Supplementary Figures
|
|
|
- Owen Robbins
- 6 years ago
- Views:
Transcription
1 Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID).
2 Supplementary Figure 2. MCF7-SIX1 tumors display an increased number of intra-tumoral lymphatic vessels when compared to MCF7-Ctrl tumors in NOD/SCID mice. Immunohistochemical staining was used to detect Lyve-1 in MCF7-Ctrl and MCF7-SIX1 tumors and sections were scored in a blinded manner by a pathologist. Error bars represent the standard error of the mean. Asterisks denote significant difference from control group **, P<0.01.
3 Supplementary Figure 3. VEGF-A and VEGF-D expression in MCF7-Ctrl and MCF7-SIX1 cells was detected by real-time PCR. Values represent the level of VEGF-A or VEGF-D transcript normalized to the level of the loading control CYCLOPHILIN B. Error bars represent the standard error of the mean.
4 Supplementary Figure 4. Examination of SIX1 binding to different sites on the VEGF-C promoter using the electrophoretic mobility shift assay (EMSA). Purified protein SIX1 and its cofactor EYA were incubated with biotin labeled 30 base pair double stranded oligonucleotides spanning the predicted SIX1 binding sites at -2034, -1755, and alone, or in the presence of unlabeled competitor DNA or mutated (scrambled the core sequence) unlabeled competitor DNA in a 200-fold excess. The EMSA was performed according to the manufacturer s protocol (Thermo scientific).
5 Supplementary Figure 5. (A) Primary tumor growth in MCF7-SIX1-scramble KD and MCF7-SIX1-VEGFC KD1/2 (two shrnas were used and combined in the figure). Tumor size in the animals was measured using calipers, and calculated according to the formula V=1/2(W) 2 (L). (B) Left panel shows representative Zsgreen positive lung from MCF7-SIX1-scramble KD tumor bearing mouse. Right panel shows histologic confirmation of lung metastasis by H& E staining.
6 Supplementary Figure 6. (A) Representative picture showing SIX1 expression levels detected in MCF7-Ctrl and MCF7-SIX1 cells by immunocytochemistry (ICC). (B) Western blotting was performed to detect SIX1 expression in MCF7-SIX1 cells compared to endogenous SIX1 expression in numerous breast cancer cell lines.
7 Supplementary Figure 7. Vegf-c expression is critical for metastasis in the 66cl4 mammary carcinoma model. (A) Detection of Vegf-c expression by real-time PCR in scramble control (scram) and two Vegf-c knockdown lines (Vegf-c KD1/2). Two different shrnas (combined as Vegf-c KD1/2) were used to knockdown Vegf-c in
8 66cl4 cells, and pooled populations were labeled with luciferase for in vivo imagining.(b) Secreted Vegf-c was measured by ELISA in conditioned medium from 66cl4-scramble or 66cl4-Vegf-c KD cells. (C)Bioluminescent imaging of Balb/c mice 40 days after injection of 66cl4-scramble or 66cl4-Vegf-c KD cells into the 4 th mammary fat pad. The luminescence signal is represented by the overlaid false-color image and the intensity of the signal is indicated by the scale. The anesthetized mice were imaged with the IVIS200, 10 minutes after intraperitoneal injection of D-luciferin. Metastatic region is highlight by the yellow box. (D) Primary tumor growth in mice bearing 66cl4-scramble or Vegf-c KD tumors. Tumor size in the animals was measured using calipers according to the formula V=1/2(W) 2 (L). Error bars represent the standard error of the mean. Asterisks denote significant difference from control group. **, P<0.01; ***, P<0.001
9 Supplementary Figure 8. (A) Vegf-a, Vegf-c and Vegf-d expression in 67NR and 66cl4 cells was detected by real-time PCR. (B)Vegf-a and Vegf-d expression in 66cl4-scramble and 66cl4-Six1 KD cells was detected by real-time PCR. Values represent the level of Vegf-a or Vegf-d transcript normalized to the level of the control Cyclophilin b mrna. Error bars represent the standard error of the mean.
10 Supplementary Figure 9. Chromatin immunoprecipitation (ChIP) was performed to detect endogenous Six1 presence on the Vegf-c promoter in 66cl4 cells.
11 Supplementary Figure 10. (A) Representative picture showing blood vessel (angiogenesis) staining by MECA-32 and CD31 in a 66cl4 tumor (Left panel). Slidebook software was utilized to transform the signal from double stained regions (Right panel). (B) Quantification of blood vessels in 66cl4-scramble and 66cl4-Six1 KD1/2 tumors. Five individual tumor sections were stained in each group.
12 Supplementary Figure 11. Primary tumor growth in mice bearing 66cl4-scramble or 66cl4-Six1 KD tumors. Tumor size in the animals was measured using calipers and calculated using the formula V=1/2(W) 2 (L). Error bars represent the standard error of the mean. Asterisks denote significant difference from control group ***, P<0.001.
13 Supplementary Figure 12. (A) Tumor sizes of 66cl4-scramble at Day 46 and 66cl4-Six KD at Day 54. Tumor size in the animals was measured using calipers and calculated using the formula V=1/2(W) 2 (L). (B) Representative pictures showing the lungs from mice bearing 66cl4-scramble and 66cl4-Six1 KD1 tumors when mice had tumors of comparable sizes.
14 Supplementary Figure 13. (A) Six1 expression was measured by real-time PCR in Six1 KD1 cells retrieved from the lungs of two individual 66cl4-Six1 KD1 tumor bearing mice, and 66cl4-scramble control cells were retrieved from the lung of one animal. (B) Six2 expression was measured by real-time PCR in Six1 KD1 cells retrieved from the lungs of two individual 66cl4-Six1 KD1 tumor bearing mice, and from 66cl4-scramble control cells retrieved from the lung of one animal.
15 Supplementary Figure 14. (A) Representative bioluminescent imaging of Balb/c mice injected with 66cl4-scramble, 66cl4-Six1 KD, and 66cl4-Six1 KD+Vegf-c cells into the 4 th mammary fat pad. (B) Quantitation of the luminescent signal in primary tumors from 66cl4-scramble, 66cl4-Six1 KD and 66cl4-Six1 KD+Vegf-c groups. Error bars represent the standard error of the mean. Asterisks denote significant difference **, P<0.01.
