SUPPLEMENTARY INFORMATION
|
|
|
- Camilla Burke
- 5 years ago
- Views:
Transcription
1 SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin 2,4,, Ashley Fritz 5, Falei Yuan 2,6, Julia Chu 2, Shyam Patel 2, David V. Schaffer 1,2,5, Song Li 1,2, Author Affiliations: 1 UC Berkeley & UCSF Joint Graduate Program in Bioengineering, Berkeley/San Francisco, CA, USA 2 Department of Bioengineering, University of California, Berkeley, B18A Stanley Hall, Berkeley, CA , USA 3 Ecole Polytechnique, Palaiseau, France 4 University Lille Nord de France, F-59 Lille, France 5 Department of Chemical and Biomolecular Engineering, University of California, Berkeley, CA , USA 6 Med-X Center, Shanghai Jiao Tong University, Shanghai 23, P.R. China These authors contributed equally. Corresponding address: Song Li, Ph.D. Department of Bioengineering University of California, Berkeley B18A Stanley Hall, Berkeley, CA Telephone: (51) Fax: (51) song_li@berkeley.edu NATURE MATERIALS 1
2 Supplemental Fig. S1 a 1. Flat 4 µm Nuclear Shape Index (NSI) µm 1 µm Angle ( ) b % EdU Incorporation Flat 4µm 2µm 1µm Supplemental Figure S1. Effect of microgrooves on fibroblast nuclear shape and proliferation. a, Nuclear shape index (NSI) and alignment of fibroblasts cultured on different microgrooved substrates. We quantified nuclei elongation by calculating NSI according to the formula, NSI = 4πA/P 2, where A is nucleus projection area and P is nucleus perimeter. b, Edu incorporation analysis showing slight but significant decreases in fibroblast proliferation on microgrooved substrates (n=6). indicates that there is statistically significant difference (p<.5) compared with the control flat surface. Differences between groups were determined by using a oneway analysis of variance (ANOVA) and a Tukey s post-hoc test. Error bars represent one standard deviation.
3 Supplemental Fig. S2 a GFP + colonies per 1 5 cells Flat 1 μm b Flat 1 μm Oct4-GFP c Phase contrast Oct4-GFP Merged Supplemental Figure S2. OSKM reprogramming on flat and microgrooved surfaces with Oct4-GFP mouse fibroblasts. a, Quantification of GFP + colonies (n=4). indicates that there is statistically significant difference (p<.5) compared with the control flat surface. Differences between groups were determined using a two-tailed, unpaired t-test. Error bars represent one standard deviation. b, Fluorescence micrographs of GFP + colonies on flat and grooved surfaces. c, Phase contrast and fluorescence micrographs of GFP + colonies on 1 μm-width grooves. (scale bars = 2 μm)
4 Supplemental Fig. S3 a Oct4 Sox2 Nanog SSEA-1 Oct4 Sox2 Nanog SSEA-1 Merge Marker DAPI Marker DAPI b Supplemental Figure S3. ipsc characterization. Immunostaining of ipsc colonies generated using OSK (a) and OSKM (b) reprogramming factors on grooves of 1 µm width. Colonies show nuclear localization of pluripotency genes Oct4, Sox2, and Nanog in generated ipscs. Colonies also stained positive for SSEA-1, a cell surface marker specific to mescs (scale bar = 1 µm).
5 Supplemental Fig. S4 a b c S1 d e f -Catenin Vimentin SMA NFM g h i CNN1 GFAP Collagen-1 j k l SOX1 TUJ1 TUJ1 m n SOX17 PDX1 Supplemental Figure S4. In vitro differentiation potential of mouse ipsc. a, ipsc embryoid body (EB) formation (scale bar = 2 µm). b-n, Immunostaining of spontaneously differentiated cells from ipsc EB. Cells express markers (shown in red or green) found in all germ layers. Cells are costained with DAPI for identification of nuclei (scale bar = 1 µm). Mouse ipscs were differentiated for 2 weeks. Gene symbol key: S1 calcium binding protein B (S1β); Cadherin associated protein, β1 (β -Catenin); Smooth muscle α-actin (SMA); Neurofilament medium polypeptide (NFM); Calponin 1 (CNN1); Glial fibrillary acidic protein (GFAP); SRY-box containing gene 1 (Sox1); SRY-box containing gene 17 (Sox17); β-iii tubulin (TUJ1); pancreatic and duodenal homeobox 1 (PDX1).
6 Supplemental Fig. S5 a Flat 1 μm c Flat 1 μm Nanog + colonies per 1 4 cells Flat 1 μm DAPI + DAPI + Oct4 Sox2 Nanog SSEA-4 TUJ1 Nestin SMA S1 DAPI + Nanog b d DAPI e f Sox17 CNN1 Sox1 NFM Supplemental Figure S5. Effects of microtopography on human cell reprogramming. a, Images show fluorescence micrograph of nucleus (DAPI, blue) and actin network (phalloidin, green) of normal neonatal human dermal fibroblasts (NHDF) on flat and microgrooved PDMS surfaces (scale bars=1 µm). b, Reprogramming efficiency of NHDF transduced with OSKM and cultured on PDMS membranes with flat and microgrooved surfaces. indicates p<.5 (n=6). Differences between groups were determined using a two-tailed, unpaired t-test. Error bars represent one standard deviation. c, Fluorescence micrographs show ipsc colonies generated on flat and microgrooved surfaces. d, Phase contrast image shows expanded colony morphology. e, Fluorescence micrograph shows expression of pluripotency markers in generated clones. (scale bar in c-e, 2 µm). f, In vitro differentiation potential of NHDF-iPSCs (scale bar = 1 µm).
