Supporting Information
|
|
|
- Carmel Osborne
- 5 years ago
- Views:
Transcription
1 Supporting Information Battisti et al /pnas SI Text SI Materials and Methods. Virus propagation and purification. Egggrown Newcastle disease virus (NDV; strain B1), harvested at 42 h postinfection, was purified as previously described (1). For cryoelectron tomography the virus was placed into a TNE buffer (0.02 M Tris, 0.12 M NaCl, and M EDTA, ph 8.0). The total protein concentration was adjusted to 1 3 mg ml (corresponding to a virus titer of to pfu ml). SDS-PAGE was used to detect the presence of viral proteins and assess purity. Cryoelectron tomography and 3D reconstruction. A suspension of BSA-conjugated 10 nm gold beads (BSA Gold Tracers; Aurion) was added to the purified NDV suspension. Next, 3.5 μl aliquots of the virus gold mixture were applied to holey carbon covered EM grids (Quantifoil Micro Tools GmbH), blotted with filter paper, and plunged into liquid ethane. The vitrified samples were imaged using a Titan Krios (FEI) field emission gun microscope operated at 300 kv. Additionally, the microscope was equipped with an energy filter (Gatan, Inc.) operated in zero-energy-loss mode with a slit width of ev. Images were recorded on a 2,048- by 2,048-pixel CCD camera at a magnification of 19;500 (0.75 nm separation between pixels). The tilt-series images were obtained over an angular range of 69 in 1.5 increments at a defocus of 8 9 μm. The cumulative electron dose for the complete series was in the range of electron Å 2. The FEI Batch Tomography software was used to perform automatically tilting, tracking, focusing, and image acquisition. Tiltseries images were aligned using the colloidal gold particles as fiducial markers and reconstructed using the weighted back-projection method as implemented in the IMOD software package (2). The resolution of tomograms is difficult to quantify and is exceedingly isotropic because of the missing wedge of data. However, the best resolution in the direction perpendicular to the electron beam is better than 7 nm, based on the ability to observe the spacing between matrix protein dimers. Alignment and averaging of matrix protein subunits. In order to improve the matrix protein density, adjoining regions of the matrix protein layer plus associated membrane were aligned and averaged using the AVE and IMP programs from the Uppsala Software Factory (3, 4). The best rotational and translational parameters relating each region to its neighbor were determined by maximizing the correlation coefficient between neighboring regions. Using a density threshold that would make the averaged membrane density about 4 nm thick (corresponding to 1.7 standard deviations above the mean density value), the volume of one subunit was measured to be about 85 nm 3 using the Chimera visualization tool (5). Assuming an average protein density of 1.35 g cm 3, the molecular mass of one subunit would be about 70 kda, consistent with each subunit being a dimer of 76 kda. Cloning, expression, and purification of newcastle disease virus matrix protein. The full-length gene of the NDV matrix protein (6) was amplified by PCR and ligated into the pet28a (Merck KGaA) expression vector, which has an N-terminal 6-histidine tag sequence. The expression plasmid was transfected into Escherichia coli, strain BL21 (Rosetta), for protein expression. The cells were grown at 37 C until the optical density reached , at which point 1 M IPTG was added to induce the expression of the protein. This culture was then grown at 18 C for 20 h, harvested by centrifugation, and suspended in Hepes buffer (500 mm NaCl, 50 mm Hepes, ph 8.0). The harvested cells were lysed and centrifuged at 15;000 g for 30 min. The supernatant was applied to a nickel affinity column and washed in an imidazole gradient. The fraction containing the matrix protein was concentrated by centrifugation and further purified by size exclusion chromatography using a S200 Superdex column (GE Healthcare). The purified matrix protein was concentrated to 3 mg ml and frozen in liquid nitrogen for further use. Crystallization, X-ray data collection, and structure determination of the matrix protein. Crystallization conditions were screened by the sitting drop vapor diffusion method using a Honeybee crystallization robot (Digilab, Inc.). Almost 500 conditions were screened using Hampton (Hampton Research) and Emerald (Emerald Biosystems) crystallization kits. Each 1 μl drop was a mixture of 0.5 μl protein and 0.5 μl well solution. Small, 0.01 mm diameter plate-like crystals were obtained initially in several conditions. These conditions were further optimized to improve the size and quality of the crystals, which eventually could be grown to about 0.3 mm in 1 week. The crystals were mounted on loops and flash frozen in liquid nitrogen after soaking in the cryoprotectant, which consisted of a mixture of the mother liquor with 25% (vol vol) glycerol. Mercury-derivatized crystals were obtained by cocrystallization with various mercury compounds. Two forms of native crystals and one form of a nonisomorphous Hg-derivative crystal were found. Diffraction data were collected at the Advanced Photon Source beamline 23 ID-B/D using crystals maintained at 100 K and a MAR 300 CCD detector (Table S1). All data were processed using the HKL2000 program (7). The Matthews coefficient for the Hg derivative was 2.4 Å 3 Da assuming two molecules in the asymmetric unit. Phases were determined with the Phenix program (8) using single-wavelength anomalous dispersion data. Twelve heavy-atom sites were found in the crystallographic asymmetric unit. The initial phases were iteratively refined using density modifications. The programs Phenix and Resolve (9) were used to build the structure into the electron density map. This structure was then refined using the program Refmac (10) in CCP4 (11) and viewed with the program Coot (12). Molecular replacement was used to determine the structure of the two native datasets using the program Molrep (13) in CCP4 (11). The final R working and R free factors for the three datasets were better than 24.5% and 28.8%, respectively (Table S1). The rmsd between noncrystallographic symmetry (NCS)-equivalent Cα atoms within each structure was 0.2 Å and the NCS rotation was within 0.3 of 180. The two domains within each monomer are related by a 67 rotation, which is coincident with the twofold NCS axis. The NDV matrix protein structure was visualized using PyMol (Schrödinger, LLC) and represented schematically using Top- Draw (14). A structure-based sequence alignment comparing the NDV matrix protein to other Mononegavirales matrix proteins was carried out using the program HOMOlogy (15). The crystallographic matrix dimer was fitted into the tomographic matrix density using the program EMfit (16). Helical reconstruction of the nucleocapsid. An averaged one-dimensional Fourier transform from 25 nucleocapsid regions in the NDV tomograms, scanned along their lengths, had one large coefficient corresponding to a 7 nm repeat. The nucleocapsid density was helically averaged assuming different pitch and twist values using a script implemented in the EMAN package (17). Plotting the correlation coefficient between the averaged and unaveraged nucleocapsid maps as a function of twist and pitch 1of8
