Supplementary Materials
|
|
|
- Buck Phillips
- 5 years ago
- Views:
Transcription
1 Supplementary Materials Table S1. Oligonucleotide sequences and PCR conditions used to amplify the indicated genes. TA = annealing temperature; gdna = genomic DNA; cdna = complementary DNA; c = concentration. Gene Oligo Sequence (5'-->3') T A in C c [MgCl] in mm PCR-Product Size EF H2A LTP1 LTP3 LTP4 AT1G12090 LTP8 AT5G05960 AT4G22610 AT3G22620 AT4G33550 AT1G62510 Pb Actin nos-terminator tcccttcaacactcctttatagc aacagtctatgcgacacgtca cggggaaaggtgctaaaggtt aaatgcctcggcgagatacgt ctccaaggtggtgtcattcc cccatataacaagaacaccacaa tgttatcacccaaaaagaagttca ttattattacgttcgtatgcgttgg tatcacccaaaagagaagagca acacaagtatacaacataacaaagc cccaattcactcacaacctagc atcaccccaatgaacaccag gcaacaacaaagaaaccccta ttaggacaagatggaccattga tcatcactcaagaatggaaacc acgtctattgctttctgtctgc taaatccaagccctcacctc cagcaacaactacgatcatgc cacacttcaaacacaaaaccac cttattccacaaagcaatgacc cggaccaaatattcgcattc ccggaatggtgtaacctataaca tcccaattcacacataccaaag ggtagaaatcatcttgtctgtcca agctggcgtacgtggcgcag ccttgacgcgcatcgacgac tatagcggccgcggatcgttcaaacatttggcaata gcgcgagctcatctagtaacataga gdna: 2354 bp cdna: 1500 bp gdna: 690 bp cdna: 282 bp gdna: 450 bp cdna: 380 bp gdna: 703 bp cdna: 610 bp gdna: 649 bp cdna: 510 bp gdna: 630 bp cdna: 630 bp gdna: 885 bp cdna: 448 bp gdna: 390 bp cdna: 479 bp gdna: 501 bp cdna: 501 bp gdna: 979 bp cdna: 790 bp gdna: 490 bp cdna: 490 bp gdna: 605 bp cdna: 605 bp gdna: 342 bp cdna: 342 bp bp
2 Table S2. Oligonucleotide sequences and PCR conditions used to verify the T-DNA integration. TA = annealing temperature; c = concentration. T-DNA Insertion Line Acc. No/ Disrupted Gene N647582/ AT1G12090 N520925/ LTP8 N648038/ AT3G22620 N527420/ AT5G06960 N500561/ LTP4 N595248/ LTP3 Oligos Used 5'-->3' cgatcctgtttcgcgtagat atcaccccaatgaacaccag tgttgaaagagcttgtagtaatgg gcaacaacaaagaaaccccta tccgaatacgacctacaatga cttattccacaaagcaatgacc ttgaggactggagaaaagga acgtctattgctttctgtctgc aaggattcttgcagtttcat acacaagtatacaacataacaaagc tgttatcacccaaaaagaagttca ttattattacgttcgtatgcgttgg Site of T-DNA Insertion T A in C c [MgCl] in mm PCR-Product Size without T-DNA Promoter 60 2, ' utr 58 2, Promoter/ 5' utr 58 2, Promoter 58 2,5 872 Promoter 58 2, Exon 58 2,5 703 Figure S1. Expression of selected LTP genes in clubroot infected A. thaliana roots. The expression for some LTP genes during clubroot infection between 7 and 26 days after inoculation is shown. For the gene AT1G62510 the expression was not detectable. For infection a field isolate [44] was used. C = control; I = inoculated.
3 Figure S2. Expression of some LTP genes in roots of LTP mutants. The expression for the indicated LTP genes (left) in comparison to the wild type (WT) is shown. The elongation factor 1B gamma (AT1G09640) and histon H2A (AT1G52740) were used as reference genes. For the semi-quantitative expression analyses the RNA was extracted from 24-day-old A. thaliana roots. The red squares mark the expression of the overexpressed (OX) or downregulated (KO) gene. For AT1G62510 and AT4G33550 the expression in the transgenic plants as well as in the wild type plants and for AT1G62510 in the T-DNA insertion lines and their wild type control the gene expression was not detectable.