7 Supplemental Fig. S6 Flat 1 µm AcH3 H3K4me2 H3K4me3 H3 GAPDH Actin Supplemental Figure S6. Biophysical regulation of epigenetics following the forced expression of OSKM. Western blotting analysis of mouse fibroblasts infected with OSKM and cultured on flat and microgrooved surfaces for 8 days.
8 Supplemental Fig. S7 a b Nanog + colonies per 1 5 cells Flat 1µm mirna 32/367 Supplemental Figure S7. mirna generated ipsc-like colonies. a, Quantification of colony formation in fibroblasts transduced with mirna 32/367 in the absence of VPA (n=4). Differences between groups were determined using a twotailed, unpaired t-test. indicates that there is statistically significant difference (p<.5) compared with the control flat surface. Error bars represent one standard deviation. b, Fluorescence micrograph shows Nanog expression in ipsc-like colonies generated from wild-type mouse ear fibroblasts on microgrooved surfaces (scale bar = 1 mm). c, Phase contrast and fluorescence micrograph of GFP + colonies generated from mouse ear fibroblasts and mouse tail fibroblasts isolated from Oct4-GFP reporter mice (scale bar = 1 μm). DAPI Nanog Merge c Phase Oct4-GFP Merged Mouse tail fibroblasts Mouse ear fibroblasts
9 Supplemental Fig. S8 Gene expression (WDR5/18s).4.2. ctrl sirna WDR5 sirna Supplemental Figure S8. WDR5 RNA interference. qrt- PCR confirms the successful sirna knockdown of WDR5 gene expression. Gene expression was normalized to 18S. p<.5 (n=3). Differences between groups were determined using a two-tailed, unpaired t-test. Error bars represent one standard deviation.
10 Supplemental Fig. S9 E-cad Fold change Flat 1 μm. Flat 1 μm AcH3 H3K4me3 Supplemental Figure S9. ChIP-qPCR analysis. ChIP-qPCR analysis shows that there is no significant fold enrichment of histone modifications, AcH3 and H3K4me3, at the promoter region of E-cad (n=3). Statistical significance was determined using a two-tailed, unpaired t-test. Error bars represent one standard deviation.
11 Supplemental Fig. S1 Aligned Random Supplemental Figure S1. ipsc colony morphology on random and aligned nanofibres. Phase contrast image shows ipsc colony elongation on aligned versus random nanofibres (scale bar = 5 µm).
12 Supplemental Fig. S11 1. Nuclear Shape Index (NSI) Cell Shape Index (CSI) Supplemental Figure S11. Effect of cell shape on nuclear shape index (NSI). Quantification of NSI for fibroblasts seeded onto microislands of round (CSI=1) and elongated (CSI=.1) morphologies. Differences between groups were determined using a two-tailed, unpaired t-test. indicates that there is statistically significant difference (p<.5) compared with the flat control. Error bars represent one standard deviation.
13 Supplemental Fig. S12 Oct4 Nanog Fold Enrichment Fold Enrichment Sox E-cad Supplemental Figure S12. ChiP antibody fold enrichment. AcH3, H3K4me2 and H3K4me3 antibody fold enrichment over normal rabbit IgG control was quantified. Substantial enrichment was observed at the promoter regions of Oct, Sox2, Nanog, and E-cad genes following immunoprecipitation with antibodies against AcH3, H3K4me2 or H3K4me3.
14 Supplemental Table S1 Supplemental Table S1. Forward and reverse primers used for ChIP-qPCR analysis. Promoter Forward Primer Reverse Primer Oct4 CCTAAGGGTTGTCCTGTCCA GTGGAAAGACGGCTCACCTA Sox2 CCCATTTATTCCCTGACAGC GGGCTCCAAACTTCTCTCCT Nanog GGAGAATAGGGGGTGGGTAG CAGCCTTCCCACAGAAAGAG E-cad TGGAATAGGAAGCTGGGAAG CTCCCACACCAGTGAGCAG
15 Supplemental Table S2 Supplemental Table S2. Antibody information for immunostaining and Western blotting analysis. Antibody Company Catalog # AcH3 Millipore H3K4me2 Abcam ab32356 H3K4me3 Millipore Dilution/ Application 1:1/WB; 1:5/IF 1:1/WB; 1:25/IF 1:1/WB; 1:5/IF HDAC1 Santa Cruz sc :1/WB HDAC2 Santa Cruz sc :1/WB; 1:5/IF HDAC3 Santa Cruz sc :1/WB Actin Santa Cruz sc :1/WB GAPDH Santa Cruz sc :1/WB WDR5 Abcam ab :1/WB; 1:5/IF H3 Abcam ab1791 1:1/WB Nanog Abcam ab7482 1:5/IF Oct4 Santa Cruz sc :5/IF SSEA-1 Santa Cruz sc :5/IF Sox2 Millipore ab563 1:5/IF
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and
 Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like
 Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like morphology in co-culture with mouse embryonic feeder fibroblasts
Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like morphology in co-culture with mouse embryonic feeder fibroblasts
bronchial epithelial cells (I). Bronchi are outlined with dashed line. Scale bars = 25 µm, if not
 Supplemental Figure S1: ronchial epithelial cell polarity and integrity is maintained in bronchi. (A-E) Staining for selected markers of bronchial cell differentiation and intracellular compartments is
Supplemental Figure S1: ronchial epithelial cell polarity and integrity is maintained in bronchi. (A-E) Staining for selected markers of bronchial cell differentiation and intracellular compartments is
Supplementary Figure 1.
 Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
0.9 5 H M L E R -C tr l in T w is t1 C M
 a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Data. Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells
 Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State
 Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
SUPPLEMENTARY FIG. S9. Morphology and DNA staining of cipsc-differentiated trophoblast cell-like cell. Scale bar: 100 mm. SUPPLEMENTARY FIG. S10.
 Supplementary Data SUPPLEMENTARY FIG. S1. Lentivirus-infected canine fibroblasts show YFP expression. (A, B) Uninfected canine skin fibroblasts (CSFs); (C, D) infected CSFs; (E, F) uninfected CTFs; (G,
Supplementary Data SUPPLEMENTARY FIG. S1. Lentivirus-infected canine fibroblasts show YFP expression. (A, B) Uninfected canine skin fibroblasts (CSFs); (C, D) infected CSFs; (E, F) uninfected CTFs; (G,
Accelerating skin wound healing by M-CSF through generating SSEA-1 and -3 stem cells. in the injured sites
 Accelerating skin wound healing by M-CSF through generating SSEA-1 and -3 stem cells in the injured sites Yunyuan Li, Reza Baradar Jalili, Aziz Ghahary Department of Surgery, University of British Columbia,
Accelerating skin wound healing by M-CSF through generating SSEA-1 and -3 stem cells in the injured sites Yunyuan Li, Reza Baradar Jalili, Aziz Ghahary Department of Surgery, University of British Columbia,
A Novel Platform to Enable the High-Throughput Derivation and Characterization of Feeder-Free Human ipscs
 Supplementary Information A Novel Platform to Enable the High-Throughput Derivation and Characterization of Feeder-Free Human ipscs Bahram Valamehr, Ramzey Abujarour, Megan Robinson, Thuy Le, David Robbins,
Supplementary Information A Novel Platform to Enable the High-Throughput Derivation and Characterization of Feeder-Free Human ipscs Bahram Valamehr, Ramzey Abujarour, Megan Robinson, Thuy Le, David Robbins,
Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and
 Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
SUPPLEMENTARY INFORMATION
 SUPPLEMENTRY INFORMTION DOI:.38/ncb Kdmb locus kb Long isoform Short isoform Long isoform Jmj XX PHD F-box LRR Short isoform XX PHD F-box LRR Target Vector 3xFlag Left H Neo Right H TK LoxP LoxP Kdmb Locus
SUPPLEMENTRY INFORMTION DOI:.38/ncb Kdmb locus kb Long isoform Short isoform Long isoform Jmj XX PHD F-box LRR Short isoform XX PHD F-box LRR Target Vector 3xFlag Left H Neo Right H TK LoxP LoxP Kdmb Locus
Supplementary Figure 1.
 Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Supplemental Figure 1 (Figure S1), related to Figure 1 Figure S1 provides evidence to demonstrate Nfatc1Cre is a mouse line that directed gene
 Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Resveratrol inhibits epithelial-mesenchymal transition of retinal. pigment epithelium and development of proliferative vitreoretinopathy
 Resveratrol inhibits epithelial-mesenchymal transition of retinal pigment epithelium and development of proliferative vitreoretinopathy Keijiro Ishikawa, 1,2 Shikun He, 2, 3 Hiroto Terasaki, 1 Hossein
Resveratrol inhibits epithelial-mesenchymal transition of retinal pigment epithelium and development of proliferative vitreoretinopathy Keijiro Ishikawa, 1,2 Shikun He, 2, 3 Hiroto Terasaki, 1 Hossein
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Generation of NSCs from hpscs in SDC medium.