16 Supplementary Figure 15. SIX1 and VEGF-C expression are correlated in ovarian cancer cell lines.
17 Supplementary Figure 16. Immunohistochemical staining for SIX1 and VEGF-C on 110 cases of invasive ductal carcinoma were quantified by a pathologist and their correlation in breast tumors was analyzed by Chi-square test.
18 Supplementary Figure 17. (A) Vegf-c levels were detected by Western blotting in WT and Six1 knock out mouse embryonic fibroblast cells (MEF). (B) SIX1 and VEGF-C expression in MC12A-Ctrl and MC12A-SIX1 cells was detected by real-time PCR. Values represent the level of SIX1 or VEGF-C transcript normalized to the level of the control CYCLOPHILIN B. Error bars represent the standard error of the mean. Asterisks denote significant difference from control group ***, P<0.001.
19 Supplementary Methods Cell lines MCF7-Ctrl and MCF7-SIX1 stable cell lines were generated as previously described 1. MCF7-Ctrl and MCF-SIX1 cells used within the study are clonal isolates, and three clonal isolates from each line (MCF7-Ctrl and MCF7-SIX1) were used in our experiments. The results of all three clonal isolates in each group are combined to show the groups as MCF7-Ctrl and MCF7-SIX1. MCF7-Ctrl and MCF7-SIX1 cells were fluorescently tagged with Zsgreen to enhance our ability to follow metastasis. To generate VEGF-C knockdown in MCF7-SIX1 cells, shrnas against VEGF-C were purchased from Open Biosystems and delivered retrovirally according to the manufacture s protocol. Cells containing the knockdown constructs were selected using puromycin (2.5ug/ml), and the two stable knockdown clones that most efficiently reduced VEGF-C levels were selected for subsequent studies. The 67NR and 66cl4 mammary carcinoma cell lines were generously provided by Fred Miller 2. 66cl4 cells were tagged with luciferase for in vivo imaging of metastases. Vegf-c and Six1 were knocked down in 66cl4 cells using two different shrnas from Open Biosystems that were lentivirally delivered according to the manufacturer s protocol. Stable knockdown cells were selected with puromycin. To restore Vegf-c expression, a Vegf-c expression vector (CMV-sport6) was purchased from Open Biosystems and
20 the full length cdna was cloned into pcdna3.1 (Invitrogen) and transfected into 66cl4 Six1 knockdown cells. Stable clones with Vegf-c restored were selected via treatment with neomycin (400ug/ml). In vitro assays TaqMan Analysis: TaqMan primers used against VEGF-A, VEGF-C, VEGF-D, and Lyve-1 were purchased from Applied Biosystems. All analyses were performed using the Bio-Rad CFX-96 real time system machine and iscript cdna synthesis kits (Bio-Rad) as well as TaqMan Fast universal PCR master mix (Applied Biosystems) according to the manufacturer s recommended protocols. Luciferase reporter assay: Using a Fugene lipid-based transfection reagent (Roche), MCF7 Cells were transfected with a pgl3-vegf-c promoter luciferase construct (0.5ug) (a kind gift from Kari Alitalo) along with increasing amounts of pcdna3.1-six1 (up to 0.9ug) and a constant amount of pcdna-eya2 (0.2ug). A Renilla luciferase construct was also transfected as an internal control. After transfection, the cells were incubated for 48 hours and then harvested in passive lysis buffer (Promega). Firefly and Renilla luciferase activity were measured using the Dual Luciferase Reporter Assay System (Promega) on a Modulus Luminometer (Turner BioSystems). Luciferase activity was normalized to renilla activity, then the reading was analyzed by the following equation:
21 (SIX1/EYA+pVEGFC)-(SIX1/EYA+pGL3)/(pcDNA+pVEGFC)-(pcDNA+PGL3). Enzyme-linked immunosorbent assay (ELISA): Cells were cultured under serum starvation for 48 hours and medium was collected and concentrated on an Amicon Ultra (Millipore) column. Equal amounts of protein were loaded and measured by ELISA plate (ELISA Tech) according to the manufacturer s recommended protocol. The VEGF-C antibody from R&D systems (BAF752 and MAB752) was used (information provided by ELISA Tech). Chromatin immunoprecipitation (ChIP): MCF7 cells were transfected with pcdna3.1-six1 and pcdna3.1-eya2. After cross-linking and sonication, Protein-DNA complexes were precipitated with a SIX1 specific antibody made as previously described 3 as well as a control Rabbit IgG antibody (cell signaling). Real-time PCR was utilized to amplify regions around predicted SIX1 binding sites, as well as one upstream region with no predicted SIX1 binding site (no ATCCTGA or TGATAC core sequence) on the VEGF-C promoter as a negative control. 66cl4 cells were used to determine if endogenous Six1 binds to the mouse Vegf-c promoter. ChIP procedures are as described above except that no transfections were performed in the 66cl4 cells. The y-axis (enrichment relative to input) is a the percentage of VEGF-C promoter IP ed using the Six1 antibody or the percentage of the VEGF-C promoter IP ed using a control IgG antibody normalized to the amount of VEGF-C promoter
22 region found in the input DNA. Histology, immunohistochemistry and immunofluorescence Tumors, lymph nodes and lungs from animals were fixed in 4% paraformaldehyde, embedded in paraffin, and cut into 5-μm sections. Sections were stained with H&E to confirm metastases as previously described 4. Primary antibodies against VEGF-C (1:200, ZYMED laboratories) and Lyve-1 (1:200, Angiobio), MECA32 (1:50, BD), Podoplanin (1:800, developmental studies hybridoma bank), CD31 (1:50, Abcam) were used on tissue sections. Tumor arrays (US Biomax) were stained using an anti-six1 antibody (1:100; Atlas antibodies) and an anti-vegf-c antibody (1:200, ZYMED laboratories). For immunofluorescence staining, corresponding secondary antibodies were labeled with AlexaFluor488 or AlexaFluor594 (Invitrogen) and images were captured using Nikon Diaphot fluorescence microscope and analyzed by Slidebook software (Intelligent Imaging Innovations, Inc., Denver, CO); images were converted to TIFF files and processed by Photoshop. ShRNA hairpin sequence used to knock down VEGF-C and Six1. shrna target Hairpin sequence VEGFC (Homo sapiens) TGCTGTTGACAGTGAGCGCCGCGACAAACACCTTCTTTAATAGTGAAGC CACAGATGTATTAAAGAAGGTGTTTGTCGCGATGCCTACTGCCTCGGA