2 resulted in a line of high correlation with a positive slope, indicative of a right-handed helix with a pitch of 7 nm. The line of high-correlation values was fairly continuous, indicating that the tomograms were not of sufficient resolution to discern independent nucleocapsid subunits. Combining the averaged matrix protein tomographic density with the averaged tomographic nucleocapsid structures. A 60 nm long averaged helical nucleocapsid structure was fitted into 25 nearly straight nucleocapsid densities from six different tomograms using the program IMP (3). Similarly, the averaged matrix protein region was fitted into the matrix densities neighboring the fitted nucleocapsids. For 18 of the 25 nucleocapsid matrix interfaces the 7 nm pitch of the nucleocapsid was found to be in register (10 ) with the distance between matrix protein subunits along the array diagonal. 1. McGinnes LW, Pantua H, Reitter J, Morrison TG (2006) Newcastle disease virus: Propagation, quantification, and storage. Curr Protoc Microbiol Chapter 15:Unit 15F Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of threedimensional image data using IMOD. J Struct Biol 116: Jones TA (1992) A set of averaging programs. Molecular Replacement, eds Dodson EJ, Gover S, Wolf W (SERC Daresbury Laboratory, Warrington, UK), pp Kleywegt GJ, Jones TA (1994) Halloween masks and bones. From First Map to Final Model, eds Bailey S, Hubbard R, Waller D (SERC Daresbury Laboratory, Warrington, UK), pp Pettersen EF, et al. (2004) UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem 25: McGinnes LW, Morrison TG (1987) The nucleotide sequence of the gene encoding the Newcastle disease virus membrane protein and comparisons of membrane protein sequences. Virology 156: Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol, ed Carter CW, Jr (Academic Press, New York), Vol 276, pp Adams PD, et al. (2002) PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr Sect D: Biol Crystallogr 58: Terwilliger TC (2003) Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr Sect D: Biol Crystallogr 59: Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D: Biol Crystallogr 53: Winn MD, et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr Sect D: Biol Crystallogr 67: Emsley P, Cowtan K (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr Sect D: Biol Crystallogr 60: Vagin A, Teplyakov A (1997) MOLREP: An automated program for molecular replacement. J Appl Crystallogr 30: Bond CS (2003) TopDraw: A sketchpad for protein structure topology cartoons. Bioinformatics 19: Rao ST, Rossmann MG (1973) Comparison of super-secondary structures in proteins. J Mol Biol 76: Rossmann MG, Bernal R, Pletnev SV (2001) Combining electron microscopic with x-ray crystallographic structures. J Struct Biol 136: Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: Fig. S1. The NDV nucleocapsid. (A) A repeating pattern along the length of NDV nucleocapsids was seen in the tomographic data. The image was generated by averaging 20 layers of voxels over a thickness of 30 nm. Black represents high density. (B) The Fourier transform of a one-dimensional scan over the length of the image has a large peak corresponding to a helical pitch of 7 nm. (C) Reconstruction of the helical nucleocapsid assuming a pitch of 7 nm and a twist of 18 subunits/turn. 2of8
3 Fig. S2. Nonreducing, silver-stained SDS-PAGE of purifiedvirus. The center and right lanes were loaded with 1and 2.5μL of purified NDV, respectively. The left lane was loaded with a molecular weight marker. The viral proteins and marker molecular weights are identified in the figure. Although the majority of virions do not have an ordered matrix protein layer, the matrix protein is present at about the same relative abundance as the other structural proteins in the virions. 3of8
4 Fig. S3. The relationship between the matrix layer and the nucleocapsid filaments. (A and B) Tomographic sections showing portions of the nucleocapsid of two virions (Top). The yellow arrowsindicateportionsofthe nucleocapsid that are roughlyinregister with theadjoiningmatrix array (Middle). The relationship between the nucleocapsids and matrix array are shown schematically (Bottom). The yellow rectangles represent portions of the nucleocapsid and the blue squares indicate the approximate orientation of the matrix array, the diagonal of which forms a roughly 45 angle with the long axis of elongated virions. High density is black and the sections represent an average of five planes of voxels (a thickness of approximately 75 nm). The scale bars represent 25 nm. (C) The averaged helical nucleocapsid structure (yellow density) was fitted into the virion depicted in A. A number of copies of the averaged matrix protein subunit (blue density) have been placed into a portion of the matrix array shown in A. The rotational and translational elements determined during the averaging procedure were used to place the matrix density. The 7 nm repeating units of the matrix array and nucleocapsid structure are oriented in roughly the same direction. The scale bar represents 25 nm. (D) The structures depicted in C, rotated approximately 55 about the horizontal axis. The scale bar represents 25 nm. (E) The matrix protein dimer structure (blue ribbon) and averaged helical nucleocapsid structure (yellow density) were fitted into the virion depictedinb. The 7 nm repeating units of the matrix array and nucleocapsid structure are oriented in roughly the same direction. (F) The structures depicted in E, rotated approximately 90 about the horizontal axis. The scale bar represents 25 nm. 4of8
5 Fig. S4. Gel filtration indicates the matrix protein is dimeric. (Left) The matrix protein was purified using a Superdex /60 GL (GE Healthcare) sizeexclusion column. The arrows indicate three different elution peaks for NDV matrix protein. The dashed vertical lines indicate the elution peaks of four molecular weight standards. A comparison between the matrix protein elution positions and the molecular weight standards indicates that the matrix protein is mainly dimeric or forming higher-order oligomers in solution, though some monomeric protein is present. (Inset, Right) SDS-PAGE of the protein eluted from the monomeric (P3), dimeric (P2), and higher-order oligomer (P1) peaks. The left lane was loaded with a molecular weight marker. Marker molecular weights are indicated in the figure. 5of8
6 Fig. S5. Conservation of residues in the matrix protein dimer interface. (A) A surface representation of the NDV matrix protein monomer. The monomer monomer interface within a dimer is outlined by a thick black line. The dark blue residues are completely conserved among 10 members of the Paramyxovirinae subfamily. Lighter blue indicates partial conservation and white indicates no significant conservation. Of residues in the dimer interface, 36.6% (15 out of 41) are completely conserved, whereas only 14.3% of other surface residues (34 out of 237) are completely conserved. (B) The residues on the opposite side of the monomer to the dimer interface are less conserved among paramyxoviruses. 6of8