 Supplementary Figure 1 Generation of NSCs from hpscs in SDC medium. (a) Q-PCR of pluripotent markers (OCT4, NANOG), neural markers (SOX1, PAX6, N-Cadherin), markers for the other germ layers (T, EOMES,
Supplementary Figure 1 Generation of NSCs from hpscs in SDC medium. (a) Q-PCR of pluripotent markers (OCT4, NANOG), neural markers (SOX1, PAX6, N-Cadherin), markers for the other germ layers (T, EOMES,
Supplementary Figure 1. ERK signaling is not activated at early hypertension. a, Western blot analysis for the level of phospho-erk (perk) and total
 Supplementary Figure 1. ERK signaling is not activated at early hypertension. a, Western blot analysis for the level of phospho-erk (perk) and total ERK in the aortic tissue from the saline- or AngII-infused
Supplementary Figure 1. ERK signaling is not activated at early hypertension. a, Western blot analysis for the level of phospho-erk (perk) and total ERK in the aortic tissue from the saline- or AngII-infused
Table S1. Components for knockout serum replacer medium (KSR medium) (500 ml) Component Supplier Catalogue number
 Table S1. Components for knockout serum replacer medium (KSR medium) (500 ml) Component Supplier Catalogue number Stock Concentration Final concentration Volume used Advanced DMEM/F12 Invitrogen 12634010-78%
Table S1. Components for knockout serum replacer medium (KSR medium) (500 ml) Component Supplier Catalogue number Stock Concentration Final concentration Volume used Advanced DMEM/F12 Invitrogen 12634010-78%
Rapid differentiation of human pluripotent stem cells into functional motor neurons by
 Supplementary Information Rapid differentiation of human pluripotent stem cells into functional motor neurons by mrnas encoding transcription factors Sravan Kumar Goparaju, Kazuhisa Kohda, Keiji Ibata,
Supplementary Information Rapid differentiation of human pluripotent stem cells into functional motor neurons by mrnas encoding transcription factors Sravan Kumar Goparaju, Kazuhisa Kohda, Keiji Ibata,
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure 1. ips_3y+mir-302b. ips_3y+mir-372. ips_3y+mir-302b + mir-372. ips_4y. ips_4y+mir-302b. ips_4y + mir-372
 Supplementary Figure 1 ips_3y+mir-32b ips_3y+ ips_3y+mir-32b + ips_4y ips_4y+mir-32b ips_4y + Nature Biotechnology: doi:1.138/nb.t.1862 TRA-1-6 DAPI Oct3/4 Supplementary Figure 1: ips cells derived from
Supplementary Figure 1 ips_3y+mir-32b ips_3y+ ips_3y+mir-32b + ips_4y ips_4y+mir-32b ips_4y + Nature Biotechnology: doi:1.138/nb.t.1862 TRA-1-6 DAPI Oct3/4 Supplementary Figure 1: ips cells derived from
 Supplemental Tables Supplemental Table 1. Phenotypic profile of different HSC lines. Marker d0 mhsc d7 mhsc 8B LX2 CD11b 3.61% 2.21% 2.16% 3.77% F4/80 4.54% 2.05% 0.127% 1.44% Elastin 3.90% 2.95% 4.23%
Supplemental Tables Supplemental Table 1. Phenotypic profile of different HSC lines. Marker d0 mhsc d7 mhsc 8B LX2 CD11b 3.61% 2.21% 2.16% 3.77% F4/80 4.54% 2.05% 0.127% 1.44% Elastin 3.90% 2.95% 4.23%
The p53-puma axis suppresses ipsc generation
 The p53-puma axis suppresses ipsc generation Yanxin Li, Haizhong Feng, Haihui Gu, Dale W. Lewis, Youzhong Yuan, Lei Zhang, Hui Yu, Peng Zhang, Haizi Cheng, Weimin Miao, Weiping Yuan, Shi-Yuan Cheng, Susanne
The p53-puma axis suppresses ipsc generation Yanxin Li, Haizhong Feng, Haihui Gu, Dale W. Lewis, Youzhong Yuan, Lei Zhang, Hui Yu, Peng Zhang, Haizi Cheng, Weimin Miao, Weiping Yuan, Shi-Yuan Cheng, Susanne
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1 Characterization of GSCs. a. Immunostaining of primary GSC spheres from GSC lines. Nestin (neural progenitor marker, red), TLX (green). Merged images of nestin,
Supplementary Figures Supplementary Fig. 1 Characterization of GSCs. a. Immunostaining of primary GSC spheres from GSC lines. Nestin (neural progenitor marker, red), TLX (green). Merged images of nestin,
a KYSE270-CON KYSE270-Id1
 a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
STAT3 signaling controls satellite cell expansion and skeletal muscle repair
 SUPPLEMENTARY INFORMATION STAT3 signaling controls satellite cell expansion and skeletal muscle repair Matthew Timothy Tierney 1 *, Tufan Aydogdu 2,3 *, David Sala 2, Barbora Malecova 2, Sole Gatto 2,
SUPPLEMENTARY INFORMATION STAT3 signaling controls satellite cell expansion and skeletal muscle repair Matthew Timothy Tierney 1 *, Tufan Aydogdu 2,3 *, David Sala 2, Barbora Malecova 2, Sole Gatto 2,
Derivation of UCSFB lines from biopsied blastomeres of cleavage-stage human. Spare embryos were obtained through the UCSF IVF Tissue Bank from donors
 Supplementary Materials and Methods Derivation of UCSFB lines from biopsied blastomeres of cleavage-stage human embryos Spare embryos were obtained through the UCSF IVF Tissue Bank from donors undergoing
Supplementary Materials and Methods Derivation of UCSFB lines from biopsied blastomeres of cleavage-stage human embryos Spare embryos were obtained through the UCSF IVF Tissue Bank from donors undergoing
gacgacgaggagaccaccgctttg aggcacattgaaggtctcaaacatg
 Supplementary information Supplementary table 1: primers for cloning and sequencing cloning for E- Ras ggg aat tcc ctt gag ctg ctg ggg aat ggc ttt gcc ggt cta gag tat aaa gga agc ttt gaa tcc Tpbp Oct3/4
Supplementary information Supplementary table 1: primers for cloning and sequencing cloning for E- Ras ggg aat tcc ctt gag ctg ctg ggg aat ggc ttt gcc ggt cta gag tat aaa gga agc ttt gaa tcc Tpbp Oct3/4