23 VEGFC (Homo sapiens) TGCTGTTGACAGTGAGCGCGCACATTATAATACAGAGATCTAGTGAAGCC ACAGATGTAGATCTCTGTATTATAATGTGCTTGCCTACTGCCTCGGA VEGFC (Mus musculus) CCGGCCCAAGTCTGTGTTTATTGAACTCGAGTTCAATAAACACAGACTTG GGTTTTTG VEGFC (Mus musculus) CCGGCCCTAATTCATGTGGAGCCAACTCGAGTTGGCTCCACATGAATTAG GGTTTTTG Six1 (Mus musculus) CCGGAGCTCAAACTATTCTCTTCCACTCGAGTGGAAGAGAATAGTTTGA GCTTTTTTG Six1 (Mus musculus) CCGGCTCCAACAAGCAGAATCAACTCTCGAGAGTTGATTCTGCTTGTTG GAGTTTTTG Color Codes: sense loop antisense (Sequence obtained from Open Biosystems) Primer sequence designed for the detection of predicted SIX1 binding sites on human VEGF-C promoter. Detected Primer sequence region -3447~ CTGTTTGCCAAAGGAAGCAC3 (forward) 5 CCCCATCTCTGCAATCAATC3 (reverse)
24 -2076~ GTTGGAAATGCCTGTGGTTC3 (forward) 5 GCCTTTCTGGCTTAACTGGTC3 (reverse) -1776~ AGCAGGAAGACAGCAAGAGAGTGA3 (forward) 5 GACTCGGTCCATGATCTAGCCTGT3 (reverse) -1547~ GTGAGTCAACACTGTGATTTGGCTGC3 (forward) 5 GCATGCAGAATGGACAGTATGCCTTTG3 (reverse) -673~ TAGCATCCATCCCAACAGC3 (forward) 5 GGTCCCCTCTCTCCCTTG3 (reverse) -4486~ TCAAGAGGCCTCAGTGATTTG3 (forward) 5 TCGGGTGTACAACAACTTGG3 (reverse)
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Convoy TM Transfection Reagent
 Convoy TM Transfection Reagent Catalog No.11103 0.25ml (40-80 transfections in 35mm dishes) Catalog No.11105 0.5 ml (80-165 transfections in 35mm dishes) Catalog No.11110 1.0 ml (165-330 transfections
Convoy TM Transfection Reagent Catalog No.11103 0.25ml (40-80 transfections in 35mm dishes) Catalog No.11105 0.5 ml (80-165 transfections in 35mm dishes) Catalog No.11110 1.0 ml (165-330 transfections
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 BALB/c LYVE1-deficient mice exhibited reduced lymphatic trafficking of all DC subsets after oxazolone-induced sensitization. (a) Schematic overview of the mouse skin oxazolone contact
Supplementary Figure 1 BALB/c LYVE1-deficient mice exhibited reduced lymphatic trafficking of all DC subsets after oxazolone-induced sensitization. (a) Schematic overview of the mouse skin oxazolone contact
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde
 Supplementary text Supplementary materials and methods Histopathological examination Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin wax with
Supplementary text Supplementary materials and methods Histopathological examination Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin wax with
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Methods of Biomaterials Testing Lesson 3-5. Biochemical Methods - Molecular Biology -
 Methods of Biomaterials Testing Lesson 3-5 Biochemical Methods - Molecular Biology - Chromosomes in the Cell Nucleus DNA in the Chromosome Deoxyribonucleic Acid (DNA) DNA has double-helix structure The
Methods of Biomaterials Testing Lesson 3-5 Biochemical Methods - Molecular Biology - Chromosomes in the Cell Nucleus DNA in the Chromosome Deoxyribonucleic Acid (DNA) DNA has double-helix structure The
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
Tropix Chemiluminescent Kits and Reagents For Cell Biology Applications
 PRODUCT FAMILY BULLETIN Tropix Chemiluminescent Kits and Reagents Tropix Chemiluminescent Kits and Reagents For Cell Biology Applications Introduction to Chemiluminescence Chemiluminescence is the conversion
PRODUCT FAMILY BULLETIN Tropix Chemiluminescent Kits and Reagents Tropix Chemiluminescent Kits and Reagents For Cell Biology Applications Introduction to Chemiluminescence Chemiluminescence is the conversion
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on
 Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
3.1.4 DNA Microarray Technology
 3.1.4 DNA Microarray Technology Scientists have discovered that one of the differences between healthy and cancer is which genes are turned on in each. Scientists can compare the gene expression patterns
3.1.4 DNA Microarray Technology Scientists have discovered that one of the differences between healthy and cancer is which genes are turned on in each. Scientists can compare the gene expression patterns
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/323/5910/124/dc1 Supporting Online Material for Regulation of Neuronal Survival Factor MEF2D by Chaperone-Mediated Autophagy Qian Yang, Hua She, Marla Gearing, Emanuela
www.sciencemag.org/cgi/content/full/323/5910/124/dc1 Supporting Online Material for Regulation of Neuronal Survival Factor MEF2D by Chaperone-Mediated Autophagy Qian Yang, Hua She, Marla Gearing, Emanuela
special offers from your protein biology resource
 special offers from your protein biology resource Pop open your cells, extract your proteins, purify, quantify and express them. Seeking knowledge about proteins with Thermo Scientific Protein Research
special offers from your protein biology resource Pop open your cells, extract your proteins, purify, quantify and express them. Seeking knowledge about proteins with Thermo Scientific Protein Research
Plasmid DNA transfection of SW480 human colorectal cancer cells with the Biontex K2 Transfection System
 Plasmid DNA transfection of human colorectal cancer cells with the Biontex K2 Transfection System Stephanie Hehlgans and Franz Rödel, Department of Radiotherapy and Oncology, Goethe- University Frankfurt,
Plasmid DNA transfection of human colorectal cancer cells with the Biontex K2 Transfection System Stephanie Hehlgans and Franz Rödel, Department of Radiotherapy and Oncology, Goethe- University Frankfurt,