7 Fig. S6. Structure-basedsequencealignment ofmononegavirales matrix proteins. The crystal structures of the respiratory syncytial virus matrix protein, Borna virus matrix protein, and Ebola virus VP40 were structurally aligned with the NDV matrix protein. Secondary structural elements for NDV are shown above the sequence alignment. Residues that are conserved for at least two of the four aligned proteins are highlighted in red. 7of8
8 Table S1. Crystallographic data collection and refinement statistics* Native 1 Native 2 Hg derivative Data collection X-ray source 23-ID-D 23-ID-D 23-ID-B Wavelength (Å) Cell dimensions a ¼ 163, b ¼ 47, c ¼ 117; β ¼ 131, α ¼ γ ¼ 90 a ¼ 247, b ¼ 45, c ¼ 58; β ¼ 103, α ¼ γ ¼ 90 a ¼ 46.8, b ¼ c ¼ 141.8;α ¼ β ¼ γ ¼ 90 Space group C2 C2 P22121 No. monomers per asymmetric unit Oscillation angle No. frames Resolution range R merge 0.088(0.612) 0.078(0.602) 0.116(0.364) Completeness 99.6(100) 93.9(94.6) 100(100) Redundancy Refinement Resolution (Å) R working (%) R free (%) Average B factor (Å 2 ) Rmsd bonds (Å) Rmsd angles ( ) Ramachandran disallowed *Values in parentheses throughout the table correspond to the last shell. R merge ¼ ΣjI hiij ΣI, where I is the measured intensity for reflections with indices hkl. R working ¼ ΣjjF obs j jf calc jj ΣjF obs j. R free has the same formula as R working, except that the calculation was made with the structure factors from the test set. Table S2. Fitting of the crystallographic matrix protein dimer into the averaged matrix protein density from cryoelectron tomography using the EMfit program Sumf* Clash -Den % 8.7% *Sumf is the mean density height averaged over all atoms where the maximum density in the electron density map is set to 100. Clash represents the percentage of atoms in the model that approach closer than 5 Å to neighboring matrix dimers. The percentage of atoms in density less than the mean density value is given by Den. Table S3. Sequence and structural comparisons between the 360 Cα atoms of the NDV matrix protein and other Mononegavirales* matrix proteins Molecule Dali score Rmsd between Cα atoms No. of aligned Cα atoms Total no. of Cα atoms PDB ID Sequence identity RSV M VQP 7.5% BDV M F1J 7.8% EBV M ES6 4.6% M, matrix protein; PDB, Protein Data Bank; RSV, respiratory syncytial virus; BDV, Borna disease virus: EBV, Ebola virus. *The structure of the vesicular stomatitis virus matrix protein was not included because it has a different fold. Alignment of the BDV matrix protein with the C-terminal domain of NDV M. Alignment of the Ebola virus matrix protein with the N-terminal domain of NDV M. 8of8
Proteins were extracted from cultured cells using a modified buffer, and immunoprecipitation and
 Materials and Methods Immunoprecipitation and immunoblot analysis Proteins were extracted from cultured cells using a modified buffer, and immunoprecipitation and immunoblot analyses with corresponding
Materials and Methods Immunoprecipitation and immunoblot analysis Proteins were extracted from cultured cells using a modified buffer, and immunoprecipitation and immunoblot analyses with corresponding
Six genes, Lsm1, Lsm2, Lsm3, Lsm5, Lsm6, and Lsm7, were amplified from the
 Supplementary information, Data S1 Methods Clones and protein preparation Six genes, Lsm1, Lsm2, Lsm3, Lsm5, Lsm6, and Lsm7, were amplified from the Saccharomyces cerevisiae genomic DNA by polymerase chain
Supplementary information, Data S1 Methods Clones and protein preparation Six genes, Lsm1, Lsm2, Lsm3, Lsm5, Lsm6, and Lsm7, were amplified from the Saccharomyces cerevisiae genomic DNA by polymerase chain
Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1.
 GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
The Skap-hom Dimerization and PH Domains Comprise
 Molecular Cell, Volume 32 Supplemental Data The Skap-hom Dimerization and PH Domains Comprise a 3 -Phosphoinositide-Gated Molecular Switch Kenneth D. Swanson, Yong Tang, Derek F. Ceccarelli, Florence Poy,
Molecular Cell, Volume 32 Supplemental Data The Skap-hom Dimerization and PH Domains Comprise a 3 -Phosphoinositide-Gated Molecular Switch Kenneth D. Swanson, Yong Tang, Derek F. Ceccarelli, Florence Poy,
BACTERIAL PRODUCTION EXPRESSION METHOD OVERVIEW: PEF # GENE NAME EXPRESSION VECTOR MOLECULAR WEIGHT kda (full-length) 34.
 BACTERIAL PRODUCTION PEF # GENE NAME EXPRESSION VECTOR MOLECULAR WEIGHT 2015-XXXX XXXX pet-32a 50.9 kda (full-length) 34.0 kda (cleaved) EXPRESSION METHOD OVERVIEW: Plasmid DNA was transformed into BL21
BACTERIAL PRODUCTION PEF # GENE NAME EXPRESSION VECTOR MOLECULAR WEIGHT 2015-XXXX XXXX pet-32a 50.9 kda (full-length) 34.0 kda (cleaved) EXPRESSION METHOD OVERVIEW: Plasmid DNA was transformed into BL21
Supporting Information
 Supporting Information Nishimura et al. 10.1073/pnas.1003553107 SI Text SI Materials and Methods. Plasmid construction. The cdnas of human Gα q were amplified and subcloned into pcmv5. Mutants of Gα q
Supporting Information Nishimura et al. 10.1073/pnas.1003553107 SI Text SI Materials and Methods. Plasmid construction. The cdnas of human Gα q were amplified and subcloned into pcmv5. Mutants of Gα q
Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid
 Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
SUPPLEMENTAL DATA Experimental procedures
 SUPPLEMENTAL DATA Experimental procedures Cloning, protein production and purification. The DNA sequences corresponding to residues R10-S250 (DBD) and E258-T351 (IPT/TIG) in human EBF1 (gi:31415878), and
SUPPLEMENTAL DATA Experimental procedures Cloning, protein production and purification. The DNA sequences corresponding to residues R10-S250 (DBD) and E258-T351 (IPT/TIG) in human EBF1 (gi:31415878), and
nature methods Enabling IMAC purification of low abundance recombinant proteins from E. coli lysates
 nature methods Enabling IMAC purification of low abundance recombinant proteins from E. coli lysates Audur Magnusdottir, Ida Johansson, Lars-Göran Dahlgren, Pär Nordlund & Helena Berglund Supplementary
nature methods Enabling IMAC purification of low abundance recombinant proteins from E. coli lysates Audur Magnusdottir, Ida Johansson, Lars-Göran Dahlgren, Pär Nordlund & Helena Berglund Supplementary
Supporting Information
 Supporting Information Slep et al. 10.1073/pnas.0801569105 SI Materials and Methods Mouse RGS16, residues 53 180, was subcloned into a modified pgex-2t vector (GE Healthcare) to yield a thrombin-cleavable
Supporting Information Slep et al. 10.1073/pnas.0801569105 SI Materials and Methods Mouse RGS16, residues 53 180, was subcloned into a modified pgex-2t vector (GE Healthcare) to yield a thrombin-cleavable
SUPPLEMENTARY INFORMATION. Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly
 SUPPLEMENTARY INFORMATION Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly Dustin J. E. Huard, Kathleen M. Kane and F. Akif Tezcan* Department of Chemistry and Biochemistry,
SUPPLEMENTARY INFORMATION Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly Dustin J. E. Huard, Kathleen M. Kane and F. Akif Tezcan* Department of Chemistry and Biochemistry,
