 3' pgadsec12b-r 5' CACCCGGGTGGAATTCTCAATGTAGAAACCTTGC 3' pgadsar1b-f 5 GAATTCATGTTCCTGTGGGACTGG 3 PGADSar1b-R 5 GGATCCCTACTTGATGTACTGGGA 3 pgadsar1c-f 5' GAATTCATGTCGTTTCTGCTGGAT 3' Y2H assay pgadsar1c-f 5' GGATCCCTACTTGATGTACTGTGA 3' pgadsec13a-f 5' GGAGGCCAGTGAATTCATGCCTCCCCATAAGATA 3' pgadsec13a-r 5' CACCCGGGTGGAATTCCTAAGCCTCAACAGTGGT 3' pgadsec13b-f 5' GGAGGCCAGTGAATTCATGTCGTCAAACAAGATA 3' pgadsec13b-r 5' CACCCGGGTGGAATTCCTATGGCTCCACCTTGGT 3' pgadsec13c-f 5' GGAGGCCAGTGAATTCATGTCATCGAAGAAAATA 3' pgadsec13c-r 5' CACCCGGGTGGAATTCCTACTGCTCAGCCTTCTT 3' pgadsec23a-f 5' GGAGGCCAGTGAATTCATGGAGGAGACGTCGACG 3' pgadsec23a-r 5' CACCCGGGTGGAATTCTTAGCTTGGTTCAGGTGG 3' 16
17 pgadsec23b-f 5' GGAGGCCAGTGAATTCATGTCTGAGTTCCTGGAT 3' pgadsec23c-r 5' CACCCGGGTGGAATTCCTACTGAACTGCCAGCCG 3' pgadsec23c-f 5' GGAGGCCAGTGAATTCATGGCGGCCGATCCCTCC 3' pgadsec23c-r 5' CACCCGGGTGGAATTCTTAGGACTGAACGGCCAG 3' pgadsec24-f 5' GGAGGCCAGTGAATTCATGCAACCACCGATGAGA 3' pgadsec24-r 5' CACCCGGGTGGAATTCTCACGTCATCTTGCTCTG 3' pgadsec31-f 5' GGAGGCCAGTGAATTCATGATCAACTCCGAGGGG 3' pgadsec31-r 5' CACCCGGGTGGAATTCTTACATTCTGAAATTCTG 3' pgbt9-got1p-f 5' ACTGTATCGCCGGAATTCATGTGGCTCACAGAGGCT 3' pgbt9-got1p-r 5' ACGGATCCCCGGGAATTCTTATACTGGCAGAACCCT 3' pgbt9-hgot1a- F 5' ACTGTATCGCCGGAATTCATGATCTCCATCACCGAA 3' pgbt9-hgot1a- R 5' ACGGATCCCCGGGAATTCTCAGACCATCGAGCTAGT3' pgbt9-hgot1b- F 5' ACTGTATCGCCGGAATTCATGATCTCCTTAACGGAC 3' pgbt9-hgot1b- R 5' ACGGATCCCCGGGAATTCTTATACCATATTGTTGCT 3' 5' pgadsec23p-f ATGGAGGCCAGTGAATTCATGGACTTCGAGACTAATGA3' 5' CCCACCCGGGTGGAATTCCTATGCCTGACCAGAGACGG pgadsec23p-r 3' pgadhsec23b-f 5' ATGGAGGCCAGTGAATTCATGGCGACATACCTGGAG 3' pgadhsec23b-r 5' CCCACCCGGGTGGAATTCTTAACAGGCACTGGAGAC 3' pbt3n-got1b-f 5 GCCGCCTCGGCCCCATGGATGGTTTCCTTCGAGATG 3 pbt3n-got1b-r 5 GTTAGCTACTTACCATGGCTACACTGGAACCCGTTT 3 5 TTCCAGATTACGCTGGATCCATGGAGGAGACGTCGACG ppr3n-sec23a-f 3 5 TGATACCACTGCTTGGATCCTTAGCTTGGTTCAGGTGG ppr3n-sec23a-r 3 ppr3n-sec23b-f 5 TTCCAGATTACGCTGGATCCATGTCTGAGTTCCTGGAT 3 5 TGATACCACTGCTTGGATCCCTACTGAACTGCCAGCCG ppr3n-sec23b-r 3 5 TTCCAGATTACGCTGGATCCATGGCGGCCGATCCCTCC ppr3n-sec23c-f 3 5 TGATACCACTGCTTGGATCCTTAGGACTGAACGGCCAG ppr3n-sec23c-r 3 qpcr primers for OsSec23 genes OsSec23a-F 5' TGGCAAGTGTGCAGGATGATGTTG 3' OsSec23a-R 5' ATAGCATCTGAGGCAGTTCTGGCA 3' OsSec23b-F 5' GATCAGCAAGGGCATGAGGCATTT 3' 17
18 OsSec23b-R 5' TGATCACACACGACCAAACGAGGA 3' OsSec23c-F 5' AGGACGATTTCCTGCTCCAAGACT 3' OsSec23c-R 5' ACCTGGAGGAACATCATGTGCTGA 3' Y N -GOT1B-F 5' GAAGGGAGGTACTAGTATGGTTTCCTTCGAGATGAA 3' Y N -GOT1B-R 5' TAGAAGCCATACTAGTCTACACTGGAACCCGTTTT 3' Y C -OsSec23c-F 5' CAAGGGCGGTACTAGTATGGCGGCCGATCCCTCC 3' BiFC assay Y C -OsSec23c-R 5' TAGAAGCCATACTAGTTTAGGACTGAACGGCCAG 3' Y C -OsSec23a-F 5' CAAGGGCGGTACTAGTTTAGCTTGGTTCAGGTGG 3 Y C -OsSec23a-R 5' TAGAAGCCATACTAGTTTAGCTTGGTTCAGGTGG 3 Y C -OsSec23b-F 5' CAAGGGCGGTACTAGTATGTCTGAGTTCCTGGAT 3 Y C -OsSec23b-R 5' TAGAAGCCATACTAGTCTACTGAACTGCCAGCCG 3 18
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
Supplemental Materials
 Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplemental Data. Sethi et al. (2014). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