4 Figure S3. Lipid composition of uninfected roots and galls of A. thaliana. Results of thin layer chromatography from non-polar lipids isolated from equal amounts of infected roots (galls) 30 days after inoculation or of healthy root material of the same age. Plant material was from wild type (WT) and LTP mutants that (A) overexpress (OX) the genes LTP1, LTP3, LTP4, AT1G12090 and LTP8 and with reduced LTP gene expression (B) from T-DNA-insertion lines (KO) for the genes AT1G12090, LTP8, AT3G22620, AT5G05960, LTP4, LTP3 and antisense lines (AS) for the genes AT4G33550 and AT1G Two biological replicates with approx. 25 plants each were analyzed. Triacylglycerol (TAG) was used as a standard. R = uninfected root; G = gall (infected root). For explanation of bands a and b see main text.
5 Figure S4. Growth reduction due to salt stress conditions. Root growth and whole plant fresh weight from wild type (WT), LTP mutants (LTP4-OX, AT1G12090-OX, LTP8-OX) and the empty vector control (EPG) in response to salt stress are shown. To calculate the root growth and fresh weight (in %) the root growth and fresh weight from unstressed plants (0 mm NaCl) were set to 100%. Therefore the graphs show the growth reduction due to salt stress. n 50. Asterisks indicates a significant difference (for ** p < 0.01; *** p < 0.001). OX: overexpression of the indicated gene.
6 Figure S5. Growth reduction due to salt stress conditions. Root growth and whole plant fresh weight from wild type (WT) and LTP mutants (AT1G12090-KO, LTP8-KO, AT3G22620, AT5G05960-KO, LTP3-KO, LTP4-KO) in response to salt stress are shown. To calculate the root growth and fresh weight (in %) the root growth and fresh weight from unstressed plants (0 mm NaCl) were set to 100%. Therefore the graphs show the growth reduction due to salt stress. Since these graphs show a ratio based on the mean value the standard deviation could not be plotted on this graph. n 30, KO: knockout or knockdown of the indicated gene. Asterisks indicates a significant difference (for * p < 0.05; ** p < 0.01; *** p < 0.001).
7 Figure S6. Growth reduction due to osmotic stress conditions. Root growth and whole plant fresh weight from wild type (WT) and LTP mutants (LTP1-OX, LTP3-OX, LTP4-OX, AT1G12090-OX, LTP8-OX and AT4G33550-AS, AT1G62510-AS) in response to mannitol treatment (300 mm, 350 mm) are shown. To calculate the root growth and fresh weight (in %) the root growth and fresh weight from unstressed plants (0 mm NaCl) were set to 100%. Because of the obvious differences between the vector control (EPG) and the wild type plants, the mutants were compared to the vector control plants (EPG). Since these graphs show a ratio based on the mean value the standard deviation could not be plottet on this graph. n 30, OX: overexpression of the indicated gene; AS: silencing of the indicated gene using antisense technique. Asterisks indicates a significant difference (for ** p < 0.01; *** p < 0.001).
8 Figure S7. Growth reduction due to osmotic stress conditions. Root growth and whole plant fresh weight from wild type (WT) and LTP mutants (AT4G33550-AS, AT1G62510-AS, AT1G12090-KO, LTP8-KO, AT3G22620-KO, AT5G05960-KO, LTP3-KO, LTP4-KO) in response to mannitol treatment (300 mm, 350 mm) are shown. To calculate the root growth and fresh weight (in %) the root growth and fresh weight from unstressed plants (0 mm NaCl) were set to 100%. Since these graphs show a ratio based on the mean value the standard deviation could not be plotted on this graph. Because of the obvious differences between the vector control (EPG) and the wildtype plants (see S7). n 30, KO: knockout or knockdown of the indicated gene, Asterisks indicates a significant difference (for ** p < 0.01).
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Two classes of silencing RNAs move between Caenorhabditis elegans tissues.
 Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification.
 Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
Arabidopsis actin depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB
 Arabidopsis actin depolymerizing factor mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB Files in this Data Supplement: Supplemental Table S1 Supplemental Table
Arabidopsis actin depolymerizing factor mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB Files in this Data Supplement: Supplemental Table S1 Supplemental Table
- 1 - Supplemental Data
 - 1-1 Supplemental Data 2 3 4 5 6 7 8 9 Supplemental Figure S1. Differential expression of AtPIP Genes in DC3000-inoculated plants. Gene expression in leaves was analyzed by real-time RT-PCR and expression
- 1-1 Supplemental Data 2 3 4 5 6 7 8 9 Supplemental Figure S1. Differential expression of AtPIP Genes in DC3000-inoculated plants. Gene expression in leaves was analyzed by real-time RT-PCR and expression
Integrated Omics Study Delineates the Dynamics of Lipid Droplets in Rhodococcus Opacus PD630
 School of Natural Sciences and Mathematics 2013-10-22 Integrated Omics Study Delineates the Dynamics of Lipid Droplets in Rhodococcus Opacus PD630 UTD AUTHOR(S): Michael Qiwei Zhang 2013 The Authors This
School of Natural Sciences and Mathematics 2013-10-22 Integrated Omics Study Delineates the Dynamics of Lipid Droplets in Rhodococcus Opacus PD630 UTD AUTHOR(S): Michael Qiwei Zhang 2013 The Authors This
SUPPLEMENTARY INFORMATION
 Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
SUPPLEMENTARY INFORMATION
 AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
7.012 Problem Set 5. Question 1
 Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase
 A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase tag insertion at the 3 end using CRISPR-Cas9 genome editing technique Sambuddha Basu 1, 3, Levi Adams 1, 3, Subhrangshu
A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase tag insertion at the 3 end using CRISPR-Cas9 genome editing technique Sambuddha Basu 1, 3, Levi Adams 1, 3, Subhrangshu
Supplementary Information
 Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc RGLG Hormonal treatment H2O B RGLG µm ABA µm ACC µm GA Time (hours) µm µm MJ µm IA
 Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Reading Lecture 8: Lecture 9: Lecture 8. DNA Libraries. Definition Types Construction
 Lecture 8 Reading Lecture 8: 96-110 Lecture 9: 111-120 DNA Libraries Definition Types Construction 142 DNA Libraries A DNA library is a collection of clones of genomic fragments or cdnas from a certain
Lecture 8 Reading Lecture 8: 96-110 Lecture 9: 111-120 DNA Libraries Definition Types Construction 142 DNA Libraries A DNA library is a collection of clones of genomic fragments or cdnas from a certain
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
Percent survival. Supplementary fig. S3 A.
 Supplementary fig. S3 A. B. 100 Percent survival 80 60 40 20 Ml 0 0 100 C. Fig. S3 Comparison of leukaemia incidence rate in the triple targeted chimaeric mice and germline-transmission translocator mice
Supplementary fig. S3 A. B. 100 Percent survival 80 60 40 20 Ml 0 0 100 C. Fig. S3 Comparison of leukaemia incidence rate in the triple targeted chimaeric mice and germline-transmission translocator mice
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
CGAGACCTTGTCACATTGGCGTCAACTC hoga Down GATGCCTCCTGTCAAGATTGCAATGAAC
 Supplemental Table 1. PCR and qrt-pcr primers used in this study Primer designation Oligonucleotide sequence (5-3 ) PCR primers 5 SphI hoga GCATGCGAAGACTGATTCAGATAATTAGTTC 5 SalI hoga GTCGACCTGATGATCTCCACTGTCTGAAC
Supplemental Table 1. PCR and qrt-pcr primers used in this study Primer designation Oligonucleotide sequence (5-3 ) PCR primers 5 SphI hoga GCATGCGAAGACTGATTCAGATAATTAGTTC 5 SalI hoga GTCGACCTGATGATCTCCACTGTCTGAAC
A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Student Learning Outcomes (SLOS)
 Student Learning Outcomes (SLOS) KNOWLEDGE AND LEARNING SKILLS USE OF KNOWLEDGE AND LEARNING SKILLS - how to use Annhyb to save and manage sequences - how to use BLAST to compare sequences - how to get
Student Learning Outcomes (SLOS) KNOWLEDGE AND LEARNING SKILLS USE OF KNOWLEDGE AND LEARNING SKILLS - how to use Annhyb to save and manage sequences - how to use BLAST to compare sequences - how to get
Supplementary information
 Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Intron distance to transcription start (nt)*
 Schwab et al. SOM: Enhanced microrna accumulation through stemloop-adjacent introns 1 Supplemental Table 1. Characteristics of introns used in this study Intron name Length intron (nt) Length cloned fragment
Schwab et al. SOM: Enhanced microrna accumulation through stemloop-adjacent introns 1 Supplemental Table 1. Characteristics of introns used in this study Intron name Length intron (nt) Length cloned fragment
Supplemental Data. Osakabe et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Phylogenetic analysis of KUP in various species. The amino acid sequences of KUPs from green algae and land plants were identified with a BLAST search and aligned using ClustalW
Supplemental Figure 1. Phylogenetic analysis of KUP in various species. The amino acid sequences of KUPs from green algae and land plants were identified with a BLAST search and aligned using ClustalW
Supplementary Material
 Fig S1. Interactions of OsDof24 with the OsC4PPDK promoter in rice protoplasts (a) Loss-of-function analysis of the OsC4PPDK promoter Effects of OsDof24 overexpression and the mapping of OsDof24-binding
Fig S1. Interactions of OsDof24 with the OsC4PPDK promoter in rice protoplasts (a) Loss-of-function analysis of the OsC4PPDK promoter Effects of OsDof24 overexpression and the mapping of OsDof24-binding
Nature Genetics: doi: /ng Supplementary Figure 1. ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets.
 Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis
 Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis seedlings expressing ERF109pro-GUS. The GUS staining
Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis seedlings expressing ERF109pro-GUS. The GUS staining
*** *** * *** *** *** * *** ** ***
 Figure S: OsMADS6 over- or down-expression is stable across generations A, OsMADS6 expression in overexpressing (OX, OX, dark bars) and corresponding control (OX, WT, white bars) T4 plants. B, OsMADS6
Figure S: OsMADS6 over- or down-expression is stable across generations A, OsMADS6 expression in overexpressing (OX, OX, dark bars) and corresponding control (OX, WT, white bars) T4 plants. B, OsMADS6
TRANSPOSON INSERTION SITE VERIFICATION
 TRANSPOSON INSERTION SITE VERIFICATION Transposon and T-DNA insertion in Arabidopsis genes can be identified using the Arabidopsis thaliana Insertion Database (ATIdb) (http://atidb.org/cgi-perl/gbrowse/atibrowse).
TRANSPOSON INSERTION SITE VERIFICATION Transposon and T-DNA insertion in Arabidopsis genes can be identified using the Arabidopsis thaliana Insertion Database (ATIdb) (http://atidb.org/cgi-perl/gbrowse/atibrowse).
WiscDsLox485 ATG < > //----- E1 E2 E3 E4 E bp. Col-0 arr7 ARR7 ACTIN7. s of mrna/ng total RNA (x10 3 ) ARR7.
 A WiscDsLox8 ATG < > -9 +0 > ---------//----- E E E E E UTR +9 UTR +0 > 00bp B S D ol-0 arr7 ol-0 arr7 ARR7 ATIN7 D s of mrna/ng total RNA opie 0. ol-0 (x0 ) arr7 ARR7 Supplemental Fig.. Genotyping and
A WiscDsLox8 ATG < > -9 +0 > ---------//----- E E E E E UTR +9 UTR +0 > 00bp B S D ol-0 arr7 ol-0 arr7 ARR7 ATIN7 D s of mrna/ng total RNA opie 0. ol-0 (x0 ) arr7 ARR7 Supplemental Fig.. Genotyping and
SUPPLEMENTAL DATA. Supplementary data
 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 SUPPLEMENTAL DATA Supplementary data Figure S1: Verification of pla-iα1 knockdown lines a: Semiquantitative
1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 SUPPLEMENTAL DATA Supplementary data Figure S1: Verification of pla-iα1 knockdown lines a: Semiquantitative
BS 50 Genetics and Genomics Week of Oct 24
 BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
Supplementary Information
 Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Supplemental Data. Lee et al. Plant Cell. (2010) /tpc Supplemental Figure 1. Protein and Gene Structures of DWA1 and DWA2.
 Supplemental Figure 1. Protein and Gene Structures of DWA1 and DWA2. (A) Protein structures of DWA1 and DWA2. WD40 region was determined based on the NCBI conserved domain databases (B, C) Schematic representation
Supplemental Figure 1. Protein and Gene Structures of DWA1 and DWA2. (A) Protein structures of DWA1 and DWA2. WD40 region was determined based on the NCBI conserved domain databases (B, C) Schematic representation
Supplemental materials
 Supplemental materials Materials and methods for supplemental figures Yeast two-hybrid assays TAP46-PP2Ac interactions I. The TAP46 was used as the bait and the full-length cdnas of the five C subunits
Supplemental materials Materials and methods for supplemental figures Yeast two-hybrid assays TAP46-PP2Ac interactions I. The TAP46 was used as the bait and the full-length cdnas of the five C subunits
Supplemental Table/Figure Legends
 MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
 http://fire.biol.wwu.edu/trent/trent/direct_detection_of_genotype.html 1 Like most other model organism Arabidopsis thaliana has a sequenced genome? What do we mean by sequenced genome? What sort of info
http://fire.biol.wwu.edu/trent/trent/direct_detection_of_genotype.html 1 Like most other model organism Arabidopsis thaliana has a sequenced genome? What do we mean by sequenced genome? What sort of info
Rassf1a -/- Sav1 +/- mice. Supplementary Figures:
 1 Supplementary Figures: Figure S1: Genotyping of Rassf1a and Sav1 mutant mice. A. Rassf1a knockout mice lack exon 1 of the Rassf1 gene. A diagram and typical PCR genotyping of wildtype and Rassf1a -/-
1 Supplementary Figures: Figure S1: Genotyping of Rassf1a and Sav1 mutant mice. A. Rassf1a knockout mice lack exon 1 of the Rassf1 gene. A diagram and typical PCR genotyping of wildtype and Rassf1a -/-
Antisense RNA Insert Design for Plasmid Construction to Knockdown Target Gene Expression
 Vol. 1:7-15 Antisense RNA Insert Design for Plasmid Construction to Knockdown Target Gene Expression Ji, Tom, Lu, Aneka, Wu, Kaylee Department of Microbiology and Immunology, University of British Columbia
Vol. 1:7-15 Antisense RNA Insert Design for Plasmid Construction to Knockdown Target Gene Expression Ji, Tom, Lu, Aneka, Wu, Kaylee Department of Microbiology and Immunology, University of British Columbia
Candidate region (0.74 Mb) ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC ATC TCT GGG ACT CAT TAG CAG GAG GCT AGC
 A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
BIOLOGY - CLUTCH CH.20 - BIOTECHNOLOGY.