The YTH domain (residues ) of human YTHDF2 (NP_ ) was subcloned
 Supplementary information, Data S1 Materials and Methods Protein Expression, Purification and Crystallization The YTH domain (residues 383-553) of human YTHDF2 (NP_057342.2) was subcloned into a modified
Supplementary information, Data S1 Materials and Methods Protein Expression, Purification and Crystallization The YTH domain (residues 383-553) of human YTHDF2 (NP_057342.2) was subcloned into a modified
Supporting Information
 Supporting Information Rahmeh et al. 1.173/pnas.1914719 SI Materials and Methods Protein Expression and Purification. was expressed in Sf1 cells with an N-terminal 6xHis tag and purified by Ni- nitrilotriacetic
Supporting Information Rahmeh et al. 1.173/pnas.1914719 SI Materials and Methods Protein Expression and Purification. was expressed in Sf1 cells with an N-terminal 6xHis tag and purified by Ni- nitrilotriacetic
Figure S1
 Supplementary Figure 1 The distribution of chlorophyll containing complexes eluted from DEAE-cellulose column in a sucrose gradient tube Six pigment-containing bands were resolved and identified as: B1,
Supplementary Figure 1 The distribution of chlorophyll containing complexes eluted from DEAE-cellulose column in a sucrose gradient tube Six pigment-containing bands were resolved and identified as: B1,
Surface modification of a DNA tetrahedron
 Surface modification of a DNA tetrahedron Chuan Zhang, Min Su, Yu He, Yujun Leng, Alexander E. Ribbe, Guansong Wang ±, Wen Jiang & Chengde Mao * Department of Chemistry, Markey Center for Structural Biology
Surface modification of a DNA tetrahedron Chuan Zhang, Min Su, Yu He, Yujun Leng, Alexander E. Ribbe, Guansong Wang ±, Wen Jiang & Chengde Mao * Department of Chemistry, Markey Center for Structural Biology
Figure S2, related to Figure 1. Stereo images of the CarD/RNAP complex and. electrostatic potential surface representation of the CarD/RNAP interface
 Structure, Volume 21 Supplemental Information Structure of the Mtb CarD/RNAP -Lobes Complex Reveals the Molecular Basis of Interaction and Presents a Distinct DNA-Binding Domain for Mtb CarD Gulcin Gulten
Structure, Volume 21 Supplemental Information Structure of the Mtb CarD/RNAP -Lobes Complex Reveals the Molecular Basis of Interaction and Presents a Distinct DNA-Binding Domain for Mtb CarD Gulcin Gulten
1 24 C63 C β- β-
 M40 Signal leaved RS1 Domain 59 110 142 Discoidin Domain 223 1 24 63 219 224 + - β- β- M M e e O O H H His 6 -Tag 250 150 100 75 50 37 * 25 20 Supplementary Figure 1 Purification of wild-type retinoschisin.
M40 Signal leaved RS1 Domain 59 110 142 Discoidin Domain 223 1 24 63 219 224 + - β- β- M M e e O O H H His 6 -Tag 250 150 100 75 50 37 * 25 20 Supplementary Figure 1 Purification of wild-type retinoschisin.
SUPPORTING INFORMATION
 SUPPORTING INFORMATION Text S1: EvoDesign profile construction Table S1 lists the pair-wise structural alignments generated by the TM-align program [1], between the XIAP scaffold protein (PDB ID: 2OPY)
SUPPORTING INFORMATION Text S1: EvoDesign profile construction Table S1 lists the pair-wise structural alignments generated by the TM-align program [1], between the XIAP scaffold protein (PDB ID: 2OPY)
Phase Contrast Cryo Electron Tomography of Ice embedded Biological Specimens. 1. Introduction.
 Phase Contrast Cryo Electron Tomography of Ice embedded Biological Specimens Radostin Danev, Yoshiyuki Fukuda, Kuniaki Nagayama 1. Introduction. This course is aimed at introducing beginner to intermediate
Phase Contrast Cryo Electron Tomography of Ice embedded Biological Specimens Radostin Danev, Yoshiyuki Fukuda, Kuniaki Nagayama 1. Introduction. This course is aimed at introducing beginner to intermediate
Supplementary Information
 Supplementary Information Structure of Active, Dimeric Human Telomerase Anselm Sauerwald, Sara Sandin, Gaël Cristofari, Sjors H.W. Scheres, Joachim Lingner, and Daniela Rhodes Table S1: Composition of
Supplementary Information Structure of Active, Dimeric Human Telomerase Anselm Sauerwald, Sara Sandin, Gaël Cristofari, Sjors H.W. Scheres, Joachim Lingner, and Daniela Rhodes Table S1: Composition of
Supporting Online Material. Av1 and Av2 were isolated and purified under anaerobic conditions according to
 Supporting Online Material Materials and Methods Av1 and Av2 were isolated and purified under anaerobic conditions according to published protocols (S1). Crystals of nf-, pcp- and adp-av2:av1 complexes
Supporting Online Material Materials and Methods Av1 and Av2 were isolated and purified under anaerobic conditions according to published protocols (S1). Crystals of nf-, pcp- and adp-av2:av1 complexes
Supporting Information
 Title Structure of bacterial cellulose synthase subunit D Hu, Song-Qing; Gao, Yong-Gui; Tajima, Kenji; Sunagaw Author(s) Yoda, Takanori; Shimura, Daisuke; Satoh, Yasuharu; M CitationProceedings of the
Title Structure of bacterial cellulose synthase subunit D Hu, Song-Qing; Gao, Yong-Gui; Tajima, Kenji; Sunagaw Author(s) Yoda, Takanori; Shimura, Daisuke; Satoh, Yasuharu; M CitationProceedings of the
Supporting Information
 Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
Steps in solving a structure. Diffraction experiment. Obtaining well-diffracting crystals. Three dimensional crystals
 Protein structure from X-ray diffraction Diffraction images: ciprocal space Protein, chemical structure: IALEFGPSLKMNE Conformation, 3D-structure: CRYST1 221.200 73.600 80.900 90.00 90.00 90.00 P 21 21
Protein structure from X-ray diffraction Diffraction images: ciprocal space Protein, chemical structure: IALEFGPSLKMNE Conformation, 3D-structure: CRYST1 221.200 73.600 80.900 90.00 90.00 90.00 P 21 21
Supplementary Online Material. Structural mimicry in transcription regulation of human RNA polymerase II by the. DNA helicase RECQL5
 Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Supplemental Online Material
 Supplemental Online Material Supplemental Figures Supplemental Figure 1. A) Crosslinking efficiencies of 5 -biotin tagged oligonucleotides screened with the FASTDXL method 1. Reactive and nonreactive cysteine
Supplemental Online Material Supplemental Figures Supplemental Figure 1. A) Crosslinking efficiencies of 5 -biotin tagged oligonucleotides screened with the FASTDXL method 1. Reactive and nonreactive cysteine
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
INSECT CELL/BACULOVIRUS PRODUCTION
 INSECT CELL/BACULOVIRUS PRODUCTION PEF # GENE NAME TRANSFER VECTOR BEVS MOLECULAR WEIGHT 2015-XXXX XXXX pbac1 flashbacultra TM 36.0 kda EXPRESSION METHOD OVERVIEW: Insect cells Spodoptera frugiperda (Sf9)
INSECT CELL/BACULOVIRUS PRODUCTION PEF # GENE NAME TRANSFER VECTOR BEVS MOLECULAR WEIGHT 2015-XXXX XXXX pbac1 flashbacultra TM 36.0 kda EXPRESSION METHOD OVERVIEW: Insect cells Spodoptera frugiperda (Sf9)
Secondary structure of hvps4b (above) and sequence alignments of VPS4 proteins from
 Supplemental Figure 1. Secondary structure of hvps4b (above) and sequence alignments of VPS4 proteins from different species as well as four other representative members of the meiotic clade of AAA ATPases.
