SUPPLEMENTAL METHODS Reverse transcriptase PCR (RT-PCR) and quantitative real-time PCR (Q-RT-PCR) RNA was isolated with TRIZOL (Invitrogen), and
|
|
|
- Doris Stephanie Cole
- 6 years ago
- Views:
Transcription
1 SUPPLEMENTAL METHODS Reverse transcriptase PCR (RT-PCR) and quantitative real-time PCR (Q-RT-PCR) RNA was isolated with TRIZOL (Invitrogen), and first strand cdna was synthesized using 1 μg of RNA and SuperScript III Reverse Transcriptase (Invitrogen). RT-PCR was performed using primer-specific annealing temperature. Q-RT-PCR was performed with SYBR Green PCR reagents on an ABI Prism 7900 detection system (Applied Biosystems, Foster City, CA). RNA levels were normalized to the level of β-actin or GAPDH and calculated as delta-delta threshold cycle (ΔΔCT). The detailed procedures of RT-PCR and Q-RT-PCR were followed as described previously (1). Primers used for both RT-PCR and Q-RT-PCR are listed in supplementary Table 1. All Q-RT-PCR reactions were performed in triplicate. Lentivirus-mediated short hairpin RNA knockdown and ectopic gene overexpression Lentiviral plasmids (PLKO.1) containing different short hairpin RNAs (shrnas) driven by the U6 promoter were obtained from the National RNAi Core Facility (Taipei, Taiwan, ( Mouse hepatoma cell line Hepar 1-6 (ATCC, Manassas, VA, which expresses EpCAM, Cldn7, and Cd9, was used to assay the knockdown efficiency of different clones using transient transfection. shrna clones with the best knockdown effect (clone TRCN for mouse EpCAM; clone TRCN for mouse Cldn7, and clone TRCN for mouse Cd9) were selected for subsequent lentivirus production using 293T cells; lentiviruses were harvested at 48 and 72 hours post-transfection. MEFs and Hepar 1-6 cells were infected by lentiviruses using 8 μg/ml polybrene (Sigma-Aldrich) according to the protocol provided by the National RNAi Core Facility. Q-RT-PCR was performed in Hepar 1-6 cells 72 hours after infection to evaluate the knockdown effect. Inducible lentiviral vectors overexpressing mouse or human EpCAM, Cldn7 (CLDN7), and EpICD cdnas were obtained by subcloning these cdnas into the TetO-FUW vector using an EcoRI site. 293T cells were used to produce viruses as described above. Co-immunoprecipitation and Western blotting Collected pluripotent cells were extracted for one hour in ice-cold CSK lysis buffer [50 mm NaCl, 300 mm Sucrose, 10 mm Pipes, ph 6.8, 3 mm MgCl2 and 0.5% (v/v) Triton X-100] containing protease inhibitor cocktail (Sigma-Aldrich) at 4 C. After centrifugation (10 min, 20,000 g), supernatant was pre-cleared by incubation with 1:10 volume Protein G-Sepharose (GE Healthcare, UK, Pre-cleared supernatants were incubated for 16 hours at 4 C with 2 g of selected antibody or control IgG. The following day, Protein G-Sepharose was added for 2 more hours to pull down the immuno-complexes. The pull-down complex was washed three times with CSK buffer. Immunoprecipitated proteins were analyzed by SDS-PAGE, followed by Western 1
2 blotting (WB) as described previously (2). The antibodies used in Co-immunoprecipitation and WB are listed in Supplementary Table 2. Cloning of the OCT4 and Oct4 promoters and Luciferase reporter assay Human OCT4 promoter-containing plasmid was a kind gift from Dr. Wei Cui, (Imperial College London, UK). The ~3- kb (-2951 ~ +26) mouse Oct4 promoter fragment was PCR-amplified using Pfu Turbo DNA polymerase (Agilent, Santa Clara, CA) and a pair of primers (Table S6) from the genomic DNA and was sequenced to verify it. Both human and mouse Oct4 promoters were subcloned into pgl3-basic luciferase reporter plasmid using MluI and XhoI sites. One day prior to transfection, 293T cells were seeded in 6-well plates at 80,000 cells per well. Inducible lentivirus plasmids that contained EpCAM or EpICD cdnas (3 μg for each) were co-transfected with M2rtTA vector, reporter plasmids, and the CMV-β-galactosidase plasmid (as a control for transfection efficiency) into cells using the calcium phosphate precipitation method. After transfection, cells were washed with phosphate-buffered saline and re-fed with the fresh medium containing 2 μg/ml of doxycycline for an additional 32 hours. Cells were then harvested and analyzed for luciferase and β-galactosidase activities in accordance with the manufacturer s instructions for the Luciferase Assay and β-galactosidase Enzyme System (Promega, Madison, WI). REFERENCES 1. Chen, H. F., Chuang, C. Y., Shieh, Y. K., Chang, H. W., Ho, H. N., and Kuo, H. C. (2009) Hum Reprod 24, Huang, H. P., Yu, C. Y., Chen, H. F., Chen, P. H., Chuang, C. Y., Lin, S. J., Huang, S. T., Chan, W. H., Ueng, T. H., Ho, H. N., and Kuo, H. C. (2010) J Biol Chem 285,