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Toll Receptor-Mediated Hippo Signaling Controls Innate Immunity in Drosophila
 Cell Supplemental Information Toll Receptor-Mediated Hippo Signaling Controls Innate Immunity in Drosophila Bo Liu, Yonggang Zheng, Feng Yin, Jianzhong Yu, Neal Silverman, and Duojia Pan Supplemental Experimental
Cell Supplemental Information Toll Receptor-Mediated Hippo Signaling Controls Innate Immunity in Drosophila Bo Liu, Yonggang Zheng, Feng Yin, Jianzhong Yu, Neal Silverman, and Duojia Pan Supplemental Experimental
ENCODE RBP Antibody Characterization Guidelines
 ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
Supplemental Data. LMO4 Controls the Balance between Excitatory. and Inhibitory Spinal V2 Interneurons
 Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Sensitivity vs Specificity
 Viral Detection Animal Inoculation Culturing the Virus Definitive Length of time Serology Detecting antibodies to the infectious agent Detecting Viral Proteins Western Blot ELISA Detecting the Viral Genome
Viral Detection Animal Inoculation Culturing the Virus Definitive Length of time Serology Detecting antibodies to the infectious agent Detecting Viral Proteins Western Blot ELISA Detecting the Viral Genome
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
Real-time PCR. TaqMan Protein Assays. Unlock the power of real-time PCR for protein analysis
 Real-time PCR TaqMan Protein Assays Unlock the power of real-time PCR for protein analysis I can use my real-time PCR instrument to quantitate protein? Do protein levels correlate with related mrna levels
Real-time PCR TaqMan Protein Assays Unlock the power of real-time PCR for protein analysis I can use my real-time PCR instrument to quantitate protein? Do protein levels correlate with related mrna levels
in-situ PCR Presented for: Presented by: Date:
 in-situ PCR Presented for: Presented by: Date: 2 in situ Hybridization - Definition in situ PCR is a method in which the polymerase chain reaction actually takes place in the cell on a slide, and the product
in-situ PCR Presented for: Presented by: Date: 2 in situ Hybridization - Definition in situ PCR is a method in which the polymerase chain reaction actually takes place in the cell on a slide, and the product
SureSilencing sirna Array Technology Overview
 SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
Custom Antibodies Services. GeneCust Europe. GeneCust Europe
 GeneCust Europe Laboratoire de Biotechnologie du Luxembourg S.A. 2 route de Remich L-5690 Ellange Luxembourg Tél. : +352 27620411 Fax : +352 27620412 Email : info@genecust.com Web : www.genecust.com Custom
GeneCust Europe Laboratoire de Biotechnologie du Luxembourg S.A. 2 route de Remich L-5690 Ellange Luxembourg Tél. : +352 27620411 Fax : +352 27620412 Email : info@genecust.com Web : www.genecust.com Custom
Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles
 Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles Long-term gene silencing shrna-specific design algorithm High titer, purified particles Thermo Scientific Dharmacon SMARTvector shrna
Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles Long-term gene silencing shrna-specific design algorithm High titer, purified particles Thermo Scientific Dharmacon SMARTvector shrna
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets.
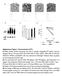 Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Globo H Monoclonal Antibody (VK9), ebioscience Catalog Number Product data sheet
 Website: thermofisher.com/ebioscience Customer Service (US): 1-888-999-1371 thermofisher.com/contactus Globo H Monoclonal Antibody (VK9), ebioscience Catalog Number 14-9700-82 Product data sheet Details
Website: thermofisher.com/ebioscience Customer Service (US): 1-888-999-1371 thermofisher.com/contactus Globo H Monoclonal Antibody (VK9), ebioscience Catalog Number 14-9700-82 Product data sheet Details
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor
 Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit
 Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Jae Myoung Suh, Daniel Zeve, Renee McKay, Jin Seo, Zack Salo, Robert Li, Michael Wang, and Jonathan M. Graff
 Cell Metabolism, Volume 6 Supplemental Data Adipose Is a Conserved, Dosage-Sensitive Antiobesity Gene Jae Myoung Suh, Daniel Zeve, Renee McKay, Jin Seo, Zack Salo, Robert Li, Michael Wang, and Jonathan
Cell Metabolism, Volume 6 Supplemental Data Adipose Is a Conserved, Dosage-Sensitive Antiobesity Gene Jae Myoung Suh, Daniel Zeve, Renee McKay, Jin Seo, Zack Salo, Robert Li, Michael Wang, and Jonathan
Purification Kits. Fast and Convenient PROSEP -A and PROSEP-G Spin Column Kits for Antibody Purification DATA SHEET
 Â Montage Antibody Purification Kits Fast and Convenient PROSEP -A and PROSEP-G Spin Column Kits for Antibody Purification DATA SHEET Available with immobilized Protein A or Protein G Easy-to-use Antibody
 Montage Antibody Purification Kits Fast and Convenient PROSEP -A and PROSEP-G Spin Column Kits for Antibody Purification DATA SHEET Available with immobilized Protein A or Protein G Easy-to-use Antibody
SOD1 as a Molecular Switch for Initiating the Homeostatic ER Stress Response under Zinc Deficiency
 Molecular Cell, Volume 52 Supplemental Information SOD1 as a Molecular Switch for Initiating the Homeostatic ER Stress Response under Zinc Deficiency Kengo Homma, Takao Fujisawa, Naomi Tsuburaya, Namiko
Molecular Cell, Volume 52 Supplemental Information SOD1 as a Molecular Switch for Initiating the Homeostatic ER Stress Response under Zinc Deficiency Kengo Homma, Takao Fujisawa, Naomi Tsuburaya, Namiko
Comparison of Commercial Transfection Reagents: Cell line optimized transfection kits for in vitro cancer research.