Supplemental Figure 1. Secondary structure of hvps4b (above) and sequence alignments of VPS4 proteins from different species as well as four other representative members of the meiotic clade of AAA ATPases.
Nature Structural & Molecular Biology: doi: /nsmb.1969
 Supplementary Methods Structure determination All the diffraction data sets were collected on BL-41XU (using ADSC Quantum 315 HE CCD detector) at SPring8 (Harima, Japan) or on BL5A (using ADSC Quantum
Supplementary Methods Structure determination All the diffraction data sets were collected on BL-41XU (using ADSC Quantum 315 HE CCD detector) at SPring8 (Harima, Japan) or on BL5A (using ADSC Quantum
Supplemental Material
 Supplemental Material Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase Shuang Yang, 1, 2 Xiangdong Zheng, 1, 2, 3 Chao Lu, 4 Guo-Min Li, 2 C. David Allis, 4 1, 2, 3, 5* and Haitao
Supplemental Material Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase Shuang Yang, 1, 2 Xiangdong Zheng, 1, 2, 3 Chao Lu, 4 Guo-Min Li, 2 C. David Allis, 4 1, 2, 3, 5* and Haitao
Supplemental Information
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supplemental Information Experimental Procedures Cloning, expression, purification and mutagenesis
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supplemental Information Experimental Procedures Cloning, expression, purification and mutagenesis
Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions
 Supporting Information Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions Lise Schoonen, b Sjors Maassen, b Roeland J. M. Nolte b and Jan C. M. van
Supporting Information Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions Lise Schoonen, b Sjors Maassen, b Roeland J. M. Nolte b and Jan C. M. van
Supplementary Figure 1 Sequence alignment of representative CbbQ sequences.
 Walker A Pore loop 1 Walker B Supplementary Figure 1 Sequence alignment of representative CbbQ sequences. Amino acid sequences of CbbQ1 and CbbQ2 proteins from selected chemoautotrophic bacteria were aligned
Walker A Pore loop 1 Walker B Supplementary Figure 1 Sequence alignment of representative CbbQ sequences. Amino acid sequences of CbbQ1 and CbbQ2 proteins from selected chemoautotrophic bacteria were aligned
The Evolution of Size, Shape, and Surface Morphology. of Gold Nanorods
 Electronic Supplementary Material (ESI) for Chemical Communications. This journal is The Royal Society of Chemistry 2018 Supporting Information The Evolution of Size, Shape, and Surface Morphology of Gold
Electronic Supplementary Material (ESI) for Chemical Communications. This journal is The Royal Society of Chemistry 2018 Supporting Information The Evolution of Size, Shape, and Surface Morphology of Gold
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system
 SUPPLEMENTARY DATA Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system Daniel Gleditzsch 1, Hanna Müller-Esparza 1, Patrick Pausch 2,3, Kundan Sharma 4, Srivatsa Dwarakanath 1,
SUPPLEMENTARY DATA Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system Daniel Gleditzsch 1, Hanna Müller-Esparza 1, Patrick Pausch 2,3, Kundan Sharma 4, Srivatsa Dwarakanath 1,
Minicollagen cysteine-rich domains encode distinct modes of. Anja Tursch, Davide Mercadante, Jutta Tennigkeit, Frauke Gräter and Suat
 Supplementary Figure S1. Conformational dynamics of partially reduced CRDs. (A) Root mean square deviation (RMSD) and (B) radius of gyration (R G ) distributions for the N-CRD (green) and C-CRD (red) domains,
Supplementary Figure S1. Conformational dynamics of partially reduced CRDs. (A) Root mean square deviation (RMSD) and (B) radius of gyration (R G ) distributions for the N-CRD (green) and C-CRD (red) domains,
Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg
![Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg](/thumbs/84/90687567.jpg) Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport
 Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport Ivan Laponogov 1,2, Dennis A. Veselkov 1, Isabelle M-T. Crevel
Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport Ivan Laponogov 1,2, Dennis A. Veselkov 1, Isabelle M-T. Crevel
SUPPLEMENTARY INFORMATION
 Molecular basis of RNA-dependent RNA polymerase II activity Elisabeth Lehmann, Florian Brueckner, and Patrick Cramer Gene Center Munich and Center for integrated Protein Science CiPS M, Department of Chemistry
Molecular basis of RNA-dependent RNA polymerase II activity Elisabeth Lehmann, Florian Brueckner, and Patrick Cramer Gene Center Munich and Center for integrated Protein Science CiPS M, Department of Chemistry
SUPPLEMENTARY INFORMATION. The nucleotide binding dynamics of MSH2/MSH3 are lesion-dependent.
 SUPPLEMENTARY INFORMATION The nucleotide binding dynamics of /MSH are lesion-dependent. Barbara A. L. Owen, Walter H. Lang, and Cynthia T. McMurray* mau 1 8 6 4 2 mau 8 6 4 2 mau 8 6 4 2 mau 8 7 6 5 4
SUPPLEMENTARY INFORMATION The nucleotide binding dynamics of /MSH are lesion-dependent. Barbara A. L. Owen, Walter H. Lang, and Cynthia T. McMurray* mau 1 8 6 4 2 mau 8 6 4 2 mau 8 6 4 2 mau 8 7 6 5 4
A(+1) A( 7) RNA hairpin
 PP7 CP dimer A(+1) A( 7) RNA hairpin 5 3 Supplemental Figure 1. Sample electron density of RNA hairpin. Cartoon showing PP7 FG CP (wheat) bound to RNA hairpin (violet) showing 2Fo-Fc electron density (blue)
PP7 CP dimer A(+1) A( 7) RNA hairpin 5 3 Supplemental Figure 1. Sample electron density of RNA hairpin. Cartoon showing PP7 FG CP (wheat) bound to RNA hairpin (violet) showing 2Fo-Fc electron density (blue)
Supplementary Figure 1 Two distinct conformational states of the HNH domain in crystal structures. a
 Supplementary Figure 1 Two distinct conformational states of the HNH domain in crystal structures. a HNH-state 1 in PDB 4OO8, in which the distance from the C atom of the HNH catalytic residue 840 to the
Supplementary Figure 1 Two distinct conformational states of the HNH domain in crystal structures. a HNH-state 1 in PDB 4OO8, in which the distance from the C atom of the HNH catalytic residue 840 to the
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1164346/dc1 Supporting Online Material for Electron Cryomicroscopy of E. coli Reveals Filament Bundles Involved in Plasmid DNA Segregation Jeanne Salje,* Benoit Zuber,
www.sciencemag.org/cgi/content/full/1164346/dc1 Supporting Online Material for Electron Cryomicroscopy of E. coli Reveals Filament Bundles Involved in Plasmid DNA Segregation Jeanne Salje,* Benoit Zuber,
Appendix B Dansyl probe syntheses and characterization and D-8-Ad:P450cam structure determination
 201 Appendix B Dansyl probe syntheses and characterization and D-8-Ad:P450cam structure determination Acknowlegements. The structure of the D-8-Ad:P450cam conjugate was determined by Anna-Maria A. Hays.