Supplemental Table 1. List of PCR primers
 Supplemental Table 1. List of PCR primers Primer Sequences (5 to 3 ) 1 TATCCATGGCGCCGGCGGCGAGGGCGGAG 2 TATAAGCTTCTTGGCGTGTCCAGCCCACGGGGCGTAGAACTCGAC 3 ATAAAGCTTGCTCCAGAGTATGAGAAAGCT 4 ATACTCGAGGAGCTCATCCTTGAGAGGCTC
Supplemental Table 1. List of PCR primers Primer Sequences (5 to 3 ) 1 TATCCATGGCGCCGGCGGCGAGGGCGGAG 2 TATAAGCTTCTTGGCGTGTCCAGCCCACGGGGCGTAGAACTCGAC 3 ATAAAGCTTGCTCCAGAGTATGAGAAAGCT 4 ATACTCGAGGAGCTCATCCTTGAGAGGCTC
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc RGLG Hormonal treatment H2O B RGLG µm ABA µm ACC µm GA Time (hours) µm µm MJ µm IA
 Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Aung et al. (2011). Plant Cell /tpc
 35S pro:pmd1-yfp 10 µm Supplemental Figure 1. -terminal YFP fusion of PMD1 (PMD1-YFP, in green) is localized to the cytosol. grobacterium cells harboring 35S pro :PMD1-YFP were infiltrated into tobacco
35S pro:pmd1-yfp 10 µm Supplemental Figure 1. -terminal YFP fusion of PMD1 (PMD1-YFP, in green) is localized to the cytosol. grobacterium cells harboring 35S pro :PMD1-YFP were infiltrated into tobacco
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Characterization of a novel gene involved in border cell migration
 Characterization of a novel gene involved in border cell migration Wei-Hao Li, He-Yen Chou and Li-Mei Pai Graduate Institute of Basic Medical Science Chang-Gung University, Tao-Yuan, Taiwan, R.O.C. Abstract
Characterization of a novel gene involved in border cell migration Wei-Hao Li, He-Yen Chou and Li-Mei Pai Graduate Institute of Basic Medical Science Chang-Gung University, Tao-Yuan, Taiwan, R.O.C. Abstract
Sequence of C-terminal tail regions of myosin heavy chains in class XI of Nicotiana benthamiana (Nb
 Fig. S1 Sequence of C-terminal tail regions of myosin heavy chains in class XI of Nicotiana benthamiana (Nb myosin XI-2, -F and K) and BY-2 cell (Nt 170-kD myosin and Nt 175-kD myosin). Amino acids identical
Fig. S1 Sequence of C-terminal tail regions of myosin heavy chains in class XI of Nicotiana benthamiana (Nb myosin XI-2, -F and K) and BY-2 cell (Nt 170-kD myosin and Nt 175-kD myosin). Amino acids identical
Supplementary Data Supplementary Figures
 Supplementary Data Supplementary Figures Supplementary Figure 1. Pi04314 is expressed during infection, each GFP-Pi04314 fusion is stable and myr GFP-Pi04314 is removed from the nucleus while NLS GFP-Pi04314
Supplementary Data Supplementary Figures Supplementary Figure 1. Pi04314 is expressed during infection, each GFP-Pi04314 fusion is stable and myr GFP-Pi04314 is removed from the nucleus while NLS GFP-Pi04314
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast. Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice
 A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
Supplemental Data. Seo et al. (2014). Plant Cell /tpc
 Supplemental Figure 1. Protein alignment of ABD1 from other model organisms. The alignment was performed with H. sapiens DCAF8, M. musculus DCAF8 and O. sativa Os10g0544500. The WD40 domains are underlined.