3 SUPPLEMENTAL DATA Supplemental Tables Table S1: Summary of the expression patterns of EpCAM complex proteins in mescs, hescs, mipscs, hipscs and somatic cells EpCAM CLDN7 (Cldn7) mescs Mem. Mem. hescs Mem. Mem. mipscs Mem. Mem. hipscs Mem. Mem. MEFs none none HFs none none Mem., membrane, Cyto., cytoplasm CD9 (Cd9) ADAM 17 (Adam17) Mem. Mem. & Cyto. Mem. Mem. & Cyto. Mem. Mem. & Cyto. Mem. Mem. &Cyto. none Cyto. none Cyto. PSEN2 (Psen2) Mem. Mem. Mem. Mem. Cyto. Cyto. 3
4 Table S2: Summary of the correlation of RNA expression levels of mouse EpCAM complex protein-encoding genes and pluripotency genes in mipscs Nanog Oct4 Sox2 Klf4 c-myc EpCAM Cldn7 Cd9 Adam 17 Psen2 Nanog ρ P Oct4 ρ P Sox2 ρ P Klf4 ρ P c-myc ρ P EpCAM ρ P Cldn7 ρ P Cd9 ρ P Adam 17 ρ P Psen2 ρ P ρ, Pearson s correlation coefficient; bold characters, P values less than 0.05, RNAs were from 17 mouse pluripotent stem cell lines (1 mouse ESC and 16 mipscs) 4
5 Table S3: Summary of the correlation of RNA expression levels of human EpCAM complex protein-encoding genes and pluripotency genes in hipscs NANOG OCT4 SOX2 KLF4 MYC EpCAM CLDN7 CD9 ADAM17 PSEN2 NANOG ρ P OCT4 ρ P SOX2 ρ P KLF4 ρ P MYC ρ P EpCAM ρ P CLDN7 ρ P CD9 ρ P ADAM17 ρ P PSEN2 ρ P ρ, Pearson s correlation coefficient; bold characters, P values less than 0.05; RNAs were from 9 hipscs 5
6 Table S4 Percentage of clones with higher levels of Nanog (> 1 fold of that in mescs) and expression in mipsc clones generated by over-expressing EpCAM, Cldn7, EpCAM plus Cldn7, or EpICD, and OSKM. ipsc clones generated with EpCAM complex protein No. of clones tested No. of clones with Nanog > 1 mescs (%) Control ipsc (OSKM only) 9 4 (44.4) ipsc-ep 9 6 (66.7) ipsc-cl (30.0) ipsc-epcl (25.0) ipsc-epi 13 7 (53.7) 6
7 Table S5 Antibodies used for IF, immunoprecipitation, and western blotting Antigen Type Company Dilution Human and mouse EpCAM Human EpCAM Human EpCAM Mouse EpCAM Human CLDN7 Human CD9 Human PSEN2 and mouse Psen2 Human ADAM17 and mouse Adam17 Human OCT4 and mouse Oct4 Human SOX2 and mouse Sox2 Human NANOG Human SSEA-4 Human TRA1-60 Mouse Nanog Mouse Cldn7 Mouse Cd9 Mouse SSEA-1 Mouse MAP2 Mouse alpha-actinin Mouse mucin 5 mouse, monoclonal rabbit, monoclonal goat, polyclonal rat, monoclonal rabbit, monoclonal mouse, monoclonal rabbit, monoclonal rabbit, polyclonal mouse, monoclonal mouse goat, polyclonal mouse, monoclonal mouse, monoclonal goat, polyclonal rabbit, polyclonal rat, monoclonal mouse, monoclonal rabbit, polyclonal mouse, monoclonal mouse, monoclonal GeneTex Abcam R&D ebiosciences Novus Biologicals Abcam GeneTex GeneTex Santa Cruz R & D R & D Chemicon Chemicon Abcam Novus Biologicals ebioscience Chemicon Millipore Sigma Sigma 2 μg for each IP 1:1000 for WB 1:500 for IF 1:200 for IF 1:1000for WB 1: 200 for IF 1:1000 for WB 1:50 for IF 1:1000 for WB 1: 100 for IF 1:1000 for WB 1:100 for IF 1:200 for IF 1:200 for IF 1:200 for IF 1:50 for IF 1:50 for IF 1:200 for IF 1:200 for IF 1:100 for IF 1:200 for IF 1:800 for IF 1:800 for IF 1:200 for IF IP, immunoprecipitation; WB, western blot; IF, immunofluorescence 7
8 Table S6: Primer sets used for RT-PCR, QRT-PCR, and PCR cloning Name of Genes mgapdh mnanog moct4 msox2 mklf4 mc-myc mepcam mcldn7 mcd9 mpsen2 madam 17 mcd44v6 mtspan8 mp53 mp21 mpcna mki67 hactb hnanog hoct4 hsox2 hklf4 hmyc hepcam hepicd Forward primer (5-3 ) Backward primer (5-3 ) ACCACAGTCCATGCCATCAC TCCACCACCCTGTTGCTGTA AGGGTCTGCTACTGAGATGCTCTG CAACCACTGGTTTTTCTGCCACCG TCTTTCCACCAGGCCCCCGGCTC TGCGGGCGGACATGGGGAGATCC TAGAGCTAGACTCCGGGCGATGA TTGCCTTAAACAAGACCACGAAA GCGAACTCACACAGGCGAGAAACC TCGCTTCCTCTTCCTCCGACACA TGACCTAACTCGAGGAGGAGCTGG AAGTTTGAGGCAGTTAAAATTATG AATC GCTGAAGC TTGCTCCAAACTGGCGTCTA ACGTGATCTCCGTGTCCTTGT CGGCATGATGAGCTGCAAA CCACAATGAAAACAATGCCTCC CTGGCATTGCAGTGCTTGCTA AACCCGAAGAACAATCCCAGC CTCAGCAAGCAAGCGTCTCTTC TCCCAGCAGTCACTGCAGAAAT CAGAAGAAGTGCCAGCAGGCTA GTTGTCAGTGTCAACGCATGC GCACCCCAGAAAGCTACATTTT CCCTTCGTCACATGGGAGTCTTC GCTTCTGTCGGACAACACTG ATTCTCCAAGCCACAGCACT AAAGAGAGCGCTGCCCACCT CTCCCGGAACATCTCGAAGC ACGTGGCCTTGTCGCTGTCT GACCAATCTGCGCTTGGAGTG TGCTCTGAGGTACCTGAACT TGCTTCCTCATCTTCAATCT CCTTTGCTGTCCCCGAAGA GGCTTCTCATCTGTTGCTTCCT GAGCACAGAGCCTCGCCTTT ACATGCCGGAGCCGTTGTC AGTCCCAAAGGCAAACAACCCACT TGCTGGAGGCTGAGGTATTTCTGTC TC TC GACAGGGGGAGGGGAGGAGCTAGG CTTCCCTCCAACCAGTTGCCCCAA AC GGGAAATGGGAGGGGTGCAAAAGA TTGCGTGAGTGTGGATGGGATTGG GG TG ACGATCGTGGCCCCGGAAAAGGAC CAACAACCGAAAATGCACCAGCCC C CAG GCGTCCTGGGAAGGGAGATCCGGA TTGAGGGGCATCGTCGCGGGAGGC GC TG AGAACCTACTGGATCATCATTGAAC CGCGTTGTGATCTCCTTCTG TAA TGGGTGAGATGCATAGGGAAC GGCATTAAAGCAGCGTATCCACA 8