A population of Nestin expressing progenitors in the cerebellum exhibits increased tumorigenicity
 A population of Nestin expressing progenitors in the cerebellum exhibits increased tumorigenicity Peng Li 1,2, Fang Du 1, Larra W. Yuelling 1, Tiffany Lin 3, Renata E. Muradimova 1, Rossella Tricarico
A population of Nestin expressing progenitors in the cerebellum exhibits increased tumorigenicity Peng Li 1,2, Fang Du 1, Larra W. Yuelling 1, Tiffany Lin 3, Renata E. Muradimova 1, Rossella Tricarico
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Wnt16 smact merge VK/AB
 A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
Mannen et al., http :// /cgi /content /full /jcb /DC1
 Supplemental material JCB Mannen et al., http ://www.jcb.org /cgi /content /full /jcb.201601024 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Characterization of SNB components. (A) SNB localization of Venus-tagged
Supplemental material JCB Mannen et al., http ://www.jcb.org /cgi /content /full /jcb.201601024 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Characterization of SNB components. (A) SNB localization of Venus-tagged
Supplemental Information Control of apico-basal epithelial polarity by the microtubule minus-end binding protein CAMSAP3 and spectraplakin ACF7
 Supplemental Information Control of apico-basal epithelial polarity by the microtubule minus-end binding protein CAMSAP3 and spectraplakin ACF7 Ivar Noordstra, Qingyang Liu, Wilco Nijenhuis, Shasha Hua,
Supplemental Information Control of apico-basal epithelial polarity by the microtubule minus-end binding protein CAMSAP3 and spectraplakin ACF7 Ivar Noordstra, Qingyang Liu, Wilco Nijenhuis, Shasha Hua,
Supplementary Figure S1. Alterations in Fzr1( / );Nestin-Cre brains. (a) P10 Cdh1-deficient brains display low levels of Myelin basic protein (MBP)
 Supplementary Figure S1. Alterations in Fzr1( / );Nestin-Cre brains. (a) P10 Cdh1-deficient brains display low levels of Myelin basic protein (MBP) in the cortex (area 1 as defined in Fig. 2a), and corpus
Supplementary Figure S1. Alterations in Fzr1( / );Nestin-Cre brains. (a) P10 Cdh1-deficient brains display low levels of Myelin basic protein (MBP) in the cortex (area 1 as defined in Fig. 2a), and corpus
WT Day 90 after injections
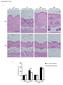 Supplementary Figure 1 a Day 1 after injections Day 9 after injections Klf5 +/- Day 1 after injections Klf5 +/- Day 9 after injections BLM PBS b Day 1 after injections Dermal thickness (μm) 3 1 Day 9 after
Supplementary Figure 1 a Day 1 after injections Day 9 after injections Klf5 +/- Day 1 after injections Klf5 +/- Day 9 after injections BLM PBS b Day 1 after injections Dermal thickness (μm) 3 1 Day 9 after
Chemically defined conditions for human ipsc derivation and culture
 Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2239 Hepatocytes (Endoderm) Foreskin fibroblasts (Mesoderm) Melanocytes (Ectoderm) +Dox rtta TetO CMV O,S,M,K & N H1 hes H7 hes H9 hes H1 hes-derived EBs H7 hes-derived EBs H9 hes-derived
DOI: 10.1038/ncb2239 Hepatocytes (Endoderm) Foreskin fibroblasts (Mesoderm) Melanocytes (Ectoderm) +Dox rtta TetO CMV O,S,M,K & N H1 hes H7 hes H9 hes H1 hes-derived EBs H7 hes-derived EBs H9 hes-derived
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb37 a mrna relative expression 1.3 1..8.5.3. CTR NGPS OCT4 SOX2 KLF4 c-myc b BANF1 mrna levels 1.2 1..8.6.4.2. plko.1 shbanf1 c Tra-1-6+ colonies 3 1 plko.1 shbanf1 d BANF1 locus Plasmid donor
DOI: 1.138/ncb37 a mrna relative expression 1.3 1..8.5.3. CTR NGPS OCT4 SOX2 KLF4 c-myc b BANF1 mrna levels 1.2 1..8.6.4.2. plko.1 shbanf1 c Tra-1-6+ colonies 3 1 plko.1 shbanf1 d BANF1 locus Plasmid donor
Supplementary Figure 1 Characterization of OKSM-iNSCs
 Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
Supplementary Fig. 1
 a FL (1-2266) NL (1-1190) CL (1191-2266) HA-ICE1: - HA-ICE1: - - - FLAG-ICE2: + + + + FLAG-ELL: + + + + + + IP: anti-ha FLAG-ICE2 HA-ICE1-FL HA-ICE1-NL HA-ICE1-CL FLAG-ICE2 b IP: anti-ha FL (1-2266) NL
a FL (1-2266) NL (1-1190) CL (1191-2266) HA-ICE1: - HA-ICE1: - - - FLAG-ICE2: + + + + FLAG-ELL: + + + + + + IP: anti-ha FLAG-ICE2 HA-ICE1-FL HA-ICE1-NL HA-ICE1-CL FLAG-ICE2 b IP: anti-ha FL (1-2266) NL
Gene name forward primer 5->3 reverse primer 5->3 product GCCCATA. Nfatc1 AGGTGCAGCCCAAGTCTCAC GTGGCCATCTGGAGCCTTC CTGT GATGC GCT ACCAA
 Supplementary able 1 is presenting the PCR primer list used. ene name forward primer 5->3 reverse primer 5->3 product size cdna VE-Cadherin AACCCAAAAA CACCCCAAAAC 260bp CCA CCCAA PECAM-1 CACCACAA CCCCCACC
Supplementary able 1 is presenting the PCR primer list used. ene name forward primer 5->3 reverse primer 5->3 product size cdna VE-Cadherin AACCCAAAAA CACCCCAAAAC 260bp CCA CCCAA PECAM-1 CACCACAA CCCCCACC
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
SUPPLEMENTARY INFORMATION
 Supplementary Figure S1 Colony-forming efficiencies using transposition of inducible mouse factors. (a) Morphologically distinct growth foci appeared from rtta-mefs 6-8 days following PB-TET-mFx cocktail
Supplementary Figure S1 Colony-forming efficiencies using transposition of inducible mouse factors. (a) Morphologically distinct growth foci appeared from rtta-mefs 6-8 days following PB-TET-mFx cocktail
Supplementary Information
 Supplementary Information Supplementary Figure 1: Identification of new regulators of MuSC by a proteome-based shrna screen. (a) FACS plots of GFP + and GFP - cells from Pax7 ICN -Z/EG (upper panel) and