201 Appendix B Dansyl probe syntheses and characterization and D-8-Ad:P450cam structure determination Acknowlegements. The structure of the D-8-Ad:P450cam conjugate was determined by Anna-Maria A. Hays.
Nature Structural & Molecular Biology: doi: /nsmb.2996
 Supplementary Figure 1 Negative-stain EM of native cytoplasmic dynein. (a) A representative micrograph of negatively stained dynein, with the flexible dimeric molecules circled in white. (b) Reference-free
Supplementary Figure 1 Negative-stain EM of native cytoplasmic dynein. (a) A representative micrograph of negatively stained dynein, with the flexible dimeric molecules circled in white. (b) Reference-free
Combine Cross-Diffusion Microbatch Method with Seeding Technique to Enhance Protein Crystallization Based on a Common Dispersing Agent
 Electronic Supplementary Material (ESI) for CrystEngComm. This journal is The Royal Society of Chemistry 2017 Supplementary Material Combine Cross-Diffusion Microbatch Method with Seeding Technique to
Electronic Supplementary Material (ESI) for CrystEngComm. This journal is The Royal Society of Chemistry 2017 Supplementary Material Combine Cross-Diffusion Microbatch Method with Seeding Technique to
Supplemental Information
 FOOT-AND-MOUTH DISEASE VIRUS PROTEIN 2C IS A HEXAMERIC AAA+ PROTEIN WITH A COORDINATED ATP HYDROLYSIS MECHANISM. Trevor R. Sweeney 1, Valentina Cisnetto 1, Daniel Bose 2, Matthew Bailey 3, Jon R. Wilson
FOOT-AND-MOUTH DISEASE VIRUS PROTEIN 2C IS A HEXAMERIC AAA+ PROTEIN WITH A COORDINATED ATP HYDROLYSIS MECHANISM. Trevor R. Sweeney 1, Valentina Cisnetto 1, Daniel Bose 2, Matthew Bailey 3, Jon R. Wilson
HEK293T. Fig. 1 in the
 Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Protocol for in vitro transcription
 Protocol for in vitro transcription Assemble the reaction at room temperature in the following order: Component 10xTranscription Buffer rntp T7 RNA Polymerase Mix grna PCR DEPC H 2 O volume 2μl 2μl 2μl
Protocol for in vitro transcription Assemble the reaction at room temperature in the following order: Component 10xTranscription Buffer rntp T7 RNA Polymerase Mix grna PCR DEPC H 2 O volume 2μl 2μl 2μl
Structural bases for N-glycan processing by mannosidephosphorylase
 Supporting information Volume 71 (2015) Supporting information for article: Structural bases for N-glycan processing by mannosidephosphorylase Simon Ladevèze, Gianluca Cioci, Pierre Roblin, Lionel Mourey,
Supporting information Volume 71 (2015) Supporting information for article: Structural bases for N-glycan processing by mannosidephosphorylase Simon Ladevèze, Gianluca Cioci, Pierre Roblin, Lionel Mourey,
Bio5325 Fall Crystal Vocabulary
 Crystals and Crystallization Bio5325 Fall 2007 Crystal Vocabulary Mosaicity (mosaic spread) Protein crystals are imperfect, consisting of a mosaic of domains that are slightly misaligned. As a result,
Crystals and Crystallization Bio5325 Fall 2007 Crystal Vocabulary Mosaicity (mosaic spread) Protein crystals are imperfect, consisting of a mosaic of domains that are slightly misaligned. As a result,
Supporting Information. Noncovalent insertion of ferrocenes into the protein shell of apo-ferritin
 Supporting Information Noncovalent insertion of ferrocenes into the protein shell of apo-ferritin Jochen Niemeyer, Satoshi Abe, Tatsuo Hikage, Takafumi Ueno, Gerhard Erker, Yoshihito Watanabe Department
Supporting Information Noncovalent insertion of ferrocenes into the protein shell of apo-ferritin Jochen Niemeyer, Satoshi Abe, Tatsuo Hikage, Takafumi Ueno, Gerhard Erker, Yoshihito Watanabe Department
Supplemental Information. The structural basis of R Spondin recognition by LGR5 and RNF43
 Supplemental Information The structural basis of R Spondin recognition by LGR5 and RNF43 Po Han Chen 1, Xiaoyan Chen 1, Deyu Fang 2, Xiaolin He 1* 1 Department of Molecular Pharmacology and Biological
Supplemental Information The structural basis of R Spondin recognition by LGR5 and RNF43 Po Han Chen 1, Xiaoyan Chen 1, Deyu Fang 2, Xiaolin He 1* 1 Department of Molecular Pharmacology and Biological
Optically Controlled Reversible Protein Hydrogels Based on Photoswitchable Fluorescent Protein Dronpa. Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Optically Controlled Reversible Protein Hydrogels Based on Photoswitchable Fluorescent Protein
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Optically Controlled Reversible Protein Hydrogels Based on Photoswitchable Fluorescent Protein
Cryoplunge 3 with GentleBlot for cryo-em
 Cryoplunge 3 with GentleBlot for cryo-em Cryoplunge 3 with GentleBlot system GentleBlot technology provides very gentle 1- and 2-side blotting for preparing frozen hydrated specimens for cryo-em Ethane
Cryoplunge 3 with GentleBlot for cryo-em Cryoplunge 3 with GentleBlot system GentleBlot technology provides very gentle 1- and 2-side blotting for preparing frozen hydrated specimens for cryo-em Ethane
Structural basis of transferrin sequestration by transferrin-binding protein B
 Supplementary Information for Structural basis of transferrin sequestration by transferrin-binding protein B Charles Calmettes, Joenel Alcantara, Rong-Hua Yu, Anthony B. Schryvers and Trevor F. Moraes*
Supplementary Information for Structural basis of transferrin sequestration by transferrin-binding protein B Charles Calmettes, Joenel Alcantara, Rong-Hua Yu, Anthony B. Schryvers and Trevor F. Moraes*
Bioinformatics & Protein Structural Analysis. Bioinformatics & Protein Structural Analysis. Learning Objective. Proteomics
 The molecular structures of proteins are complex and can be defined at various levels. These structures can also be predicted from their amino-acid sequences. Protein structure prediction is one of the
The molecular structures of proteins are complex and can be defined at various levels. These structures can also be predicted from their amino-acid sequences. Protein structure prediction is one of the
SUPPLEMENTARY INFORMATION
 Results Construct purification and coupling. Two A1-GP1bα ReaLiSM constructs, with and without cysteine residues near the N and C-termini (Fig. S2a), were expressed and purified by Ni affinity chromatography
Results Construct purification and coupling. Two A1-GP1bα ReaLiSM constructs, with and without cysteine residues near the N and C-termini (Fig. S2a), were expressed and purified by Ni affinity chromatography
A Molecular Replacement Pipeline