 Comparison of Commercial Transfection Reagents: Cell line optimized transfection kits for in vitro cancer research. by Altogen Labs, 11200 Manchaca Road, Suite 203 Austin TX 78748 USA Tel. (512) 433-6177
Comparison of Commercial Transfection Reagents: Cell line optimized transfection kits for in vitro cancer research. by Altogen Labs, 11200 Manchaca Road, Suite 203 Austin TX 78748 USA Tel. (512) 433-6177
Galina Gabriely, Ph.D. BWH/HMS
 Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
T7-Based RNA Amplification Protocol (in progress)
 T7-Based RNA Amplification Protocol (in progress) Jacqueline Ann Lopez (modifications) Amy Cash & Justen Andrews INTRODUCTION T7 RNA Amplification, a technique originally developed in the laboratory of
T7-Based RNA Amplification Protocol (in progress) Jacqueline Ann Lopez (modifications) Amy Cash & Justen Andrews INTRODUCTION T7 RNA Amplification, a technique originally developed in the laboratory of
FFPE DNA Extraction kit
 FFPE DNA Extraction kit Instruction Manual FFPE DNA Extraction kit Cat. No. C20000030, Format 50 rxns Version 1 / 14.08.13 Content Introduction 4 Overview of FFPE DNA extraction workflow 4 Kit contents
FFPE DNA Extraction kit Instruction Manual FFPE DNA Extraction kit Cat. No. C20000030, Format 50 rxns Version 1 / 14.08.13 Content Introduction 4 Overview of FFPE DNA extraction workflow 4 Kit contents
Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo
 Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo Rampyari Raja Walia and Bakhos A. Tannous 1 2 1 Pluristem Innovations, 1453
Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo Rampyari Raja Walia and Bakhos A. Tannous 1 2 1 Pluristem Innovations, 1453
Myers Lab ChIP-seq Protocol v Modified January 10, 2014
 Myers Lab ChIP-seq Protocol V011014 1 Contact information: Dr. Florencia Pauli Behn HudsonAlpha Institute for Biotechnology 601 Genome Way Huntsville, AL 35806 Telephone: 256-327-5229 Email: fpauli@hudsonalpha.org
Myers Lab ChIP-seq Protocol V011014 1 Contact information: Dr. Florencia Pauli Behn HudsonAlpha Institute for Biotechnology 601 Genome Way Huntsville, AL 35806 Telephone: 256-327-5229 Email: fpauli@hudsonalpha.org
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing
 Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing Chase L. Beisel, Yvonne Y. Chen, Stephanie J. Culler, Kevin G. Hoff, & Christina
Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing Chase L. Beisel, Yvonne Y. Chen, Stephanie J. Culler, Kevin G. Hoff, & Christina
Title: Production and characterisation of monoclonal antibodies against RAI3 and its expression in human breast cancer
 Author's response to reviews Title: Production and characterisation of monoclonal antibodies against RAI3 and its expression in human breast cancer Authors: Hannah Jörißen (hannah.joerissen@molbiotech.rwth-aachen.de)
Author's response to reviews Title: Production and characterisation of monoclonal antibodies against RAI3 and its expression in human breast cancer Authors: Hannah Jörißen (hannah.joerissen@molbiotech.rwth-aachen.de)
To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well
 Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
Supplementary Figure 1
 Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Technical tips Session 5
 Technical tips Session 5 Chromatine Immunoprecipitation (ChIP): This is a powerful in vivo method to quantitate interaction of proteins associated with specific regions of the genome. It involves the immunoprecipitation
Technical tips Session 5 Chromatine Immunoprecipitation (ChIP): This is a powerful in vivo method to quantitate interaction of proteins associated with specific regions of the genome. It involves the immunoprecipitation
Supplementary Figure 1
 Supplementary Figure A Name Location Start position End position Oct4P Chr 3 29433673 29435 Annotation Mus musculus predicted gene 572 (Gm572), noncoding RNA RefSeq accession number NR_33594. Oct4P2 Chr
Supplementary Figure A Name Location Start position End position Oct4P Chr 3 29433673 29435 Annotation Mus musculus predicted gene 572 (Gm572), noncoding RNA RefSeq accession number NR_33594. Oct4P2 Chr
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and
 Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Generation of ips-derived model cells for analyses of hair shaft differentiation
 Supplementary Material Generation of ips-derived model cells for analyses of hair shaft differentiation Takumi Kido, Tomoatsu Horigome, Minori Uda, Naoki Adachi, Yohei Hirai Department of Biomedical Chemistry,
Supplementary Material Generation of ips-derived model cells for analyses of hair shaft differentiation Takumi Kido, Tomoatsu Horigome, Minori Uda, Naoki Adachi, Yohei Hirai Department of Biomedical Chemistry,
Technical Review. Real time PCR
 Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
TransIT-TKO Transfection Reagent
 Quick Reference Protocol, MSDS and Certificate of Analysis available at mirusbio.com/2150 INTRODUCTION TransIT-TKO is a broad spectrum sirna transfection reagent that enables high efficiency sirna delivery
Quick Reference Protocol, MSDS and Certificate of Analysis available at mirusbio.com/2150 INTRODUCTION TransIT-TKO is a broad spectrum sirna transfection reagent that enables high efficiency sirna delivery
PV92 PCR Bio Informatics
 Purpose of PCR Chromosome 16 PV92 PV92 PCR Bio Informatics Alu insert, PV92 locus, chromosome 16 Introduce the polymerase chain reaction (PCR) technique Apply PCR to population genetics Directly measure