Supplemental Figure 1. Protein alignment of ABD1 from other model organisms. The alignment was performed with H. sapiens DCAF8, M. musculus DCAF8 and O. sativa Os10g0544500. The WD40 domains are underlined.
Figure S1. DELLA Proteins Act as Positive Regulators to Mediate GA-Regulated Anthocyanin
 Supplemental Information Figure S1. DELLA Proteins Act as Positive Regulators to Mediate GA-Regulated Anthocyanin Biosynthesis. (A) Effect of GA on anthocyanin content in WT and ga1-3 seedlings. Mock,
Supplemental Information Figure S1. DELLA Proteins Act as Positive Regulators to Mediate GA-Regulated Anthocyanin Biosynthesis. (A) Effect of GA on anthocyanin content in WT and ga1-3 seedlings. Mock,
Supplementary Information
 Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues.
 Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
ATL1-GFP Marker-mCherry/RFP Merge B C D
 Input IP:α-H EDR1-H stedr1-h EV EDR1-H stedr1-h EV α-h α-mcherry TL1-GFP Marker-mCherry/RFP Merge C D GmMan49-mCherry mcherry-syp21 VH-a1-RFP E F G H I J EDR1-nYFP+TL1-cYFP ra6-mcherry Merge K L M Supplemental
Input IP:α-H EDR1-H stedr1-h EV EDR1-H stedr1-h EV α-h α-mcherry TL1-GFP Marker-mCherry/RFP Merge C D GmMan49-mCherry mcherry-syp21 VH-a1-RFP E F G H I J EDR1-nYFP+TL1-cYFP ra6-mcherry Merge K L M Supplemental
Supplementary information
 Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
To investigate the heredity of the WFP gene, we selected plants that were homozygous
 Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supporting Information
 Supporting Information Üstün and Börnke, 2015 Figure S1: XopJ does not display acetyltransferase activity. Autoacetylation activity in vitro. Acetylation reactions using MBP, MBP-XopJ, GST and GST-HopZ1a
Supporting Information Üstün and Börnke, 2015 Figure S1: XopJ does not display acetyltransferase activity. Autoacetylation activity in vitro. Acetylation reactions using MBP, MBP-XopJ, GST and GST-HopZ1a
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana.
 SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
Nature Structural & Molecular Biology: doi: /nsmb.3308
 Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplemental materials
 Supplemental materials Materials and methods for supplemental figures Yeast two-hybrid assays TAP46-PP2Ac interactions I. The TAP46 was used as the bait and the full-length cdnas of the five C subunits
Supplemental materials Materials and methods for supplemental figures Yeast two-hybrid assays TAP46-PP2Ac interactions I. The TAP46 was used as the bait and the full-length cdnas of the five C subunits
Supplemental Data. Tilbrook et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. Protein alignment of with. Identical aligned residues highlighted in black and similar and non-similar residues highlighted in grey and white, respectively. Position of Trp residues
Supplemental Figure 1. Protein alignment of with. Identical aligned residues highlighted in black and similar and non-similar residues highlighted in grey and white, respectively. Position of Trp residues
Supplemental Data. Hu et al. Plant Cell (2017) /tpc
 1 2 3 4 Supplemental Figure 1. DNA gel blot analysis of homozygous transgenic plants. (Supports Figure 1.) 5 6 7 8 Rice genomic DNA was digested with the restriction enzymes EcoRⅠ and BamHⅠ. Lanes in the
1 2 3 4 Supplemental Figure 1. DNA gel blot analysis of homozygous transgenic plants. (Supports Figure 1.) 5 6 7 8 Rice genomic DNA was digested with the restriction enzymes EcoRⅠ and BamHⅠ. Lanes in the
Supplemental Data. Zhang et al. Plant Cell (2014) /tpc
 Supplemental Data. Zhang et al. Plant Cell (214) 1.115/tpc.114.134163 55 - T C N SDIRIP1-GFP 35-25 - Psb 18 - Histone H3 Supplemental Figure 1. Detection of SDIRIP1-GFP in the nuclear fraction by Western
Supplemental Data. Zhang et al. Plant Cell (214) 1.115/tpc.114.134163 55 - T C N SDIRIP1-GFP 35-25 - Psb 18 - Histone H3 Supplemental Figure 1. Detection of SDIRIP1-GFP in the nuclear fraction by Western
α-mannosidase II (HGL1/MANII) β1,2-n-acetyl-glucosaminyltransferase II (GNTII)
 Oligosaccharyltransferase (OST) (STT3a, STT3b, DD1, DD2, DGL1) Glucosidase I, Glucosidase II ER-Mannosidase α-mannosidase I (MNI) β1,2-n-acetyl-glucosaminyltransferase I (CGL1/GNTI) α-mannosidase II (HGL1/MNII)
Oligosaccharyltransferase (OST) (STT3a, STT3b, DD1, DD2, DGL1) Glucosidase I, Glucosidase II ER-Mannosidase α-mannosidase I (MNI) β1,2-n-acetyl-glucosaminyltransferase I (CGL1/GNTI) α-mannosidase II (HGL1/MNII)
Supplementary Information
 Supplementary Information ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis Supplemental Figures a 8 6 4 2 Absolute Expression b Absolute Expression
Supplementary Information ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis Supplemental Figures a 8 6 4 2 Absolute Expression b Absolute Expression
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Figure S1. USP-46 is expressed in several tissues including the nervous system
 Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/5/244/ra72/dc1 Supplementary Materials for An Interaction Between BZR1 and DELLAs Mediates Direct Signaling Crosstalk Between Brassinosteroids and Gibberellins
www.sciencesignaling.org/cgi/content/full/5/244/ra72/dc1 Supplementary Materials for An Interaction Between BZR1 and DELLAs Mediates Direct Signaling Crosstalk Between Brassinosteroids and Gibberellins
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15
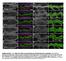 Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and
 Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts. (A) Shoot morphology of 19-day-old nla mutants under Pi-sufficient conditions. (B) [ 33 P]Pi uptake
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts. (A) Shoot morphology of 19-day-old nla mutants under Pi-sufficient conditions. (B) [ 33 P]Pi uptake
Gene specific primers. Left border primer (638) ala3-4 (Salk_082157) Gene specific primers. Left border primer (1343)
 Supplemental Data. Poulsen et al. (2008) The Arabidopsis P4-ATPase pump ALA3 localizes to the Golgi and requires a β subunit to function in lipid translocation and secretory vesicle formation. A ala3-1
Supplemental Data. Poulsen et al. (2008) The Arabidopsis P4-ATPase pump ALA3 localizes to the Golgi and requires a β subunit to function in lipid translocation and secretory vesicle formation. A ala3-1
Supplemental Data. Steiner et al. Plant Cell. (2012) /tpc
 Supplemental Figure 1. SPY does not interact with free GST. Invitro pull-down assay using E. coli-expressed MBP-SPY and GST, GST-TCP14 and GST-TCP15. MBP-SPY was used as bait and incubated with equal amount
Supplemental Figure 1. SPY does not interact with free GST. Invitro pull-down assay using E. coli-expressed MBP-SPY and GST, GST-TCP14 and GST-TCP15. MBP-SPY was used as bait and incubated with equal amount
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
PIE1 ARP6 SWC6 KU70 ARP6 PIE1. HSA SNF2_N HELICc SANT. pie1-3 A1,A2 K1,K2 K1,K3 K3,LB2 A3, A4 A3,LB1 A1,A2 K1,K2 K1,K3. swc6-1 A3,A4.