 α A Molecular Replacement Pipeline Garib Murshudov Chemistry Department, University of York 1) Introduction 2) Organisation of BALBES 3) Search model preparation 4) Updating BALBES 5) Warnings: Twin 6)
α A Molecular Replacement Pipeline Garib Murshudov Chemistry Department, University of York 1) Introduction 2) Organisation of BALBES 3) Search model preparation 4) Updating BALBES 5) Warnings: Twin 6)
His-Spin Protein Miniprep
 INSTRUCTIONS His-Spin Protein Miniprep Catalog No. P2001 (10 purifications) and P2002 (50 purifications). Highlights Fast 5 minute protocol to purify His-tagged proteins from cell-free extracts Screen
INSTRUCTIONS His-Spin Protein Miniprep Catalog No. P2001 (10 purifications) and P2002 (50 purifications). Highlights Fast 5 minute protocol to purify His-tagged proteins from cell-free extracts Screen
Dynamic High Capacity Mustang Q Membrane Units for Scaleable Anion Exchange Chromatography Purification of Adenoviral Vectors
 Contact Us: www.pall.com/contact Dynamic High Capacity Mustang Q Membrane Units for Scaleable Anion Exchange Chromatography Purification of Adenoviral Vectors Dynamic High Capacity Mustang Q Membrane Units
Contact Us: www.pall.com/contact Dynamic High Capacity Mustang Q Membrane Units for Scaleable Anion Exchange Chromatography Purification of Adenoviral Vectors Dynamic High Capacity Mustang Q Membrane Units
Protein expression and purification from Hi5 insect cells is as described (1, 2).
 Materials & methods Protein expression and purification Protein expression and purification from Hi5 insect cells is as described (1, 2). The purified complex exhibited a 2:2:2 stoichiometry as determined
Materials & methods Protein expression and purification Protein expression and purification from Hi5 insect cells is as described (1, 2). The purified complex exhibited a 2:2:2 stoichiometry as determined
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Location of the mab 6F10 epitope in capsids.
 Supplementary Figure 1 Location of the mab 6F10 epitope in capsids. The epitope of monoclonal antibody, mab 6F10, was mapped to residues A862 H880 of the HSV-1 major capsid protein VP5 by immunoblotting
Supplementary Figure 1 Location of the mab 6F10 epitope in capsids. The epitope of monoclonal antibody, mab 6F10, was mapped to residues A862 H880 of the HSV-1 major capsid protein VP5 by immunoblotting
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10258 Supplementary Figure 1 Reconstitution of the CENP-A nucleosome with recombinant human histones H2A, H2B, H4, and CENP-A. a, Purified recombinant human
SUPPLEMENTARY INFORMATION doi:10.1038/nature10258 Supplementary Figure 1 Reconstitution of the CENP-A nucleosome with recombinant human histones H2A, H2B, H4, and CENP-A. a, Purified recombinant human
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
fibrils, however, oligomeric structures and amorphous protein aggregates were
 Supplementary Figure 1: Effect of equimolar EGCG on αs and Aβ aggregate formation. A D B E αs C F Figure S1: (A-C) Analysis of EGCG treated αs (1 µm) aggregation reactions by EM. A 1:1 molar ratio of EGCG
Supplementary Figure 1: Effect of equimolar EGCG on αs and Aβ aggregate formation. A D B E αs C F Figure S1: (A-C) Analysis of EGCG treated αs (1 µm) aggregation reactions by EM. A 1:1 molar ratio of EGCG
Supporting Information
 Supporting Information Chan et al. 10.1073/pnas.0903849106 SI Text Protein Purification. PCSK9 proteins were expressed either transiently in 2936E cells (1), or stably in HepG2 cells. Conditioned culture
Supporting Information Chan et al. 10.1073/pnas.0903849106 SI Text Protein Purification. PCSK9 proteins were expressed either transiently in 2936E cells (1), or stably in HepG2 cells. Conditioned culture
N173A. residue number
 a 0.3 Δω 1 ( 1 H Ν ) [ppm] 0-0.3 N173A -0.6 Y177A b Δω 3 ( 13 C α ) [ppm] 2 0-2 c Δω 3 ( 13 C β ) [ppm] 2 0-2 -4 Supplementary Figure 1 140 150 160 170 180 190 200 residue number Comparison of chemical
a 0.3 Δω 1 ( 1 H Ν ) [ppm] 0-0.3 N173A -0.6 Y177A b Δω 3 ( 13 C α ) [ppm] 2 0-2 c Δω 3 ( 13 C β ) [ppm] 2 0-2 -4 Supplementary Figure 1 140 150 160 170 180 190 200 residue number Comparison of chemical
Supplementary Information For. A genetically encoded tool for manipulation of NADP + /NADPH in living cells
 Supplementary Information For A genetically encoded tool for manipulation of NADP + /NADPH in living cells Valentin Cracan 1,2,3, Denis V. Titov 1,2,3, Hongying Shen 1,2,3, Zenon Grabarek 1* and Vamsi
Supplementary Information For A genetically encoded tool for manipulation of NADP + /NADPH in living cells Valentin Cracan 1,2,3, Denis V. Titov 1,2,3, Hongying Shen 1,2,3, Zenon Grabarek 1* and Vamsi
Automated Protocol for High-throughput Recombinant Protein Purification Using the Biomek FX (Beckman Coulter)
 Automated Protocol for High-throughput Recombinant Protein Purification Using the Biomek FX (Beckman Coulter) HIS-Select HF Nickel Affinity Gel Catalog Number H0537 Automation Guide 2 I. Description 2
Automated Protocol for High-throughput Recombinant Protein Purification Using the Biomek FX (Beckman Coulter) HIS-Select HF Nickel Affinity Gel Catalog Number H0537 Automation Guide 2 I. Description 2
Superdex 200 Increase columns
 GE Healthcare Life Sciences Data file 29-452-69 AA Size exclusion chromatography Superdex 2 Increase columns Superdex 2 Increase prepacked columns (Fig 1) are designed for size exclusion chromatography
GE Healthcare Life Sciences Data file 29-452-69 AA Size exclusion chromatography Superdex 2 Increase columns Superdex 2 Increase prepacked columns (Fig 1) are designed for size exclusion chromatography
Superdex 200 Increase columns
 Data file 29-42-69 AC Size exclusion chromatography Superdex 2 Increase columns Superdex 2 Increase prepacked columns (Fig 1) are designed for size exclusion chromatography (SEC)/high resolution gel filtration
Data file 29-42-69 AC Size exclusion chromatography Superdex 2 Increase columns Superdex 2 Increase prepacked columns (Fig 1) are designed for size exclusion chromatography (SEC)/high resolution gel filtration
Deep sequencing reveals global patterns of mrna recruitment
 Supplementary information for: Deep sequencing reveals global patterns of mrna recruitment during translation initiation Rong Gao 1#*, Kai Yu 1#, Ju-Kui Nie 1,Teng-Fei Lian 1, Jian-Shi Jin 1, Anders Liljas
Supplementary information for: Deep sequencing reveals global patterns of mrna recruitment during translation initiation Rong Gao 1#*, Kai Yu 1#, Ju-Kui Nie 1,Teng-Fei Lian 1, Jian-Shi Jin 1, Anders Liljas