 !! www.clutchprep.com CONCEPT: DNA CLONING DNA cloning is a technique that inserts a foreign gene into a living host to replicate the gene and produce gene products. Transformation the process by which
!! www.clutchprep.com CONCEPT: DNA CLONING DNA cloning is a technique that inserts a foreign gene into a living host to replicate the gene and produce gene products. Transformation the process by which
Identification and characterization of a pathogenicity-related gene VdCYP1 from Verticillium dahliae
 Identification and characterization of a pathogenicity-related gene VdCYP1 from Verticillium dahliae Dan-Dan Zhang*, Xin-Yan Wang*, Jie-Yin Chen*, Zhi-Qiang Kong, Yue-Jing Gui, Nan-Yang Li, Yu-Ming Bao,
Identification and characterization of a pathogenicity-related gene VdCYP1 from Verticillium dahliae Dan-Dan Zhang*, Xin-Yan Wang*, Jie-Yin Chen*, Zhi-Qiang Kong, Yue-Jing Gui, Nan-Yang Li, Yu-Ming Bao,
Regulation of ARE transcript 3 end processing by the. yeast Cth2 mrna decay factor
 Regulation of ARE transcript 3 end processing by the yeast Cth2 mrna decay factor Manoël Prouteau, Marie-Claire Daugeron and Bertrand Séraphin Supplementary Information Material and Methods Plasmid construction
Regulation of ARE transcript 3 end processing by the yeast Cth2 mrna decay factor Manoël Prouteau, Marie-Claire Daugeron and Bertrand Séraphin Supplementary Information Material and Methods Plasmid construction
SUPPLEMENTARY INFORMATION
 6 Relative levels of ma3 RNA 5 4 3 2 1 LN MG SG PG Spl Thy ma3 ß actin Lymph node Mammary gland Prostate gland Salivary gland Spleen Thymus MMTV target tissues Fig. S1: MMTV target tissues express ma3.
6 Relative levels of ma3 RNA 5 4 3 2 1 LN MG SG PG Spl Thy ma3 ß actin Lymph node Mammary gland Prostate gland Salivary gland Spleen Thymus MMTV target tissues Fig. S1: MMTV target tissues express ma3.
Supplemental Figure 1
 Supplemental Figure 1 Supplemental Figure 1 (previous page): Heavy chain gene content of ARS/Igh66 transgenic lines. A. Tail DNA from the indicated transgenic lines was amplified for the indicated gene
Supplemental Figure 1 Supplemental Figure 1 (previous page): Heavy chain gene content of ARS/Igh66 transgenic lines. A. Tail DNA from the indicated transgenic lines was amplified for the indicated gene
6/256 1/256 0/256 1/256 2/256 7/256 10/256. At3g06290 (SAC3B)
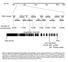 Chr.III M 5M 1M 15M 2M 23M BAC clones F22F7 F1A16 F24F17 F24P17 T8E24 F17A9 F21O3 F17A17 F18C1 F2O1 F28L1 F5E6 F3E22 T1B9 MLP3 Number of recombinants 6/256 1/256 /256 1/256 2/256 7/256 1/256 At3g629 (SAC3B)
Chr.III M 5M 1M 15M 2M 23M BAC clones F22F7 F1A16 F24F17 F24P17 T8E24 F17A9 F21O3 F17A17 F18C1 F2O1 F28L1 F5E6 F3E22 T1B9 MLP3 Number of recombinants 6/256 1/256 /256 1/256 2/256 7/256 1/256 At3g629 (SAC3B)
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
TRANSGENIC ANIMALS. transient. stable. - Two methods to produce transgenic animals:
 Only for teaching purposes - not for reproduction or sale CELL TRANSFECTION transient stable TRANSGENIC ANIMALS - Two methods to produce transgenic animals: 1- DNA microinjection 2- embryonic stem cell-mediated
Only for teaching purposes - not for reproduction or sale CELL TRANSFECTION transient stable TRANSGENIC ANIMALS - Two methods to produce transgenic animals: 1- DNA microinjection 2- embryonic stem cell-mediated
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
GENETICS - CLUTCH CH.15 GENOMES AND GENOMICS.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF GENOMICS Genomics is the study of genomes in their entirety Bioinformatics is the analysis of the information content of genomes - Genes, regulatory sequences,
!! www.clutchprep.com CONCEPT: OVERVIEW OF GENOMICS Genomics is the study of genomes in their entirety Bioinformatics is the analysis of the information content of genomes - Genes, regulatory sequences,
Table S1. List of primers used in this study.