Purpose of PCR Chromosome 16 PV92 PV92 PCR Bio Informatics Alu insert, PV92 locus, chromosome 16 Introduce the polymerase chain reaction (PCR) technique Apply PCR to population genetics Directly measure
RayBio Human NF-κB p65 Transcription Factor Activity Assay Kit
 RayBio Human NF-κB p65 Transcription Factor Activity Assay Kit Catalog #: TFEH-p65 User Manual Mar 13, 2017 3607 Parkway Lane, Suite 200 Norcross, GA 30092 Tel: 1-888-494-8555 (Toll Free) or 770-729-2992,
RayBio Human NF-κB p65 Transcription Factor Activity Assay Kit Catalog #: TFEH-p65 User Manual Mar 13, 2017 3607 Parkway Lane, Suite 200 Norcross, GA 30092 Tel: 1-888-494-8555 (Toll Free) or 770-729-2992,
GENE EXPRESSION AND EXPERIMENTAL READOUTS ON THE TURNER BIOSYSTEMS FAMILY OF INSTRUMENTS
 GENE EXPRESSION AND EXPERIMENTAL READOUTS ON THE TURNER BIOSYSTEMS FAMILY OF INSTRUMENTS Prepared by: Jeff Quast, Associate Product Manager Keywords: Gene Expression, Functional Gene Product, Protein Synthesis,
GENE EXPRESSION AND EXPERIMENTAL READOUTS ON THE TURNER BIOSYSTEMS FAMILY OF INSTRUMENTS Prepared by: Jeff Quast, Associate Product Manager Keywords: Gene Expression, Functional Gene Product, Protein Synthesis,
Mir-X mirna First-Strand Synthesis and SYBR qrt-pcr
 User Manual Mir-X mirna First-Strand Synthesis and SYBR qrt-pcr User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories,
User Manual Mir-X mirna First-Strand Synthesis and SYBR qrt-pcr User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories,
SuperScript IV Reverse Transcriptase as a better alternative to AMV-based enzymes
 WHITE PAPER SuperScript IV Reverse Transcriptase SuperScript IV Reverse Transcriptase as a better alternative to AMV-based enzymes Abstract Reverse transcriptases (RTs) from avian myeloblastosis virus
WHITE PAPER SuperScript IV Reverse Transcriptase SuperScript IV Reverse Transcriptase as a better alternative to AMV-based enzymes Abstract Reverse transcriptases (RTs) from avian myeloblastosis virus
In vitro Human Umbilical Vein Endothelial Cells (HUVEC) Tube-formation Assay. Josephine MY Ko and Maria Li Lung *
 In vitro Human Umbilical Vein Endothelial Cells (HUVEC) Tube-formation Assay Josephine MY Ko and Maria Li Lung * Clinical Oncology Department, The Univerisity of Hong Kong, Hong Kong, Hong Kong SAR *For
In vitro Human Umbilical Vein Endothelial Cells (HUVEC) Tube-formation Assay Josephine MY Ko and Maria Li Lung * Clinical Oncology Department, The Univerisity of Hong Kong, Hong Kong, Hong Kong SAR *For
Supplemental Data. Regulating Gene Expression. through RNA Nuclear Retention
 Supplemental Data Regulating Gene Expression through RNA Nuclear Retention Kannanganattu V. Prasanth, Supriya G. Prasanth, Zhenyu Xuan, Stephen Hearn, Susan M. Freier, C. Frank Bennett, Michael Q. Zhang,
Supplemental Data Regulating Gene Expression through RNA Nuclear Retention Kannanganattu V. Prasanth, Supriya G. Prasanth, Zhenyu Xuan, Stephen Hearn, Susan M. Freier, C. Frank Bennett, Michael Q. Zhang,
Tandem E2F Binding Sites in the Promoter of the p107 Cell Cycle Regulator Control p107 Expression and Its Cellular Functions
 Tandem E2F Binding Sites in the Promoter of the p107 Cell Cycle Regulator Control p107 Expression and Its Cellular Functions Deborah L. Burkhart 1,2, Stacey E. Wirt 1,2, Anne-Flore Zmoos 1, Michael S.
Tandem E2F Binding Sites in the Promoter of the p107 Cell Cycle Regulator Control p107 Expression and Its Cellular Functions Deborah L. Burkhart 1,2, Stacey E. Wirt 1,2, Anne-Flore Zmoos 1, Michael S.
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna
 Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Development of a Tissue Slice Culture Model for Prostate Cancer
 Development of a Tissue Slice Culture Model for Prostate Cancer Hanneke van Zoggel, PhD Experimental Urology Erasmus MC ; JNI, Be 330 j.vanzoggel@erasmusmc.nl 3D workshop 23/24-09-2013 Create novel models
Development of a Tissue Slice Culture Model for Prostate Cancer Hanneke van Zoggel, PhD Experimental Urology Erasmus MC ; JNI, Be 330 j.vanzoggel@erasmusmc.nl 3D workshop 23/24-09-2013 Create novel models
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
IMMUNOPRECIPITATION (IP)
 1 IMMUNOPRECIPITATION (IP) Overview and Technical Tips 2 CONTENTS 3 7 8 9 12 13 17 18 19 20 Introduction Factors Influencing IP General Protocol Modifications Of IP Protocols Troubleshooting Contact Us
1 IMMUNOPRECIPITATION (IP) Overview and Technical Tips 2 CONTENTS 3 7 8 9 12 13 17 18 19 20 Introduction Factors Influencing IP General Protocol Modifications Of IP Protocols Troubleshooting Contact Us
Data and Metadata Models Recommendations Version 1.2 Developed by the IHEC Metadata Standards Workgroup
 Data and Metadata Models Recommendations Version 1.2 Developed by the IHEC Metadata Standards Workgroup 1. Introduction The data produced by IHEC is illustrated in Figure 1. Figure 1. The space of epigenomic
Data and Metadata Models Recommendations Version 1.2 Developed by the IHEC Metadata Standards Workgroup 1. Introduction The data produced by IHEC is illustrated in Figure 1. Figure 1. The space of epigenomic
Reverse transcription-pcr (rt-pcr) Dr. Hani Alhadrami
 Reverse transcription-pcr (rt-pcr) Dr. Hani Alhadrami hanialhadrami@kau.edu.sa www.hanialhadrami.kau.edu.sa Overview Several techniques are available to detect and analyse RNA. Examples of these techniques
Reverse transcription-pcr (rt-pcr) Dr. Hani Alhadrami hanialhadrami@kau.edu.sa www.hanialhadrami.kau.edu.sa Overview Several techniques are available to detect and analyse RNA. Examples of these techniques