9 h CLDN7 hcd9 hadam17 hpsen2 htspan8 hcd44v6 FUW-TetO FUW-TetO-O SKM FUW-TetO-E pcam FUW-TetO-Cl dn7 FUW-TetO-E picd Mouse Oct4 promoter AGGCATAATTTTCATCGTGG ATGATGCTGGTGGGCTTC GTCGTGGTGGTGGATGGTAAAA GACGTCCCTAATGTCGGCTGAG GATTGCTGTAGGTGCCATCA AGTACAACGGAAGAAACAGC AGCTCGTTTAGTGAACCGTCAGATC CAGAGGAGGAACGAGCTGAAGCGC TTGCTCCAAACTGGCGTCTA CGGCATGATGAGCTGCAAA AGCTCGTTTAGTGAACCGTCAGATC AAATACAGGTGGTTTGTGGC GAGTTGGACTTAGGGTAAGAGCG GCTCATCCTTGGTTTTCAGC GCCCCATCTGTGTTGATTCTGA ACATAGCGGTCAGGGTCCTCCT TTTTTCACTTTCCCCTGTGG TTGGGTTGAAGAAATCAGTC CATAGCGTAAAAGGAGCAACA CATAGCGTAAAAGGAGCAACA CATAGCGTAAAAGGAGCAACA CATAGCGTAAAAGGAGCAACA CATAGCGTAAAAGGAGCAACA GGAAAGACGGCTCACCTA 9
10 SUPPLEMENTAL FIGURE LEGENDS Figure S1 Derivation and characterization of mipscs. mipsc clones (No. 2, 4, 5, 6, 8, 10, 11) were generated by lentiviruses carrying tetracycline-inducible OSKM genes. (A) RT-PCR analysis showed the reactivation of endogenous pluripotency genes (upper panel); genomic PCR showed positive integration of exogenous OSKM transgenes (bottom panel). (B) Phase contrast images of a mipsc colony and an AP-expressing ipsc colony (PC, phase contrast; AP, alkaline phosphatase). Confocal microscopy showed that the representative mipsc clones expressed mesc-related markers, Nanog, Oct4, Sox2, and SSEA-1. Cell nuclei were stained with DAPI. Scale bar, 30 μm. (C) Chimera mice (indicated by white arrow head) derived from mouse inducible ipsc clone No. 6. (D) Q-RT-PCR showed that the genes encoding EpCAM complex proteins were expressed in both mipsc clones (as in Fig. 2A), but not in MEF, with the levels comparable to those of mescs. The RNA level of each gene was normalized to that of mescs. Data are the mean of 3 independent assays ± SD. RT-N, reverse transcriptase-negative; N.D., not detectable. Figure S2 Characterization of mipsc clones reprogrammed from MEFs using the retrovirus system. (A) RT-PCR showed the expression of endogenous pluripotency genes in most mipsc clones; the expression of exogenous Oct4, Sox2, Klf4, and c-myc was either silenced or weak. RNAs isolated from 9 newly derived mipsc clones, mescs, MEFs, and mipsc clones (ips-mef-ng-20d-17, a gift from Dr. Shinya Yamanaka). (B) PCR analysis (right panel) of retroviral integration and transgenic expression using genomic DNA isolated from putative mipsc clones with transgene-specific primers. Figure S3 Derivation and characterization of hipscs. (A) RT-PCR (left panel) showed reactivation of endogenous (endo) pluripotency genes and silencing of exogenous (exo) pluripotency genes in 5 hipsc clones derived from foreskin fibroblasts (ipsc-cfb-54 and ips-c3) and granulosa cells (igra-01-03). PCR analysis (right panel) of retroviral integration and transgenic expression using genomic DNA isolated from putative hipsc clones with transgene-specific primers. (B) A phase-contrast image (upper left panel) and AP staining (upper middle panel) of the colony from a representative hipsc clone, and immunostaining of hipscs with antibodies against human ESC-related markers including OCT4, NANOG, SSEA4, and TRA Cell nuclei were stained with DAPI. Scale bar, 100 μm. (C) H&E staining of the teratoma from the in vivo differentiation of a representative hipsc clone in NOD/SCID mice showed differentiated derivatives of ectodermal, 10
11 mesodermal and endodermal lineages. (D) Q-RT-PCR analysis showed that the genes encoding EpCAM complex proteins were expressed in all four hipsc clones (as in Fig. 2A), but not in HFs, with the levels comparable to those of hescs. The RNA level of each gene was normalized to that of hesc line H9. Data are the mean of 3 independent assays ± SD. RT-N, reverse transcriptase-negative; N.D., not detectable Figure S4 Confirmation of the expression of transgenes in MEFs transduced with lentiviral vectors encoding EpCAM complex proteins. (A) PCR analysis of lentiviral integration using genomic DNA isolated from MEFs transduced with lentiviral vectors encoding EpCAM complex protein genes with transgene-specific primers. (B) RT-PCR analysis of the expression of transgenes using cdnas from MEFs transduced with lentiviral vectors encoding EpCAM complex protein genes with transgene-specific primers. Overexpression of transcripts of EpCAM, Cldn7 and EpICD, was confirmed. (C) Western blotting confirmed the overexpression of EpCAM complex proteins. Ctrl., control Figure S5 Characterization of EpCAM complex protein-mediated mipscs. (A) Q-RT-PCR on cdnas derived from all EpCAM complex