 Gene/Construct/ mutant Primer name Sequence (5 ->3 ) Confimation of T-DNA insertion clh1-1 LBb1 5 GCGTGGACCGCTTGCTGCAACT3 1LP1 5 CCGAAAATGATAAATGCATGG3 1RP1 5 ATGTCCAGCTCGAAAGATTCC3 clh2-1 Lb4 5 CTACAAATTGCCTTTTCTTATCGAC3
Gene/Construct/ mutant Primer name Sequence (5 ->3 ) Confimation of T-DNA insertion clh1-1 LBb1 5 GCGTGGACCGCTTGCTGCAACT3 1LP1 5 CCGAAAATGATAAATGCATGG3 1RP1 5 ATGTCCAGCTCGAAAGATTCC3 clh2-1 Lb4 5 CTACAAATTGCCTTTTCTTATCGAC3
Chapter 8: Recombinant DNA. Ways this technology touches us. Overview. Genetic Engineering
 Chapter 8 Recombinant DNA and Genetic Engineering Genetic manipulation Ways this technology touches us Criminal justice The Justice Project, started by law students to advocate for DNA testing of Death
Chapter 8 Recombinant DNA and Genetic Engineering Genetic manipulation Ways this technology touches us Criminal justice The Justice Project, started by law students to advocate for DNA testing of Death
Characterization of RNA silencing components in the plant
 1 2 3 4 5 6 Supplementary data Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum Yun Chen, Qixun Gao, Mengmeng Huang, Ye Liu, Zunyong Liu, Xin Liu, Zhonghua
1 2 3 4 5 6 Supplementary data Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum Yun Chen, Qixun Gao, Mengmeng Huang, Ye Liu, Zunyong Liu, Xin Liu, Zhonghua
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination.
 Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb
 Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
GENOTYPING BY PCR PROTOCOL FORM MUTANT MOUSE REGIONAL RESOURCE CENTER North America, International
 Please provide the following information required for genetic analysis of your mutant mice. Please fill in form electronically by tabbing through the text fields. The first 2 pages are protected with gray
Please provide the following information required for genetic analysis of your mutant mice. Please fill in form electronically by tabbing through the text fields. The first 2 pages are protected with gray
Technical Review. Real time PCR
 Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Reprogramming of the Genome by Toxic Injury Ken Ramos, MD, PhD, ATS
 Reprogramming of the Genome by Toxic Injury Ken Ramos, MD, PhD, ATS Professor of dicine, University of Arizona Health Sciences Center & Associate Vice President for Precision Health Sciences Office of
Reprogramming of the Genome by Toxic Injury Ken Ramos, MD, PhD, ATS Professor of dicine, University of Arizona Health Sciences Center & Associate Vice President for Precision Health Sciences Office of
Virus-induced gene complementation reveals a transcription factor network in modulation of tomato fruit ripening
 Supplementary Information Virus-induced gene complementation reveals a transcription factor network in modulation of tomato fruit ripening Tao Zhou 2,3, Hang Zhang 2,4, Tongfei Lai 1, Cheng Qin 1, Nongnong
Supplementary Information Virus-induced gene complementation reveals a transcription factor network in modulation of tomato fruit ripening Tao Zhou 2,3, Hang Zhang 2,4, Tongfei Lai 1, Cheng Qin 1, Nongnong
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Genetics and Genomics in Medicine Chapter 3. Questions & Answers
 Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Recombinant DNA Technology
 History of recombinant DNA technology Recombinant DNA Technology (DNA cloning) Majid Mojarrad Recombinant DNA technology is one of the recent advances in biotechnology, which was developed by two scientists