 A B N-terminal SWC2 H2A.Z SWC6 ARP6 PIE1 HSA SNF2_N HELICc SANT C pie1-3 D PIE1 ARP6 5 Kb A1 200 bp A3 A2 LB1 arp6-3 A4 E A1,A2 A3, A4 A3,LB1 K1,K2 K1,K3 K3,LB2 SWC6 swc6-1 A1,A2 A3,A4 K1,K2 K1,K3 100
A B N-terminal SWC2 H2A.Z SWC6 ARP6 PIE1 HSA SNF2_N HELICc SANT C pie1-3 D PIE1 ARP6 5 Kb A1 200 bp A3 A2 LB1 arp6-3 A4 E A1,A2 A3, A4 A3,LB1 K1,K2 K1,K3 K3,LB2 SWC6 swc6-1 A1,A2 A3,A4 K1,K2 K1,K3 100
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Data. Benstein et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
T H E J O U R N A L O F C E L L B I O L O G Y
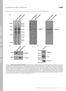 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Bays et al., http://www.jcb.org/cgi/content/full/jcb.201309092/dc1 Figure S1. Specificity of the phospho-y822 antibody. (A) Total cell
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Bays et al., http://www.jcb.org/cgi/content/full/jcb.201309092/dc1 Figure S1. Specificity of the phospho-y822 antibody. (A) Total cell
Nature Genetics: doi: /ng Supplementary Figure 1. ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets.
 Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplemental Data. Ko et al. (2014). Plant Cell /tpc Supplemental Figure 1. Evidence of T-DNA Insertion in ms142 Mutant.
 Supplemental Figure 1. Evidence of T-DNA Insertion in ms142 Mutant. (A) DNA gel blotting to hptii probe confirmed single T-DNA insertion (marked with arrow) in ms142 mutant. (B) T-DNA tagged construction
Supplemental Figure 1. Evidence of T-DNA Insertion in ms142 Mutant. (A) DNA gel blotting to hptii probe confirmed single T-DNA insertion (marked with arrow) in ms142 mutant. (B) T-DNA tagged construction
Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis
 Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis seedlings expressing ERF109pro-GUS. The GUS staining
Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis seedlings expressing ERF109pro-GUS. The GUS staining
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Supplemental Figure 1
 Supplemental Figure 1 A LK sls1 lks1-2 F 1 sls1 B LK sls1 lks1-2 F 1 lks1-2 sls1 F 1 lks1-2 sls1 F 2 Col lks1-2 Col+LKS1 Col Col+LKS1 Supplemental Figure 1. Genetic analysis of sls1 mutant. (A) and (B)
Supplemental Figure 1 A LK sls1 lks1-2 F 1 sls1 B LK sls1 lks1-2 F 1 lks1-2 sls1 F 1 lks1-2 sls1 F 2 Col lks1-2 Col+LKS1 Col Col+LKS1 Supplemental Figure 1. Genetic analysis of sls1 mutant. (A) and (B)
sides of the aleurone (Al) but it is excluded from the basal endosperm transfer layer
 Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Sperm cells are passive cargo of the pollen tube in plant fertilization
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
Supplemental Data. Zhang et al. (2013). Plant Cell /tpc
 SDLTXgal SDLTH SDLTHA ADSV4 BD BDP53 BD BDP53 BD BDP53 ADAt1g1567(aa36358) ADAt1g844(aa9354) ADAt1g844(aa51354) ADAt1g844(full) Prey ADAt1g844(aa11354) ADAt3g5994(aa48418) BD BDPAL2 BD BDPAL2 BD BDPAL2
SDLTXgal SDLTH SDLTHA ADSV4 BD BDP53 BD BDP53 BD BDP53 ADAt1g1567(aa36358) ADAt1g844(aa9354) ADAt1g844(aa51354) ADAt1g844(full) Prey ADAt1g844(aa11354) ADAt3g5994(aa48418) BD BDPAL2 BD BDPAL2 BD BDPAL2
Supplemental Figure 1. Immunoblot analysis of wild-type and mutant forms of JAZ3
 Supplemental Data. Chung and Howe (2009). A Critical Role for the TIFY Motif in Repression of Jasmonate Signaling by a Stabilized Splice Variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis.