protein-mediated mipsc clones (Fig. 6A) showed expression of endogenous Nanog, Oct4, Sox2, Klf4, c-myc, and the five genes encoding EpCAM complex proteins, with the levels comparable to those of mescs. The RNA level of each genes was normalized to vector control. Data are the mean of 3 independent assays ± SD. RT-N, reverse transcriptase-negative; N.D., not detectable. (B) Immunofluorescence analysis of teratomas derived from representative EpCAM complex protein-mediated mipsc clones (Fig. 6) showed the differentiation of MAP2-positive neural epithelial cells, alpha-actinin-positive mesodermal cells, and mucin 5-positive endodermal cells. Nuclei were stained with DAPI. Scale bar, 50 μm 11
12 Suppl. Fig. S1 D
13 Suppl. Fig. S2
14 Suppl. Fig. S3 D
15 Suppl. Fig. S4
16 Suppl. Fig. S5 A B
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplemental Methods Cell lines and culture
 Supplemental Methods Cell lines and culture AGS, CL5, BT549, and SKBR were propagated in RPMI 64 medium (Mediatech Inc., Manassas, VA) supplemented with % fetal bovine serum (FBS, Atlanta Biologicals,
Supplemental Methods Cell lines and culture AGS, CL5, BT549, and SKBR were propagated in RPMI 64 medium (Mediatech Inc., Manassas, VA) supplemented with % fetal bovine serum (FBS, Atlanta Biologicals,
Isolation of human ips cells using EOS lentiviral vectors to select for pluripotency
 nature methods Isolation of human ips cells using EOS lentiviral vectors to select for pluripotency Akitsu Hotta, Aaron Y L Cheung, Natalie Farra, Kausalia Vijayaragavan, Cheryle A Séguin, Jonathan S Draper,
nature methods Isolation of human ips cells using EOS lentiviral vectors to select for pluripotency Akitsu Hotta, Aaron Y L Cheung, Natalie Farra, Kausalia Vijayaragavan, Cheryle A Séguin, Jonathan S Draper,
RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the
 Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Data. Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells
 Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
SUPPLEMENTARY INFORMATION
 Supplementary Figure S1 Colony-forming efficiencies using transposition of inducible mouse factors. (a) Morphologically distinct growth foci appeared from rtta-mefs 6-8 days following PB-TET-mFx cocktail
Supplementary Figure S1 Colony-forming efficiencies using transposition of inducible mouse factors. (a) Morphologically distinct growth foci appeared from rtta-mefs 6-8 days following PB-TET-mFx cocktail
A Novel Platform to Enable the High-Throughput Derivation and Characterization of Feeder-Free Human ipscs
 Supplementary Information A Novel Platform to Enable the High-Throughput Derivation and Characterization of Feeder-Free Human ipscs Bahram Valamehr, Ramzey Abujarour, Megan Robinson, Thuy Le, David Robbins,
Supplementary Information A Novel Platform to Enable the High-Throughput Derivation and Characterization of Feeder-Free Human ipscs Bahram Valamehr, Ramzey Abujarour, Megan Robinson, Thuy Le, David Robbins,
SUPPLEMENTARY INFORMATION
 doi:.38/nature08267 Figure S: Karyotype analysis of ips cell line One hundred individual chromosomal spreads were counted for each of the ips samples. Shown here is an spread at passage 5, predominantly
doi:.38/nature08267 Figure S: Karyotype analysis of ips cell line One hundred individual chromosomal spreads were counted for each of the ips samples. Shown here is an spread at passage 5, predominantly
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2239 Hepatocytes (Endoderm) Foreskin fibroblasts (Mesoderm) Melanocytes (Ectoderm) +Dox rtta TetO CMV O,S,M,K & N H1 hes H7 hes H9 hes H1 hes-derived EBs H7 hes-derived EBs H9 hes-derived
DOI: 10.1038/ncb2239 Hepatocytes (Endoderm) Foreskin fibroblasts (Mesoderm) Melanocytes (Ectoderm) +Dox rtta TetO CMV O,S,M,K & N H1 hes H7 hes H9 hes H1 hes-derived EBs H7 hes-derived EBs H9 hes-derived
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like
 Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like morphology in co-culture with mouse embryonic feeder fibroblasts
Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like morphology in co-culture with mouse embryonic feeder fibroblasts
Protocol Using the Reprogramming Ecotropic Retrovirus Set: Mouse OSKM to Reprogram MEFs into ips Cells
 STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblast (MEF) cells into induced pluripotent stem (ips) cells in a 6-well format. Transduction
STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblast (MEF) cells into induced pluripotent stem (ips) cells in a 6-well format. Transduction
Supplementary Figure 1.
 Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State
 Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Chemically defined conditions for human ipsc derivation and culture
 Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
The p53-puma axis suppresses ipsc generation
 The p53-puma axis suppresses ipsc generation Yanxin Li, Haizhong Feng, Haihui Gu, Dale W. Lewis, Youzhong Yuan, Lei Zhang, Hui Yu, Peng Zhang, Haizi Cheng, Weimin Miao, Weiping Yuan, Shi-Yuan Cheng, Susanne
The p53-puma axis suppresses ipsc generation Yanxin Li, Haizhong Feng, Haihui Gu, Dale W. Lewis, Youzhong Yuan, Lei Zhang, Hui Yu, Peng Zhang, Haizi Cheng, Weimin Miao, Weiping Yuan, Shi-Yuan Cheng, Susanne
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Supplementary Information: Materials and Methods. Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered
 Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplemental Information
 Cell Stem Cell, Volume 15 Supplemental Information Generation of Naive Induced Pluripotent Stem Cells from Rhesus Monkey Fibroblasts Riguo Fang, Kang Liu, Yang Zhao, Haibo Li, Dicong Zhu, Yuanyuan Du,
Cell Stem Cell, Volume 15 Supplemental Information Generation of Naive Induced Pluripotent Stem Cells from Rhesus Monkey Fibroblasts Riguo Fang, Kang Liu, Yang Zhao, Haibo Li, Dicong Zhu, Yuanyuan Du,
Protocol Reprogramming MEFs using the Dox Inducible Reprogramming Lentivirus Set: Mouse OKSM
 STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblasts (MEFs) into induced pluripotent stem (ips) cells in a 6-well format. Transduction
STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblasts (MEFs) into induced pluripotent stem (ips) cells in a 6-well format. Transduction
Supplementary Material
 Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
SUPPLEMENTARY INFORMATION
 The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
Naked Mole Rat Induced Pluripotent Stem Cells and Their Contribution
 Stem Cell eports, Volume 9 Supplemental Information Naked Mole at Induced Pluripotent Stem Cells and Their Contribution to Interspecific Chimera Sang-Goo Lee, Aleksei E. Mikhalchenko, Sun Hee Yim, Alexei
Stem Cell eports, Volume 9 Supplemental Information Naked Mole at Induced Pluripotent Stem Cells and Their Contribution to Interspecific Chimera Sang-Goo Lee, Aleksei E. Mikhalchenko, Sun Hee Yim, Alexei
LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS.
 Supplemental Data: LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS. Scott Jepson, Bryan Vought, Christian H.
Supplemental Data: LINGO-1, A TRANSMEMBRANE SIGNALING PROTEIN, INHIBITS OLIGODENDROCYTE DIFFERENTIATION AND MYELINATION THROUGH INTERCELLULAR SELF- INTERACTIONS. Scott Jepson, Bryan Vought, Christian H.
Supplemental Information
 Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Document S1. Supplemental Experimental Procedures and Three Figures (see next page)
 Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
hipscs were derived from human skin fibroblasts (CRL-2097) by ectopic
 Generation and characterization of hipscs hipscs were derived from human skin fibroblasts (CRL-2097) by ectopic expression of OCT4, SOX2, KLF4, and C-MYC as previously described 1. hipscs showed typical
Generation and characterization of hipscs hipscs were derived from human skin fibroblasts (CRL-2097) by ectopic expression of OCT4, SOX2, KLF4, and C-MYC as previously described 1. hipscs showed typical
Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila
 Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Premade Lentiviral Particles for ips Stem Factors: Single vector system. For generation of induced pluripotent stem (ips) cells and other applications
 Premade Lentiviral Particles for ips Stem Factors: Single vector system For generation of induced pluripotent stem (ips) cells and other applications RESEARCH USE ONLY, not for diagnostics or therapeutics
Premade Lentiviral Particles for ips Stem Factors: Single vector system For generation of induced pluripotent stem (ips) cells and other applications RESEARCH USE ONLY, not for diagnostics or therapeutics
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb37 a mrna relative expression 1.3 1..8.5.3. CTR NGPS OCT4 SOX2 KLF4 c-myc b BANF1 mrna levels 1.2 1..8.6.4.2. plko.1 shbanf1 c Tra-1-6+ colonies 3 1 plko.1 shbanf1 d BANF1 locus Plasmid donor
DOI: 1.138/ncb37 a mrna relative expression 1.3 1..8.5.3. CTR NGPS OCT4 SOX2 KLF4 c-myc b BANF1 mrna levels 1.2 1..8.6.4.2. plko.1 shbanf1 c Tra-1-6+ colonies 3 1 plko.1 shbanf1 d BANF1 locus Plasmid donor
Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and
 SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Protocol Reprogramming Human Fibroblasts into ips Cells using the Stemgent Reprogramming Lentivirus Set: Human OKSM
 STEMGENT Page 1 Reprogramming Lentivirus Set: Human OKSM OVERVIEW The following procedure describes the reprogramming of BJ Human Fibroblasts (BJ cells) to induced pluripotent stem (ips) cells using the
STEMGENT Page 1 Reprogramming Lentivirus Set: Human OKSM OVERVIEW The following procedure describes the reprogramming of BJ Human Fibroblasts (BJ cells) to induced pluripotent stem (ips) cells using the
Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice
 Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice Supplementary Material Supplementary Methods Materials Spironolactone, aldosterone and β-glycerophosphate were
Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice Supplementary Material Supplementary Methods Materials Spironolactone, aldosterone and β-glycerophosphate were
Fig. S1 TGF RI inhibitor SB effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of
 Fig. S1 TGF RI inhibitor SB525334 effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of different concentrations of SB525334. Cells were lysed and
Fig. S1 TGF RI inhibitor SB525334 effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of different concentrations of SB525334. Cells were lysed and
Cell extracts and western blotting RNA isolation and real-time PCR Chromatin immunoprecipitation (ChIP)
 Cell extracts and western blotting Cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed with lysis buffer. 1 Total cell extracts were separated by SDS-PAGE and transferred to nitrocellulose
Cell extracts and western blotting Cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed with lysis buffer. 1 Total cell extracts were separated by SDS-PAGE and transferred to nitrocellulose
Biology Open (2015): doi: /bio : Supplementary information
 Fig. S1. Verification of optimal conditions for the generation of LIF-dependent inscs Representative images (from five repeat experiments) of human primary fibroblasts undergoing reprogramming for 21 days
Fig. S1. Verification of optimal conditions for the generation of LIF-dependent inscs Representative images (from five repeat experiments) of human primary fibroblasts undergoing reprogramming for 21 days
Protocol Reprogramming Human Fibriblasts using the Dox Inducible Reprogramming Polycistronic Lentivirus Set: Human 4F2A LoxP
 STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of BJ Human Fibroblasts (BJ cells) into induced pluripotent stem (ips) cells in a 6-well format. Transduction efficiency
STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of BJ Human Fibroblasts (BJ cells) into induced pluripotent stem (ips) cells in a 6-well format. Transduction efficiency
Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction
 Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
T-iPSC. Gra-iPSC. B-iPSC. TTF-iPSC. Supplementary Figure 1. Nature Biotechnology: doi: /nbt.1667
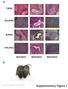 a T-iPSC Gra-iPSC B-iPSC TTF-iPSC Ectoderm Endoderm Mesoderm b Nature Biotechnology: doi:.38/nbt.667 Supplementary Figure Klf4 transgene expression Oct4 transgene expression.7 Fold GAPDH.6.5.4.3 Fold GAPDH
a T-iPSC Gra-iPSC B-iPSC TTF-iPSC Ectoderm Endoderm Mesoderm b Nature Biotechnology: doi:.38/nbt.667 Supplementary Figure Klf4 transgene expression Oct4 transgene expression.7 Fold GAPDH.6.5.4.3 Fold GAPDH
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1 - Characterization of rbag3 binding on macrophages cell surface.