Quick reference guide
 Quick reference guide Our Invitrogen GeneArt CRISPR Search and Design Tool allows you to search our database of >600,000 predesigned CRISPR guide RNA (grna) sequences or analyze your sequence of interest
Quick reference guide Our Invitrogen GeneArt CRISPR Search and Design Tool allows you to search our database of >600,000 predesigned CRISPR guide RNA (grna) sequences or analyze your sequence of interest
FEBRUARY 12, 2010 VOLUME 285 NUMBER 7 JOURNAL OF BIOLOGICAL CHEMISTRY 4847
 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 285, NO. 7, pp. 4847 4858, February 12, 2010 2010 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. The Tumor Suppressor
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 285, NO. 7, pp. 4847 4858, February 12, 2010 2010 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. The Tumor Suppressor
Supplementary Information
 Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Materials and Methods
 Materials and Methods Construction of noxa / mice and genotyping The targeting vector (see Fig. S) was prepared from a C57BL/6 DNA λ phage library (Stratagene) by replacing a 2.7 kb region encompassing
Materials and Methods Construction of noxa / mice and genotyping The targeting vector (see Fig. S) was prepared from a C57BL/6 DNA λ phage library (Stratagene) by replacing a 2.7 kb region encompassing
Thermo Scientific DharmaFECT Transfection Reagents sirna Transfection Protocol
 Protocol Thermo Scientific Transfection Reagents sirna Transfection Protocol The following is a general protocol for use of Thermo Scientific transfection reagents to deliver sirna into cultured mammalian
Protocol Thermo Scientific Transfection Reagents sirna Transfection Protocol The following is a general protocol for use of Thermo Scientific transfection reagents to deliver sirna into cultured mammalian
Lentiviral shrna expression Cloning Kit User Manual for generating shrna expression lentivectors
 Lentiviral shrna expression Cloning Kit User Manual for generating s Cat# Product Name Amount Application LTSH-H1-GB peco-lenti-h1-shrna(gfp-bsd) cloning kit with GFP-Blasticidin selection LTSH-H1-GP peco-lenti-h1-shrna(gfp-puro)
Lentiviral shrna expression Cloning Kit User Manual for generating s Cat# Product Name Amount Application LTSH-H1-GB peco-lenti-h1-shrna(gfp-bsd) cloning kit with GFP-Blasticidin selection LTSH-H1-GP peco-lenti-h1-shrna(gfp-puro)
Globin Block Modules for QuantSeq Instruction Manual
 Globin Block Modules for QuantSeq Instruction Manual Catalog Numbers: 070 (RS-Globin Block, Homo sapiens, 96 rxn) 071 (RS-Globin Block, Sus scrofa, 96 rxn) 015 (QuantSeq 3 mrna-seq Library Prep Kit for
Globin Block Modules for QuantSeq Instruction Manual Catalog Numbers: 070 (RS-Globin Block, Homo sapiens, 96 rxn) 071 (RS-Globin Block, Sus scrofa, 96 rxn) 015 (QuantSeq 3 mrna-seq Library Prep Kit for
DNA Arrays Affymetrix GeneChip System
 DNA Arrays Affymetrix GeneChip System chip scanner Affymetrix Inc. hybridization Affymetrix Inc. data analysis Affymetrix Inc. mrna 5' 3' TGTGATGGTGGGAATTGGGTCAGAAGGACTGTGGGCGCTGCC... GGAATTGGGTCAGAAGGACTGTGGC
DNA Arrays Affymetrix GeneChip System chip scanner Affymetrix Inc. hybridization Affymetrix Inc. data analysis Affymetrix Inc. mrna 5' 3' TGTGATGGTGGGAATTGGGTCAGAAGGACTGTGGGCGCTGCC... GGAATTGGGTCAGAAGGACTGTGGC
Applied Biosystems Real-Time PCR Rapid Assay Development Guidelines
 Applied Biosystems Real-Time PCR Rapid Assay Development Guidelines Description This tutorial will discuss recommended guidelines for designing and running real-time PCR quantification and SNP Genotyping
Applied Biosystems Real-Time PCR Rapid Assay Development Guidelines Description This tutorial will discuss recommended guidelines for designing and running real-time PCR quantification and SNP Genotyping
Gene Regulation Solutions. Microarrays and Next-Generation Sequencing
 Gene Regulation Solutions Microarrays and Next-Generation Sequencing Gene Regulation Solutions The Microarrays Advantage Microarrays Lead the Industry in: Comprehensive Content SurePrint G3 Human Gene
Gene Regulation Solutions Microarrays and Next-Generation Sequencing Gene Regulation Solutions The Microarrays Advantage Microarrays Lead the Industry in: Comprehensive Content SurePrint G3 Human Gene
Supplemental Information. Lysine-5 Acetylation Negatively Regulates. Lactate Dehydrogenase A and Is Decreased. in Pancreatic Cancer
 Cancer Cell, Volume 23 Supplemental Information Lysine-5 Acetylation Negatively Regulates Lactate Dehydrogenase A and Is Decreased in Pancreatic Cancer Di Zhao, Shao-Wu Zou, Ying Liu, Xin Zhou, Yan Mo,
Cancer Cell, Volume 23 Supplemental Information Lysine-5 Acetylation Negatively Regulates Lactate Dehydrogenase A and Is Decreased in Pancreatic Cancer Di Zhao, Shao-Wu Zou, Ying Liu, Xin Zhou, Yan Mo,
Mayumi Egawa, Kaori Mukai, Soichiro Yoshikawa, Misako Iki, Naofumi Mukaida, Yohei Kawano, Yoshiyuki Minegishi, and Hajime Karasuyama
 Immunity, Volume 38 Supplemental Information Inflammatory Monocytes Recruited to Allergic Skin Acquire an Anti-inflammatory M2 Phenotype via Basophil-Derived Interleukin-4 Mayumi Egawa, Kaori Mukai, Soichiro
Immunity, Volume 38 Supplemental Information Inflammatory Monocytes Recruited to Allergic Skin Acquire an Anti-inflammatory M2 Phenotype via Basophil-Derived Interleukin-4 Mayumi Egawa, Kaori Mukai, Soichiro
TransAM Kits are DNA-binding ELISAs that facilitate the study of transcription factor activation in mammalian tissue and cell culture extracts.