Supplemental Data. Chung and Howe (2009). A Critical Role for the TIFY Motif in Repression of Jasmonate Signaling by a Stabilized Splice Variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis.
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplemental Data. Hachez et al. Plant Cell (2014) /tpc Suppl. Figure 1A
 Suppl. Figure 1A Suppl. Figure 1B Supplemental Figure 1: Results of the commercial screening of interactants using split ubiquitin technique. (A) Isolated preys (192) using the bait construct pbt3-n- as
Suppl. Figure 1A Suppl. Figure 1B Supplemental Figure 1: Results of the commercial screening of interactants using split ubiquitin technique. (A) Isolated preys (192) using the bait construct pbt3-n- as
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplemental Figure 1
 Supplemental Figure 1 A gta2-1 gta2-2 1kb AT4G08350 B Col-0 gta2-1 LP+RP+LBb1.3 C Col-0 gta2-2 LP+RP+LB1 D Col-0 gta2-2 gta2-1 GTA2 TUBLIN E Col0 gta2-1 gta2-2 Supplemental Figure 1. Phenotypic analysis
Supplemental Figure 1 A gta2-1 gta2-2 1kb AT4G08350 B Col-0 gta2-1 LP+RP+LBb1.3 C Col-0 gta2-2 LP+RP+LB1 D Col-0 gta2-2 gta2-1 GTA2 TUBLIN E Col0 gta2-1 gta2-2 Supplemental Figure 1. Phenotypic analysis
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
AD BD TOC1. Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between
 AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplemental Materials
 Supplemental Materials Supplemental Figure S. Phenotypic assessment of alb4 mutant plants under different stress conditions. (A) High-light stress and drought stress. Wild-type (WT) and alb4 mutant plants
Supplemental Materials Supplemental Figure S. Phenotypic assessment of alb4 mutant plants under different stress conditions. (A) High-light stress and drought stress. Wild-type (WT) and alb4 mutant plants
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb54 sensitivity (+/-) 3 det- WS bri-5 BL (nm) bzr-dbri- bzr-d/ bri- g h i a b c d e f Hypcocotyl length(cm) 5 5 PAC Hypocotyl length (cm)..8..4. 5 5 PPZ - - + + + + - + - + - + PPZ - - + + +
DOI:.38/ncb54 sensitivity (+/-) 3 det- WS bri-5 BL (nm) bzr-dbri- bzr-d/ bri- g h i a b c d e f Hypcocotyl length(cm) 5 5 PAC Hypocotyl length (cm)..8..4. 5 5 PPZ - - + + + + - + - + - + PPZ - - + + +
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
Supplementary information Supplementary Figure 1. Comparison of BPH resistance in RH and Taichung Native 1.
 Supplementary information Supplementary Figure 1. Comparison of BPH resistance in RH and Taichung Native 1. a, Representative image of RH and Taichung Native 1 (TN1) infested with BPH. b, Seedling mortality
Supplementary information Supplementary Figure 1. Comparison of BPH resistance in RH and Taichung Native 1. a, Representative image of RH and Taichung Native 1 (TN1) infested with BPH. b, Seedling mortality
ABI3 Controls Embryo De-greening Through Mendel's I locus
 Supporting Online Material for ABI3 Controls Embryo De-greening Through Mendel's I locus Frédéric Delmas a,b,c, Subramanian Sankaranarayanan d, Srijani Deb d, Ellen Widdup d, Céline Bournonville b,c,norbert
Supporting Online Material for ABI3 Controls Embryo De-greening Through Mendel's I locus Frédéric Delmas a,b,c, Subramanian Sankaranarayanan d, Srijani Deb d, Ellen Widdup d, Céline Bournonville b,c,norbert
Supplemental Data. Jing et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Characterization of epp1 Mutants. (A) Cotyledon angles of 5-d-old Col wild-type (gray bars) and epp1-1 (black bars) seedlings under red (R), far-red (FR) and blue (BL) light conditions,
Supplemental Figure 1. Characterization of epp1 Mutants. (A) Cotyledon angles of 5-d-old Col wild-type (gray bars) and epp1-1 (black bars) seedlings under red (R), far-red (FR) and blue (BL) light conditions,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
Supplemental Figure 1. VLN5 retains conserved residues at both type 1 and type 2 Ca 2+ -binding
 Supplemental Figure 1. VLN5 retains conserved residues at both type 1 and type 2 Ca 2+ -binding sites in the G1 domain. Multiple sequence alignment was performed with DNAMAN6.0.40. Secondary structural