 Supplementary Figure 1 - Characterization of rbag3 binding on macrophages cell surface. (a) Human PDAC cell lines were treated as indicated in Figure 1 panel F. Cells were analyzed for FITC-rBAG3 binding
Supplementary Figure 1 - Characterization of rbag3 binding on macrophages cell surface. (a) Human PDAC cell lines were treated as indicated in Figure 1 panel F. Cells were analyzed for FITC-rBAG3 binding
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Gene Forward Primer Reverse Primer GAPDH ATCATCCCTGCCTCTACTGG GTCAGGTCCACCACTGACAC SSB1 AACTTCAGTGAGCCAAACCC GTTCTCAGAGGCTGGAGAGG
 Supplemental Data EXPERIMENTAL PROCEDURES Plasmids and Antibodies- Full length cdna of INT11 or INT12 were cloned into ps- Flag-SBP vector respectively. Anti-RNA pol II (RPB1) was purchased from Santa
Supplemental Data EXPERIMENTAL PROCEDURES Plasmids and Antibodies- Full length cdna of INT11 or INT12 were cloned into ps- Flag-SBP vector respectively. Anti-RNA pol II (RPB1) was purchased from Santa
Jing Liao, Chun Cui, Siye Chen, Jiangtao Ren, Jijun Chen, Yuan Gao, Hui Li, Nannan Jia, Lu Cheng, Huasheng Xiao, and Lei Xiao
 Cell Stem Cell, Volume 4 Supplemental Data Generation of Induced Pluripotent Stem Cell Lines from Adult Rat Cells Jing Liao, Chun Cui, Siye Chen, Jiangtao Ren, Jijun Chen, Yuan Gao, Hui Li, Nannan Jia,
Cell Stem Cell, Volume 4 Supplemental Data Generation of Induced Pluripotent Stem Cell Lines from Adult Rat Cells Jing Liao, Chun Cui, Siye Chen, Jiangtao Ren, Jijun Chen, Yuan Gao, Hui Li, Nannan Jia,
gacgacgaggagaccaccgctttg aggcacattgaaggtctcaaacatg
 Supplementary information Supplementary table 1: primers for cloning and sequencing cloning for E- Ras ggg aat tcc ctt gag ctg ctg ggg aat ggc ttt gcc ggt cta gag tat aaa gga agc ttt gaa tcc Tpbp Oct3/4
Supplementary information Supplementary table 1: primers for cloning and sequencing cloning for E- Ras ggg aat tcc ctt gag ctg ctg ggg aat ggc ttt gcc ggt cta gag tat aaa gga agc ttt gaa tcc Tpbp Oct3/4
SUPPLEMENTARY EXPEMENTAL PROCEDURES
 SUPPLEMENTARY EXPEMENTAL PROCEDURES Plasmids- Total RNAs were extracted from HeLaS3 cells and reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) to obtain DNA template for the
SUPPLEMENTARY EXPEMENTAL PROCEDURES Plasmids- Total RNAs were extracted from HeLaS3 cells and reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) to obtain DNA template for the
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1154040/dc1 Supporting Online Material for Selective Blockade of MicroRNA Processing by Lin-28 Srinivas R. Viswanathan, George Q. Daley,* Richard I. Gregory* *To whom
www.sciencemag.org/cgi/content/full/1154040/dc1 Supporting Online Material for Selective Blockade of MicroRNA Processing by Lin-28 Srinivas R. Viswanathan, George Q. Daley,* Richard I. Gregory* *To whom
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p53 activity
 IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p5 activity H6 5 5 H58 A59 H6 H58 A59 anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p5 activity H6 5 5 H58 A59 H6 H58 A59 anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα
The retroviral vectors encoding WT human SIRT1 or a mutant of SIRT in which a
 Supporting online material Constructs The retroviral vectors encoding WT human SIRT1 or a mutant of SIRT in which a critical histidine in the deacetylase domain of SIRT1 has been replaced by a tyrosine
Supporting online material Constructs The retroviral vectors encoding WT human SIRT1 or a mutant of SIRT in which a critical histidine in the deacetylase domain of SIRT1 has been replaced by a tyrosine
Supplementary Figure 1 Characterization of OKSM-iNSCs
 Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
used at a final concentration of 5 ng/ml. Rabbit anti-bim and mouse anti-mkp2 antibodies were
 1 Supplemental Methods Reagents and chemicals: TGFβ was a generous gift from Genzyme Inc. (Cambridge, MA) and was used at a final concentration of 5 ng/ml. Rabbit anti-bim and mouse anti-mkp2 antibodies
1 Supplemental Methods Reagents and chemicals: TGFβ was a generous gift from Genzyme Inc. (Cambridge, MA) and was used at a final concentration of 5 ng/ml. Rabbit anti-bim and mouse anti-mkp2 antibodies
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Mammosphere formation assay. Mammosphere culture was performed as previously described (13,
 Supplemental Text Materials and Methods Mammosphere formation assay. Mammosphere culture was performed as previously described (13, 17). For co-culture with fibroblasts and treatment with CM or CCL2, fibroblasts