Supplementary Information Supplementary Figure 1: Identification of new regulators of MuSC by a proteome-based shrna screen. (a) FACS plots of GFP + and GFP - cells from Pax7 ICN -Z/EG (upper panel) and
Isolation of human ips cells using EOS lentiviral vectors to select for pluripotency
 nature methods Isolation of human ips cells using EOS lentiviral vectors to select for pluripotency Akitsu Hotta, Aaron Y L Cheung, Natalie Farra, Kausalia Vijayaragavan, Cheryle A Séguin, Jonathan S Draper,
nature methods Isolation of human ips cells using EOS lentiviral vectors to select for pluripotency Akitsu Hotta, Aaron Y L Cheung, Natalie Farra, Kausalia Vijayaragavan, Cheryle A Séguin, Jonathan S Draper,
immunofluorescence. Name of antibodies Manufacturer Catalog Number Rabbit anti-pdyn Rabbit anti-kor-1
 Supplemental Tables Table S1. List of primary antibodies used for immunohistochemistry, FACS, and immunofluorescence. Name of antibodies Manufacturer Catalog Number Rabbit anti-pdyn Bioss USA bs-13041r
Supplemental Tables Table S1. List of primary antibodies used for immunohistochemistry, FACS, and immunofluorescence. Name of antibodies Manufacturer Catalog Number Rabbit anti-pdyn Bioss USA bs-13041r
Biology Open (2015): doi: /bio : Supplementary information
 Fig. S1. Verification of optimal conditions for the generation of LIF-dependent inscs Representative images (from five repeat experiments) of human primary fibroblasts undergoing reprogramming for 21 days
Fig. S1. Verification of optimal conditions for the generation of LIF-dependent inscs Representative images (from five repeat experiments) of human primary fibroblasts undergoing reprogramming for 21 days
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in
 Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Int. J. Mol. Sci. 2016, 17, 1259; doi: /ijms
 S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Functional assays of hepatocyte-like cells induced from H1 hescs (a) P450 and AAT staining, PAS staining, LDL uptake assay, and ICG uptake and release assay
Supplementary Figure 1 Supplementary Figure 1. Functional assays of hepatocyte-like cells induced from H1 hescs (a) P450 and AAT staining, PAS staining, LDL uptake assay, and ICG uptake and release assay
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
SUPPLEMENTARY INFORMATION
 a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
Post-translational modification
 Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Paul et al.,
 Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplementary Information
 Supplementary Information Supplementary Figure S1 (a) P-cRAF colocalizes with LC3 puncta. Immunofluorescence (IF) depicting colocalization of P-cRAF (green) and LC3 puncta (red) in NIH/3T3 cells treated
Supplementary Information Supplementary Figure S1 (a) P-cRAF colocalizes with LC3 puncta. Immunofluorescence (IF) depicting colocalization of P-cRAF (green) and LC3 puncta (red) in NIH/3T3 cells treated
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Protocol Using the Reprogramming Ecotropic Retrovirus Set: Mouse OSKM to Reprogram MEFs into ips Cells
 STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblast (MEF) cells into induced pluripotent stem (ips) cells in a 6-well format. Transduction
STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblast (MEF) cells into induced pluripotent stem (ips) cells in a 6-well format. Transduction
Post-expansion antibody delivery, after epitope-preserving homogenization.
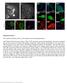 Supplementary Figure 1 Post-expansion antibody delivery, after epitope-preserving homogenization. (a, b) Wide-field fluorescence images of Thy1-YFP-expressing mouse brain hemisphere slice before expansion
Supplementary Figure 1 Post-expansion antibody delivery, after epitope-preserving homogenization. (a, b) Wide-field fluorescence images of Thy1-YFP-expressing mouse brain hemisphere slice before expansion
Figure S1. Specificity of immunofluorescence staining in STC-1 cells. STC-1 cells
 Supplementary Figures and Tables Figure S1. Figure S1. Specificity of immunofluorescence staining in STC-1 cells. STC-1 cells were treated with donkey-anti rabbit antibody conjugated with Dylight488 or
Supplementary Figures and Tables Figure S1. Figure S1. Specificity of immunofluorescence staining in STC-1 cells. STC-1 cells were treated with donkey-anti rabbit antibody conjugated with Dylight488 or
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Effect of timing of DE dissociation and RA concentration on lung field specification in hpscs. (a) Effect of duration of endoderm induction on expression of
Supplementary Figures Supplementary Figure 1. Effect of timing of DE dissociation and RA concentration on lung field specification in hpscs. (a) Effect of duration of endoderm induction on expression of
Supplementary Figures and supplementary figure legends
 Supplementary Figures and supplementary figure legends Figure S1. Effect of different percentage of FGF signaling knockdown on TGF signaling and EndMT marker gene expression. HUVECs were subjected to different
Supplementary Figures and supplementary figure legends Figure S1. Effect of different percentage of FGF signaling knockdown on TGF signaling and EndMT marker gene expression. HUVECs were subjected to different
TRIM31 is recruited to mitochondria after infection with SeV.
 Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
hipscs were derived from human skin fibroblasts (CRL-2097) by ectopic
 Generation and characterization of hipscs hipscs were derived from human skin fibroblasts (CRL-2097) by ectopic expression of OCT4, SOX2, KLF4, and C-MYC as previously described 1. hipscs showed typical
Generation and characterization of hipscs hipscs were derived from human skin fibroblasts (CRL-2097) by ectopic expression of OCT4, SOX2, KLF4, and C-MYC as previously described 1. hipscs showed typical
Supplementary Figure 1. Western analysis of p-smad1/5/8 of differentiated hescs.
 kda 65 45 45 35 Supplementary Figure 1. Western analysis of p-smad1/5/8 of differentiated hescs. H9 hescs were differentiated with or without BMP4+BMP8A and cell lysates were collected for Western analysis
kda 65 45 45 35 Supplementary Figure 1. Western analysis of p-smad1/5/8 of differentiated hescs. H9 hescs were differentiated with or without BMP4+BMP8A and cell lysates were collected for Western analysis
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
Supplementary Figure S1 Evi/Wls and Wnt3 are overexpressed in epithelial cells in colon cancers. (a) Validation of the specificity of the Wnt3
 Supplementary Figure S1 Evi/Wls and Wnt3 are overexpressed in epithelial cells in colon cancers. (a) Validation of the specificity of the Wnt3 antibody. HCT116 cells were reverse transfected with sicontrol
Supplementary Figure S1 Evi/Wls and Wnt3 are overexpressed in epithelial cells in colon cancers. (a) Validation of the specificity of the Wnt3 antibody. HCT116 cells were reverse transfected with sicontrol
Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice
 Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice Supplementary Material Supplementary Methods Materials Spironolactone, aldosterone and β-glycerophosphate were
Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice Supplementary Material Supplementary Methods Materials Spironolactone, aldosterone and β-glycerophosphate were
TITLE: Induced Pluripotent Stem Cells as Potential Therapeutic Agents in NF1
 AD Award Number: W81XWH-10-1-0181 TITLE: Induced Pluripotent Stem Cells as Potential Therapeutic Agents in NF1 PRINCIPAL INVESTIGATOR: Jonathan Chernoff, M.D., Ph.D. CONTRACTING ORGANIZATION: Institute
AD Award Number: W81XWH-10-1-0181 TITLE: Induced Pluripotent Stem Cells as Potential Therapeutic Agents in NF1 PRINCIPAL INVESTIGATOR: Jonathan Chernoff, M.D., Ph.D. CONTRACTING ORGANIZATION: Institute
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well
 Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
a Lamtor1 (gene) b Lamtor1 (mrna) c WT Lamtor1 Lamtor1 flox Lamtor2 A.U. p = LysM-Cre Lamtor3 Lamtor4 Lamtor5 BMDMs: Φ WT Φ KO β-actin WT BMDMs
 a Lamtor (gene) b Lamtor (mrna) c BMDMs: Φ WT Φ KO Lamtor flox 8 bp LysM-Cre 93 bp..5 p =.4 WT BMDMs: Φ WT Φ KO Lamtor (protein) BMDMs: Φ WT Φ KO Lamtor 8 kda Lamtor 4 kda Lamtor3 4 kda Lamtor4 kda Lamtor5.5
a Lamtor (gene) b Lamtor (mrna) c BMDMs: Φ WT Φ KO Lamtor flox 8 bp LysM-Cre 93 bp..5 p =.4 WT BMDMs: Φ WT Φ KO Lamtor (protein) BMDMs: Φ WT Φ KO Lamtor 8 kda Lamtor 4 kda Lamtor3 4 kda Lamtor4 kda Lamtor5.5
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix
 Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix Authors Kerim B. Kaylan, 1,2 Viktoriya Ermilova, 1,2 Ravi Chandra Yada,
Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix Authors Kerim B. Kaylan, 1,2 Viktoriya Ermilova, 1,2 Ravi Chandra Yada,
Human induced Pluripotent Stem Cell (ipsc) Line: GM23225*B
 Certificate of Analysis NIGMS Human tic Cell Repository Human induced Pluripotent Stem Cell (ipsc) Line: GM23225*B Diagnosis Mutation Reprogramming method Publication(s) describing ipsc line establishment
Certificate of Analysis NIGMS Human tic Cell Repository Human induced Pluripotent Stem Cell (ipsc) Line: GM23225*B Diagnosis Mutation Reprogramming method Publication(s) describing ipsc line establishment
Nature Medicine: doi: /nm.4169
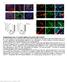 Supplementary Fig.1. EC-specific deletion of Ccm3 by Cdh5-CreERT2. a. mt/mg reporter mice were bred with Cdh5CreERT2 deleter mice followed by tamoxifen feeding from P1 to P3. mg expression was specifically
Supplementary Fig.1. EC-specific deletion of Ccm3 by Cdh5-CreERT2. a. mt/mg reporter mice were bred with Cdh5CreERT2 deleter mice followed by tamoxifen feeding from P1 to P3. mg expression was specifically