Supplemental Figure 1. VLN5 retains conserved residues at both type 1 and type 2 Ca 2+ -binding sites in the G1 domain. Multiple sequence alignment was performed with DNAMAN6.0.40. Secondary structural
Supplementary Figures 1-12
 Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
Identification and Characterization of a Plastidic Adenine. Nucleotide Uniporter (OsBT1-3) Required for Chloroplast
 Identification and Characterization of a Plastidic Adenine Nucleotide Uniporter (OsBT1-3) Required for Chloroplast Development in the Early Leaf Stage of Rice Authors: Daoheng Hu 1, Yang Li 1, Wenbin Jin
Identification and Characterization of a Plastidic Adenine Nucleotide Uniporter (OsBT1-3) Required for Chloroplast Development in the Early Leaf Stage of Rice Authors: Daoheng Hu 1, Yang Li 1, Wenbin Jin
Nature Genetics: doi: /ng.3556 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE. Supplementary Figure 1
 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1 is related to Figure 1B, showing more details of outer segment of
 Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Figure 1. Phos-Tag mobility shift detection of in vivo phosphorylated ERF6 in 35S:ERF6 WT plants after B. cinerea inoculation.
 Supplemental Figure 1. Phos-Tag mobility shift detection of in vivo phosphorylated ERF6 in 35S:ERF6 WT plants after B. cinerea inoculation. Protein extracts from 35S:ERF6 WT seedlings treated with B. cinerea
Supplemental Figure 1. Phos-Tag mobility shift detection of in vivo phosphorylated ERF6 in 35S:ERF6 WT plants after B. cinerea inoculation. Protein extracts from 35S:ERF6 WT seedlings treated with B. cinerea
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
SUPPLEMENTARY INFORMATION
 AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
Supplemental Data. Furlan et al. Plant Cell (2017) /tpc
 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure.
Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Supplemental Figure S1. Validation of auxin-related gene expression from Fig. 1A. Relative gene expression in 2x and 3x seeds 6 days after
 Supplemental Figure S1. Validation of auxin-related gene expression from Fig. 1A. Relative gene expression in 2x and 3x seeds 6 days after pollination (6 DAP), as determined by RT-qPCR for YUC10 (A), YUC11
Supplemental Figure S1. Validation of auxin-related gene expression from Fig. 1A. Relative gene expression in 2x and 3x seeds 6 days after pollination (6 DAP), as determined by RT-qPCR for YUC10 (A), YUC11
The Arabidopsis Transcription Factor BES1 Is a Direct Substrate of MPK6 and Regulates Immunity
 Supplemental Data The Arabidopsis Transcription Factor BES1 Is a Direct Substrate of MPK6 and Regulates Immunity Authors: Sining Kang, Fan Yang, Lin Li, Huamin Chen, She Chen and Jie Zhang The following
Supplemental Data The Arabidopsis Transcription Factor BES1 Is a Direct Substrate of MPK6 and Regulates Immunity Authors: Sining Kang, Fan Yang, Lin Li, Huamin Chen, She Chen and Jie Zhang The following
Supplemental Figure S1. Nucleotide and deduced amino acid sequences of pepper CaHSP70a (Capsicum annuum heat shock protein 70a) cdna.
 Supplemental Figure S1. Nucleotide and deduced amino acid sequences of pepper (Capsicum annuum heat shock protein 70a) cdna. Translation initiation codon is shown in bold typeface; termination codon is
Supplemental Figure S1. Nucleotide and deduced amino acid sequences of pepper (Capsicum annuum heat shock protein 70a) cdna. Translation initiation codon is shown in bold typeface; termination codon is
Supplemental Data. Borg et al. Plant Cell (2014) /tpc
 Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplemental Data. Li et al. (2015). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1: Characterization of asr3 T-DNA knockout lines and complementation transgenic lines. (A) The scheme of At2G33550 (ASR3) with gray boxes indicating exons and dash
Supplemental Data Supplemental Figure 1: Characterization of asr3 T-DNA knockout lines and complementation transgenic lines. (A) The scheme of At2G33550 (ASR3) with gray boxes indicating exons and dash
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
7.012 Problem Set 5. Question 1
 Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Supplementary Information. Supplementary Figure S1. Phenotypic comparison of the wild type and mutants.