Supplemental Text Materials and Methods Mammosphere formation assay. Mammosphere culture was performed as previously described (13, 17). For co-culture with fibroblasts and treatment with CM or CCL2, fibroblasts
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Functional assays of hepatocyte-like cells induced from H1 hescs (a) P450 and AAT staining, PAS staining, LDL uptake assay, and ICG uptake and release assay
Supplementary Figure 1 Supplementary Figure 1. Functional assays of hepatocyte-like cells induced from H1 hescs (a) P450 and AAT staining, PAS staining, LDL uptake assay, and ICG uptake and release assay
SUPPLEMENTAL MATERIALS SIRTUIN 1 PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF2 ACTIVATION
 SUPPLEMENTAL MATERIALS SIRTUIN PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF ACTIVATION Haranatha R. Potteti*, Subbiah Rajasekaran*, Senthilkumar B. Rajamohan*, Chandramohan R. Tamatam,
SUPPLEMENTAL MATERIALS SIRTUIN PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF ACTIVATION Haranatha R. Potteti*, Subbiah Rajasekaran*, Senthilkumar B. Rajamohan*, Chandramohan R. Tamatam,
Protocol Using a Dox-Inducible Polycistronic m4f2a Lentivirus to Reprogram MEFs into ips Cells
 STEMGENT Page 1 OVERVIEW The following protocol describes the transduction and reprogramming of one well of Oct4-GFP mouse embryonic fibroblasts (MEF) using the Dox Inducible Reprogramming Polycistronic
STEMGENT Page 1 OVERVIEW The following protocol describes the transduction and reprogramming of one well of Oct4-GFP mouse embryonic fibroblasts (MEF) using the Dox Inducible Reprogramming Polycistronic
Supplementary Figure 1. GST pull-down analysis of the interaction of GST-cIAP1 (A, B), GSTcIAP1
 Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
SUPPLEMENTAL MATERIAL. Supplemental Methods. Cumate solution was from System Biosciences. Human complement C1q and complement
 SUPPLEMENTAL MATERIAL Supplemental Methods Reagents Cumate solution was from System Biosciences. Human complement Cq and complement C-esterase inhibitor (C-INH) were from Calbiochem. C-INH (Berinert) for
SUPPLEMENTAL MATERIAL Supplemental Methods Reagents Cumate solution was from System Biosciences. Human complement Cq and complement C-esterase inhibitor (C-INH) were from Calbiochem. C-INH (Berinert) for
Transcriptional regulation of BRCA1 expression by a metabolic switch: Di, Fernandez, De Siervi, Longo, and Gardner. H3K4Me3
 ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
BF EGFP DsRed. Relative Expression. Day 10 SOX Log2(FPKM + 1)_BCL2L1. Log2(FPKM + 1)_Control
 A B C D L-hESCs TetO L ADsRed IRESNEO TetO ADsRed IRESNEO OCT/DNA Ubc rtta NANOG/DNA E BF EGFP DsRed F Relative Expression..... SOX7 Endoderm..... CXCR..... T Mesoderm..... Relative Expression.6....8.6..
A B C D L-hESCs TetO L ADsRed IRESNEO TetO ADsRed IRESNEO OCT/DNA Ubc rtta NANOG/DNA E BF EGFP DsRed F Relative Expression..... SOX7 Endoderm..... CXCR..... T Mesoderm..... Relative Expression.6....8.6..
SUPPLEMENTARY INFORMATION. LIN-28 co-transcriptionally binds primary let-7 to regulate mirna maturation in C. elegans
 SUPPLEMENTARY INFORMATION LIN-28 co-transcriptionally binds primary let-7 to regulate mirna maturation in C. elegans Priscilla M. Van Wynsberghe 1, Zoya S. Kai 1, Katlin B. Massirer 2-4, Victoria H. Burton
SUPPLEMENTARY INFORMATION LIN-28 co-transcriptionally binds primary let-7 to regulate mirna maturation in C. elegans Priscilla M. Van Wynsberghe 1, Zoya S. Kai 1, Katlin B. Massirer 2-4, Victoria H. Burton
TITLE: Induced Pluripotent Stem Cells as Potential Therapeutic Agents in NF1
 AD Award Number: W81XWH-10-1-0181 TITLE: Induced Pluripotent Stem Cells as Potential Therapeutic Agents in NF1 PRINCIPAL INVESTIGATOR: Jonathan Chernoff, M.D., Ph.D. CONTRACTING ORGANIZATION: Institute
AD Award Number: W81XWH-10-1-0181 TITLE: Induced Pluripotent Stem Cells as Potential Therapeutic Agents in NF1 PRINCIPAL INVESTIGATOR: Jonathan Chernoff, M.D., Ph.D. CONTRACTING ORGANIZATION: Institute
Nanog-Luc. R-Luc. 40 HA-Klf Relative protein level of Klf USP21. Vector USP2 USP21 *** *** *** ***
 :Flag Relative protein level of Relative protein level of : Flag Vec 21LV 21SV 2 : HA HA- HA- HA- HA-Klf4 Relative mrna expression Relative protein level of Relative protein level of Relative protein level
:Flag Relative protein level of Relative protein level of : Flag Vec 21LV 21SV 2 : HA HA- HA- HA- HA-Klf4 Relative mrna expression Relative protein level of Relative protein level of Relative protein level
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas.
 Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
SUPPLEMENTARY FIG. S9. Morphology and DNA staining of cipsc-differentiated trophoblast cell-like cell. Scale bar: 100 mm. SUPPLEMENTARY FIG. S10.