 Transcription Factor ELISAs TransAM sensitive quantitative transcription factor ELISAs TransAM Kits are DNA-binding ELISAs that facilitate the study of transcription factor activation in mammalian tissue
Transcription Factor ELISAs TransAM sensitive quantitative transcription factor ELISAs TransAM Kits are DNA-binding ELISAs that facilitate the study of transcription factor activation in mammalian tissue
Protein Purification Products. Complete Solutions for All of Your Protein Purification Applications
 Protein Purification Products Complete Solutions for All of Your Protein Purification Applications FLAG-Tagged Protein Products EXPRESS with the pcmv-dykddddk Vector Set Fuse your protein of interest to
Protein Purification Products Complete Solutions for All of Your Protein Purification Applications FLAG-Tagged Protein Products EXPRESS with the pcmv-dykddddk Vector Set Fuse your protein of interest to
mir-24-mediated down-regulation of H2AX suppresses DNA repair
 Supplemental Online Material mir-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells Ashish Lal 1,4, Yunfeng Pan 2,4, Francisco Navarro 1,4, Derek M. Dykxhoorn
Supplemental Online Material mir-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells Ashish Lal 1,4, Yunfeng Pan 2,4, Francisco Navarro 1,4, Derek M. Dykxhoorn
Xeno-Free Systems for hesc & hipsc. Facilitating the shift from Stem Cell Research to Clinical Applications
 Xeno-Free Systems for hesc & hipsc Facilitating the shift from Stem Cell Research to Clinical Applications NutriStem Defined, xeno-free (XF), serum-free media (SFM) specially formulated for growth and
Xeno-Free Systems for hesc & hipsc Facilitating the shift from Stem Cell Research to Clinical Applications NutriStem Defined, xeno-free (XF), serum-free media (SFM) specially formulated for growth and
Product: Arrest-In TM Transfection Reagent for RNAi
 Product: Arrest-In TM Transfection Reagent for RNAi Catalog #: ATR1740, ATR1741, ATR1742, ATR1743 Product Description Arrest-In transfection reagent is a proprietary polymeric formulation, developed and
Product: Arrest-In TM Transfection Reagent for RNAi Catalog #: ATR1740, ATR1741, ATR1742, ATR1743 Product Description Arrest-In transfection reagent is a proprietary polymeric formulation, developed and
Xfect Protein Transfection Reagent
 Xfect Protein Transfection Reagent Mammalian Expression Systems Rapid, high-efficiency, low-toxicity protein transfection Transfect a large amount of active protein Virtually no cytotoxicity, unlike lipofection
Xfect Protein Transfection Reagent Mammalian Expression Systems Rapid, high-efficiency, low-toxicity protein transfection Transfect a large amount of active protein Virtually no cytotoxicity, unlike lipofection
QIAGEN s NGS Solutions for Biomarkers NGS & Bioinformatics team QIAGEN (Suzhou) Translational Medicine Co.,Ltd
 QIAGEN s NGS Solutions for Biomarkers NGS & Bioinformatics team QIAGEN (Suzhou) Translational Medicine Co.,Ltd 1 Our current NGS & Bioinformatics Platform 2 Our NGS workflow and applications 3 QIAGEN s
QIAGEN s NGS Solutions for Biomarkers NGS & Bioinformatics team QIAGEN (Suzhou) Translational Medicine Co.,Ltd 1 Our current NGS & Bioinformatics Platform 2 Our NGS workflow and applications 3 QIAGEN s
Yue Wang, Zhenyu Xu, Junfeng Jiang, Chen Xu, Jiuhong Kang, Lei Xiao, Minjuan Wu, Jun Xiong, Xiaocan Guo, and Houqi Liu
 Developmental Cell, Volume 25 Supplemental Information Endogenous mirna Sponge lincrna-ror Regulates Oct4, Nanog, and Sox2 in Human Embryonic Stem Cell Self-Renewal Yue Wang, Zhenyu Xu, Junfeng Jiang,
Developmental Cell, Volume 25 Supplemental Information Endogenous mirna Sponge lincrna-ror Regulates Oct4, Nanog, and Sox2 in Human Embryonic Stem Cell Self-Renewal Yue Wang, Zhenyu Xu, Junfeng Jiang,
SureSilencing shrna Plasmid Handbook
 March 2011 SureSilencing shrna Plasmid Handbook For genomewide, plasmid-based RNA interference Sample & Assay Technologies QIAGEN Sample and Assay Technologies QIAGEN is the leading provider of innovative
March 2011 SureSilencing shrna Plasmid Handbook For genomewide, plasmid-based RNA interference Sample & Assay Technologies QIAGEN Sample and Assay Technologies QIAGEN is the leading provider of innovative
64 CuCl 2 in 50 µl 0.1N NaOAc buffer, and 20 µg of each DOTA-antibody conjugate in 40 µl
 Number of DOTA per antibody The average number of DOTA chelators per antibody was measured using a reported procedure with modifications (1,2). Briefly, nonradioactive CuCl 2 (80-fold excess of DOTA antibodies)
Number of DOTA per antibody The average number of DOTA chelators per antibody was measured using a reported procedure with modifications (1,2). Briefly, nonradioactive CuCl 2 (80-fold excess of DOTA antibodies)
Figure S1. Purity of primary cultures of renal proximal tubular epithelial culture ascertained by cytokeratin staining.
 Supplementary information Supplementary figures Figure S1. Purity of primary cultures of renal proximal tubular epithelial culture ascertained by cytokeratin staining. Figure S2. Induction of Nur77 in
Supplementary information Supplementary figures Figure S1. Purity of primary cultures of renal proximal tubular epithelial culture ascertained by cytokeratin staining. Figure S2. Induction of Nur77 in
XactEdit Cas9 Nuclease with NLS User Manual
 XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
Supplemental Online Material. The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/-
 #1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
#1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