T-iPSC. Gra-iPSC. B-iPSC. TTF-iPSC. Supplementary Figure 1. Nature Biotechnology: doi: /nbt.1667
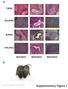 a T-iPSC Gra-iPSC B-iPSC TTF-iPSC Ectoderm Endoderm Mesoderm b Nature Biotechnology: doi:.38/nbt.667 Supplementary Figure Klf4 transgene expression Oct4 transgene expression.7 Fold GAPDH.6.5.4.3 Fold GAPDH
a T-iPSC Gra-iPSC B-iPSC TTF-iPSC Ectoderm Endoderm Mesoderm b Nature Biotechnology: doi:.38/nbt.667 Supplementary Figure Klf4 transgene expression Oct4 transgene expression.7 Fold GAPDH.6.5.4.3 Fold GAPDH
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Table S1. Primer sequences
 Table S1. Primer sequences Primers for quantitative PCR Tgf 1 Forward Tgf 1 Reverse Tgf 2Forward Tgf 2Reverse Tgf 3 Forward Tgf 3 Reverse Tgf r1 Forward Tgf r1 Reverse Tgf r2 Forward Tgf r2 Reverse Thbs1
Table S1. Primer sequences Primers for quantitative PCR Tgf 1 Forward Tgf 1 Reverse Tgf 2Forward Tgf 2Reverse Tgf 3 Forward Tgf 3 Reverse Tgf r1 Forward Tgf r1 Reverse Tgf r2 Forward Tgf r2 Reverse Thbs1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
H3K36me3 polyclonal antibody
 H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
Product Datasheet. Vinculin Antibody (hvin-1) NB Unit Size: 0.1 ml
 Product Datasheet Vinculin Antibody (hvin-1) NB600-1293 Unit Size: 0.1 ml Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Reviews: 1 Publications: 2 Protocols, Publications,
Product Datasheet Vinculin Antibody (hvin-1) NB600-1293 Unit Size: 0.1 ml Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Reviews: 1 Publications: 2 Protocols, Publications,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3164 Supplementary Figure 1 Validation of effective Gnas deletion and epithelial thickness. a, Representative genotyping in mice treated or not with tamoxifen to show Gnas deletion. To
DOI: 10.1038/ncb3164 Supplementary Figure 1 Validation of effective Gnas deletion and epithelial thickness. a, Representative genotyping in mice treated or not with tamoxifen to show Gnas deletion. To
Supplementary Data. Supplementary Table S1.
 Supplementary Data Supplementary Table S1. Primer Sequences for Real-Time Reverse Transcription-Polymerase Chain Reaction Gene Forward 5?3 Reverse 3?5 Housekeeping gene 36B4 TCCAGGCTTTGGGCATCA CTTTATCAGCTGCACATCACTCAGA
Supplementary Data Supplementary Table S1. Primer Sequences for Real-Time Reverse Transcription-Polymerase Chain Reaction Gene Forward 5?3 Reverse 3?5 Housekeeping gene 36B4 TCCAGGCTTTGGGCATCA CTTTATCAGCTGCACATCACTCAGA
Gene Expression Microarrays. For microarrays, purity of the RNA was further assessed by
 Supplemental Methods Gene Expression Microarrays. For microarrays, purity of the RNA was further assessed by an Agilent 2100 Bioanalyzer. 500 ng of RNA was reverse transcribed into crna and biotin-utp
Supplemental Methods Gene Expression Microarrays. For microarrays, purity of the RNA was further assessed by an Agilent 2100 Bioanalyzer. 500 ng of RNA was reverse transcribed into crna and biotin-utp
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in
 Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
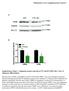 Merkestein et al. Supplementary Figure 1 A PLIN WT FTO KO 72 kda FABP4 17 kda HSC70 72 kda B Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
Merkestein et al. Supplementary Figure 1 A PLIN WT FTO KO 72 kda FABP4 17 kda HSC70 72 kda B Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
Influencing cell fate decisions using physical and chemical cues
 Influencing cell fate decisions using physical and chemical cues by Junren Sia A dissertation submitted in partial satisfaction of the requirements for the degree of Joint Doctor of Philosophy with the
Influencing cell fate decisions using physical and chemical cues by Junren Sia A dissertation submitted in partial satisfaction of the requirements for the degree of Joint Doctor of Philosophy with the
Supplementary Figures and Legends.
 Supplementary Figures and Legends. Supplementary Figure 1: Impact of injury on Rb1 and PPARϒ expression. Following ipsilateral axotomy injury, adult DRG expression of Rb1 mrna declined (*p
Supplementary Figures and Legends. Supplementary Figure 1: Impact of injury on Rb1 and PPARϒ expression. Following ipsilateral axotomy injury, adult DRG expression of Rb1 mrna declined (*p
RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the
 Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