 Supplementary Information Supplementary Figure S1. Phenotypic comparison of the wild type and mutants. Supplementary Figure S2. Transverse sections of anthers. Supplementary Figure S3. DAPI staining and
Supplementary Information Supplementary Figure S1. Phenotypic comparison of the wild type and mutants. Supplementary Figure S2. Transverse sections of anthers. Supplementary Figure S3. DAPI staining and
Institute of Biological Chemistry, Washington State University, Pullman, Washington, USA
 Plant and Cell Physiology Advance Access published July 15, 2009 Running title: The glutelin RNA ER zip code signals Name and address for editorial corresponding: Thomas W Okita Institute of Biological
Plant and Cell Physiology Advance Access published July 15, 2009 Running title: The glutelin RNA ER zip code signals Name and address for editorial corresponding: Thomas W Okita Institute of Biological
Division of Plant Sciences, National Institute of Agrobiological Sciences, Tsukuba, Ibaraki , Japan b
 This article is a Plant Cell Advance Online Publication. The date of its first appearance online is the official date of publication. The article has been edited and the authors have corrected proofs,
This article is a Plant Cell Advance Online Publication. The date of its first appearance online is the official date of publication. The article has been edited and the authors have corrected proofs,
Supplemental Data. Dai et al. (2013). Plant Cell /tpc Absolute FyPP3. Absolute
 A FyPP1 Absolute B FyPP3 Absolute Dry seeds Imbibed 24 hours Dry seeds Imbibed 24 hours C ABI5 Absolute Dry seeds Imbibed 24 hours Supplemental Figure 1. Expression of FyPP1, FyPP3 and ABI5 during seed
A FyPP1 Absolute B FyPP3 Absolute Dry seeds Imbibed 24 hours Dry seeds Imbibed 24 hours C ABI5 Absolute Dry seeds Imbibed 24 hours Supplemental Figure 1. Expression of FyPP1, FyPP3 and ABI5 during seed
Supplemental Information
 Supplemental Information Supplemental Figure 1. The Heterologous Yeast System for Screening of Arabidopsis JA Transporters. (A) Exogenous JA inhibited yeast cell growth. Yeast cells were diluted to various
Supplemental Information Supplemental Figure 1. The Heterologous Yeast System for Screening of Arabidopsis JA Transporters. (A) Exogenous JA inhibited yeast cell growth. Yeast cells were diluted to various
Supplementary Figure S1. DYF-19 is highly conserved and expressed exclusively in
 Supplementary Figure S1. DYF-19 is highly conserved and expressed exclusively in ciliated cells in C. elegans. a, Sequence alignment of DYF-19 homologs. b, A GFP reporter gene driven by the dyf-19 promoter
Supplementary Figure S1. DYF-19 is highly conserved and expressed exclusively in ciliated cells in C. elegans. a, Sequence alignment of DYF-19 homologs. b, A GFP reporter gene driven by the dyf-19 promoter
Superexpression of tuberculosis antigens in plant leaves
 Superexpression of tuberculosis antigens in plant leaves Tuberculosis Volume 87, Issue 3, May 2007, Pages 218-224 Yuri L. Dorokhov, a,, Anna A. Shevelevaa, Olga Y. Frolovaa, Tatjana V. Komarovaa, Anna
Superexpression of tuberculosis antigens in plant leaves Tuberculosis Volume 87, Issue 3, May 2007, Pages 218-224 Yuri L. Dorokhov, a,, Anna A. Shevelevaa, Olga Y. Frolovaa, Tatjana V. Komarovaa, Anna
SUPPLEMENTARY INFORMATION
 Supplementary Methods Protein expression and in vitro binding studies. Recombinant baculovirus carrying GST, GST-mCC, GST-mSec, GST-mSecA and GST-mSecD were generated according to the manufacturer s instructions
Supplementary Methods Protein expression and in vitro binding studies. Recombinant baculovirus carrying GST, GST-mCC, GST-mSec, GST-mSecA and GST-mSecD were generated according to the manufacturer s instructions
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