 Supplementary Data SUPPLEMENTARY FIG. S1. Lentivirus-infected canine fibroblasts show YFP expression. (A, B) Uninfected canine skin fibroblasts (CSFs); (C, D) infected CSFs; (E, F) uninfected CTFs; (G,
Supplementary Data SUPPLEMENTARY FIG. S1. Lentivirus-infected canine fibroblasts show YFP expression. (A, B) Uninfected canine skin fibroblasts (CSFs); (C, D) infected CSFs; (E, F) uninfected CTFs; (G,
Cat# Product Name amounts h NANOG (RFP-Bsd) inducible particles
 Premade Lentiviral Particles for ips Stem Factors For generating induced pluripotent stem (ips) cells or other applications. LVP003 LVP004 LVP005 LVP006 LVP007 LVP008 RESEARCH USE ONLY, not intend for
Premade Lentiviral Particles for ips Stem Factors For generating induced pluripotent stem (ips) cells or other applications. LVP003 LVP004 LVP005 LVP006 LVP007 LVP008 RESEARCH USE ONLY, not intend for
Supplemental Figure 1 (Figure S1), related to Figure 1 Figure S1 provides evidence to demonstrate Nfatc1Cre is a mouse line that directed gene
 Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
USP19 modulates autophagy and antiviral immune responses by. deubiquitinating Beclin-1
 USP19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1 Shouheng Jin 1,2,, Shuo Tian 1,, Yamei Chen 1,, Chuanxia Zhang 1,2, Weihong Xie, 1 Xiaojun Xia 3,4, Jun Cui 1,3* &
USP19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1 Shouheng Jin 1,2,, Shuo Tian 1,, Yamei Chen 1,, Chuanxia Zhang 1,2, Weihong Xie, 1 Xiaojun Xia 3,4, Jun Cui 1,3* &
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
SUPPLEMENTARY INFORMATION
 Supplementary Figure a 3 factors 4 factors Nanog-GFP phase b c.5 Endogenous Oct3/4 2 Total Oct3/4.5 Relative expression (ES = ).5.5 6 3 Endogenous Sox2 Nanog Relative expression (ES = ) 5 25 2 Total Sox2
Supplementary Figure a 3 factors 4 factors Nanog-GFP phase b c.5 Endogenous Oct3/4 2 Total Oct3/4.5 Relative expression (ES = ).5.5 6 3 Endogenous Sox2 Nanog Relative expression (ES = ) 5 25 2 Total Sox2
Supplemental Figure Legends:
 Supplemental Figure Legends: Fig S1. GFP-ABRO1 localization. U2OS cells were infected with retrovirus expressing GFP- ABRO1. The cells were fixed with 3.6% formaldehyde and stained with antibodies against
Supplemental Figure Legends: Fig S1. GFP-ABRO1 localization. U2OS cells were infected with retrovirus expressing GFP- ABRO1. The cells were fixed with 3.6% formaldehyde and stained with antibodies against
Supplementary Materials and Methods
 Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
APPLICATION NOTE Page 1
 APPLICATION NOTE Page 1 Generation of ips Cells from Oct4-Neo Reporter Embryonic Fibroblasts Dongmei Wu, Yan Gao, Charles Martin, Xun Cheng, Wen Xiong, Shuyuan Yao 1 Stemgent, Inc., 10575 Roselle St.,
APPLICATION NOTE Page 1 Generation of ips Cells from Oct4-Neo Reporter Embryonic Fibroblasts Dongmei Wu, Yan Gao, Charles Martin, Xun Cheng, Wen Xiong, Shuyuan Yao 1 Stemgent, Inc., 10575 Roselle St.,
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification.
 Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Supplemental Materials
 Supplemental Materials Materials and Methods Histopathologic and immunohistochemical evaluation Liver tissue was divided and fixed in phosphate-buffered neutral formalin, embedded in paraffin, and cut
Supplemental Materials Materials and Methods Histopathologic and immunohistochemical evaluation Liver tissue was divided and fixed in phosphate-buffered neutral formalin, embedded in paraffin, and cut
Partial list of differentially expressed genes from cdna microarray, comparing MUC18-
 Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
Supplementary Figure 1.
 Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera
 SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Quantitative and non-quantitative RT-PCR. cdna was generated from 500ng RNA (iscript;
 Supplemental Methods Quantitative and non-quantitative RT-PCR. cdna was generated from 500ng RNA (iscript; Bio-Rad, Hercules, CA, USA) and standard RT-PCR experiments were carried out using the 2X GoTaq
Supplemental Methods Quantitative and non-quantitative RT-PCR. cdna was generated from 500ng RNA (iscript; Bio-Rad, Hercules, CA, USA) and standard RT-PCR experiments were carried out using the 2X GoTaq
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Percent survival. Supplementary fig. S3 A.
 Supplementary fig. S3 A. B. 100 Percent survival 80 60 40 20 Ml 0 0 100 C. Fig. S3 Comparison of leukaemia incidence rate in the triple targeted chimaeric mice and germline-transmission translocator mice
Supplementary fig. S3 A. B. 100 Percent survival 80 60 40 20 Ml 0 0 100 C. Fig. S3 Comparison of leukaemia incidence rate in the triple targeted chimaeric mice and germline-transmission translocator mice
Alpha or beta human chorionic gonadotropin knockdown decrease BeWo cell fusion by
 Alpha or beta human chorionic gonadotropin knockdown decrease BeWo cell fusion by down-regulating PKA and CREB activation Sudha Saryu Malhotra 1, Pankaj Suman 2 and Satish Kumar Gupta 1 * 1 Reproductive
Alpha or beta human chorionic gonadotropin knockdown decrease BeWo cell fusion by down-regulating PKA and CREB activation Sudha Saryu Malhotra 1, Pankaj Suman 2 and Satish Kumar Gupta 1 * 1 Reproductive
Supplemental Information
 Supplemental Information Genetic and Functional Studies Implicate HIF1α as a 14q Kidney Cancer Suppressor Gene Chuan Shen, Rameen Beroukhim, Steven E. Schumacher, Jing Zhou, Michelle Chang, Sabina Signoretti,
Supplemental Information Genetic and Functional Studies Implicate HIF1α as a 14q Kidney Cancer Suppressor Gene Chuan Shen, Rameen Beroukhim, Steven E. Schumacher, Jing Zhou, Michelle Chang, Sabina Signoretti,
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