History of recombinant DNA technology Recombinant DNA Technology (DNA cloning) Majid Mojarrad Recombinant DNA technology is one of the recent advances in biotechnology, which was developed by two scientists
SUPPLEMENTARY INFORMATION
 R2A MASNSEKNPLL-SDEKPKSTEENKSS-KPESASGSSTSSAMP---GLNFNAFDFSNMASIL 56 R2B MASSSEKTPLIPSDEKNDTKEESKSTTKPESGSGAPPSPS-PTDPGLDFNAFDFSGMAGIL 60 R2A NDPSIREMAEQIAKDPAFNQLAEQLQRSIPNAGQEGGFPNFDPQQYVNTMQQVMHNPEFK
R2A MASNSEKNPLL-SDEKPKSTEENKSS-KPESASGSSTSSAMP---GLNFNAFDFSNMASIL 56 R2B MASSSEKTPLIPSDEKNDTKEESKSTTKPESGSGAPPSPS-PTDPGLDFNAFDFSGMAGIL 60 R2A NDPSIREMAEQIAKDPAFNQLAEQLQRSIPNAGQEGGFPNFDPQQYVNTMQQVMHNPEFK
A (ng) Protocol. Phenol: 87.1 Kit: 37.7 SPION: Phenol: 23.8 Kit: 14.7 SPION: 6.46
 Table 1. A super paramagnetic iron oxide nanoparticle (SPION) based genomic DNA extraction from soil method was compared to a standard phenol-based method and a commercial kit. Approximately 10 second-stage
Table 1. A super paramagnetic iron oxide nanoparticle (SPION) based genomic DNA extraction from soil method was compared to a standard phenol-based method and a commercial kit. Approximately 10 second-stage
Supplementary Materials: 1. Supplementary Figures S1-S9. 2. Supplementary Tables S1-S2
 Supplementary Materials: 1. Supplementary Figures S1-S9 2. Supplementary Tables S1-S2 S1 1. Supplementary Figures Fig. S1. Genotypes of tcp20 mutants. Fig. S2. Root phenotypes of tcp20 mutants. Fig. S3.
Supplementary Materials: 1. Supplementary Figures S1-S9 2. Supplementary Tables S1-S2 S1 1. Supplementary Figures Fig. S1. Genotypes of tcp20 mutants. Fig. S2. Root phenotypes of tcp20 mutants. Fig. S3.
Generation of App knock-in mice reveals deletion mutations protective against Alzheimer s. disease-like pathology. Nagata et al.
 Generation of App knock-in mice reveals deletion mutations protective against Alzheimer s disease-like pathology Nagata et al. Supplementary Fig 1. Previous App knock-in model did not show Aβ accumulation
Generation of App knock-in mice reveals deletion mutations protective against Alzheimer s disease-like pathology Nagata et al. Supplementary Fig 1. Previous App knock-in model did not show Aβ accumulation
Easi CRISPR for conditional and insertional alleles
 Easi CRISPR for conditional and insertional alleles C.B Gurumurthy, University Of Nebraska Medical Center Omaha, NE cgurumurthy@unmc.edu Types of Genome edits Gene disruption/inactivation Types of Genome
Easi CRISPR for conditional and insertional alleles C.B Gurumurthy, University Of Nebraska Medical Center Omaha, NE cgurumurthy@unmc.edu Types of Genome edits Gene disruption/inactivation Types of Genome
PCR analysis was performed to show the presence and the integrity of the var1csa and var-
 Supplementary information: Methods: Table S1: Primer Name Nucleotide sequence (5-3 ) DBL3-F tcc ccg cgg agt gaa aca tca tgt gac tg DBL3-R gac tag ttt ctt tca ata aat cac tcg c DBL5-F cgc cct agg tgc ttc
Supplementary information: Methods: Table S1: Primer Name Nucleotide sequence (5-3 ) DBL3-F tcc ccg cgg agt gaa aca tca tgt gac tg DBL3-R gac tag ttt ctt tca ata aat cac tcg c DBL5-F cgc cct agg tgc ttc
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast. Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice
 A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
Supplemental Figure legends Figure S1. (A) (B) (C) (D) Figure S2. Figure S3. (A-E) Figure S4. Figure S5. (A, C, E, G, I) (B, D, F, H, Figure S6.
 Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supporting Information
 Supporting Information Deng et al. 10.1073/pnas.1102117108 Fig. S1. Predicted structure of Arabidopsis bzip60 RNA. Lowest free energy form (ΔG = 309.72 (initially 343.10) of bzip60 mrna folded by M-Fold
Supporting Information Deng et al. 10.1073/pnas.1102117108 Fig. S1. Predicted structure of Arabidopsis bzip60 RNA. Lowest free energy form (ΔG = 309.72 (initially 343.10) of bzip60 mrna folded by M-Fold
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplemental Data. Benstein et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Supporting Information
 Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
PROGRESS REPORT. igem June 21, 2010
 PROGRESS REPORT igem June 21, 2010 TEAM VECTOR Aim: biobrick agrobacterium vector We are working with 6 different plasmids known as pore: O1 and O2- backbone + multiple cloning site (MCS) E3 and E4- backbone
PROGRESS REPORT igem June 21, 2010 TEAM VECTOR Aim: biobrick agrobacterium vector We are working with 6 different plasmids known as pore: O1 and O2- backbone + multiple cloning site (MCS) E3 and E4- backbone
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kawai and Amano, http://www.jcb.org/cgi/content/full/jcb.201110008/dc1 Figure S1. Regulation of mirna expression by BRCA1. (A) Confirmation
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kawai and Amano, http://www.jcb.org/cgi/content/full/jcb.201110008/dc1 Figure S1. Regulation of mirna expression by BRCA1. (A) Confirmation
Edexcel (B) Biology A-level
 Edexcel (B) Biology A-level Topic 7: Modern Genetics Notes Using Gene Sequencing Genome = all of an organism s DNA, including mitochondrial/chloroplast DNA. Polymerase chain reaction (PCR) is used to amplify
Edexcel (B) Biology A-level Topic 7: Modern Genetics Notes Using Gene Sequencing Genome = all of an organism s DNA, including mitochondrial/chloroplast DNA. Polymerase chain reaction (PCR) is used to amplify
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplementary Information. c d e
 Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
ABI3 Controls Embryo De-greening Through Mendel's I locus
 Supporting Online Material for ABI3 Controls Embryo De-greening Through Mendel's I locus Frédéric Delmas a,b,c, Subramanian Sankaranarayanan d, Srijani Deb d, Ellen Widdup d, Céline Bournonville b,c,norbert
Supporting Online Material for ABI3 Controls Embryo De-greening Through Mendel's I locus Frédéric Delmas a,b,c, Subramanian Sankaranarayanan d, Srijani Deb d, Ellen Widdup d, Céline Bournonville b,c,norbert
SUPPLEMENTARY INFORMATION
 Materials and Methods Transgenic Plant Materials and DNA Constructs. VEX1::H2B-GFP, ACA3::H2B-GFP, KRP6::H2B-GFP, KRP6::mock21ts-GFP, KRP6::TE21ts- GFP and KRP6::miR161ts-GFP constructs were generated
Materials and Methods Transgenic Plant Materials and DNA Constructs. VEX1::H2B-GFP, ACA3::H2B-GFP, KRP6::H2B-GFP, KRP6::mock21ts-GFP, KRP6::TE21ts- GFP and KRP6::miR161ts-GFP constructs were generated
LS4 final exam. Problem based, similar in style and length to the midterm. Articles: just the information covered in class
 LS4 final exam Problem based, similar in style and length to the midterm Articles: just the information covered in class Complementation and recombination rii and others Neurospora haploid spores, heterokaryon,
LS4 final exam Problem based, similar in style and length to the midterm Articles: just the information covered in class Complementation and recombination rii and others Neurospora haploid spores, heterokaryon,
pdsipher and pdsipher -GFP shrna Vector User s Guide
 pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and
 1 Supplemental information 2 3 Materials and Methods 4 Reagents and animals 5 8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and 6 Silencer Select Pre-designed sirna
1 Supplemental information 2 3 Materials and Methods 4 Reagents and animals 5 8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and 6 Silencer Select Pre-designed sirna
PLNT2530 (2018) Unit 6b Sequence Libraries
 PLNT2530 (2018) Unit 6b Sequence Libraries Molecular Biotechnology (Ch 4) Analysis of Genes and Genomes (Ch 5) Unless otherwise cited or referenced, all content of this presenataion is licensed under the
PLNT2530 (2018) Unit 6b Sequence Libraries Molecular Biotechnology (Ch 4) Analysis of Genes and Genomes (Ch 5) Unless otherwise cited or referenced, all content of this presenataion is licensed under the
Supplemental Data. Guo et al. (2015). Plant Cell /tpc
 Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Lecture 22: Molecular techniques DNA cloning and DNA libraries
 Lecture 22: Molecular techniques DNA cloning and DNA libraries DNA cloning: general strategy -> to prepare large quantities of identical DNA Vector + DNA fragment Recombinant DNA (any piece of DNA derived
Lecture 22: Molecular techniques DNA cloning and DNA libraries DNA cloning: general strategy -> to prepare large quantities of identical DNA Vector + DNA fragment Recombinant DNA (any piece of DNA derived