TECHNICAL BULLETIN. HIS-Select HF Nickel Affinity Gel. Catalog Number H0537 Storage Temperature 2 8 C
 HIS-Select HF Nickel Affinity Gel Catalog Number H0537 Storage Temperature 2 8 C TECHNICAL BULLETIN Product Description HIS-Select High Flow (HF) is an immobilized metal-ion affinity chromatography (IMAC)
HIS-Select HF Nickel Affinity Gel Catalog Number H0537 Storage Temperature 2 8 C TECHNICAL BULLETIN Product Description HIS-Select High Flow (HF) is an immobilized metal-ion affinity chromatography (IMAC)
Nature Structural and Molecular Biology: doi: /nsmb.2937
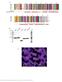 Supplementary Figure 1 Multiple sequence alignment of the CtIP N-terminal domain, purified CtIP protein constructs and details of the 2F o F c electron density map of CtIP-NTD. (a) Multiple sequence alignment,
Supplementary Figure 1 Multiple sequence alignment of the CtIP N-terminal domain, purified CtIP protein constructs and details of the 2F o F c electron density map of CtIP-NTD. (a) Multiple sequence alignment,
Fundamentals of X-ray diffraction and scattering
 Fundamentals of X-ray diffraction and scattering Don Savage dsavage@wisc.edu 1231 Engineering Research Building (608) 263-0831 X-ray diffraction and X-ray scattering Involves the elastic scattering of
Fundamentals of X-ray diffraction and scattering Don Savage dsavage@wisc.edu 1231 Engineering Research Building (608) 263-0831 X-ray diffraction and X-ray scattering Involves the elastic scattering of
DNA-templated protein arrays for single-molecule imaging. Supporting information
 DNA-templated protein arrays for single-molecule imaging Supporting information Methods Protein expression, receptor activity Gα i1 was expressed in E. coli from an IPTG-inducible pet15 plasmid. The coding
DNA-templated protein arrays for single-molecule imaging Supporting information Methods Protein expression, receptor activity Gα i1 was expressed in E. coli from an IPTG-inducible pet15 plasmid. The coding
Solutions to 7.02 Quiz II 10/27/05
 Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
A Brief Introduction to Structural Biology and Protein Crystallography
 A Brief Introduction to Structural Biology and Protein Crystallography structural biology of H2O http://courses.cm.utexas.edu/jrobertus/ch339k/overheads-1/water-structure.jpg Protein polymers fold up into
A Brief Introduction to Structural Biology and Protein Crystallography structural biology of H2O http://courses.cm.utexas.edu/jrobertus/ch339k/overheads-1/water-structure.jpg Protein polymers fold up into
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary Information. Supplementary Figures:
 Supplementary Information Supplementary Figures: Supplementary Figure 1: RCT reconstructions of Platform, PAZ and RNase III labeled Dicer. For each streptavidin labeled sample, the eight RCT reconstructions
Supplementary Information Supplementary Figures: Supplementary Figure 1: RCT reconstructions of Platform, PAZ and RNase III labeled Dicer. For each streptavidin labeled sample, the eight RCT reconstructions
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Mechanism for signal-induced opening of the DNA box. a, An atomic model of the DNA box held closed by locks (orange and blue) that are double helices formed by two short strands
Supplementary Figure 1. Mechanism for signal-induced opening of the DNA box. a, An atomic model of the DNA box held closed by locks (orange and blue) that are double helices formed by two short strands
Supplementary Figure 1. Peak assignments for V L- and O K-edge X-ray absorption near edge structure (XANES) Spectra (a) Assignments of V LIII-edge
 Supplementary Figure 1. Peak assignments for V L- and O K-edge X-ray absorption near edge structure (XANES) Spectra (a) Assignments of V LIII-edge spectral features to specific transitions measured for
Supplementary Figure 1. Peak assignments for V L- and O K-edge X-ray absorption near edge structure (XANES) Spectra (a) Assignments of V LIII-edge spectral features to specific transitions measured for
Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein. Application
 Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein Application Petra Sebastian Meike Kuschel Stefan Schmidt Abstract This Application Note describes
Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein Application Petra Sebastian Meike Kuschel Stefan Schmidt Abstract This Application Note describes
MagExtactor -His-tag-
 Instruction manual MagExtractor-His-tag-0905 F0987K MagExtactor -His-tag- Contents NPK-701 100 preparations Store at Store at 4 C [1] Introduction [2] Components [3] Materials required [4] Protocol3 1.
Instruction manual MagExtractor-His-tag-0905 F0987K MagExtactor -His-tag- Contents NPK-701 100 preparations Store at Store at 4 C [1] Introduction [2] Components [3] Materials required [4] Protocol3 1.
SI GUIDE. File Name: Supplementary Information Description: Supplementary Figures, Supplementary Notes and Supplementary References.
 SI GUIDE File Name: Supplementary Information Description: Supplementary Figures, Supplementary Notes and Supplementary References. File Name: Supplementary Movie 1 Description: (the movie from which Figs.
SI GUIDE File Name: Supplementary Information Description: Supplementary Figures, Supplementary Notes and Supplementary References. File Name: Supplementary Movie 1 Description: (the movie from which Figs.
Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron
 Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron density map obtained after SAD-phasing and density modification
Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron density map obtained after SAD-phasing and density modification
Institute of Physics, Academy of Sciences of the Czech Republic, v.v.i., Na Slovance 2, CZ , Prague 8, Czech Republic 2
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Theoretical and experimental study of antifreeze protein AFP752, trehalose and dimethyl sulfoxide
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Theoretical and experimental study of antifreeze protein AFP752, trehalose and dimethyl sulfoxide
RNP purification, components and activity.
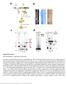 Supplementary Figure 1 RNP purification, components and activity. (a) Intein-mediated RNP production and purification. The Ll.LtrB intron RNA (red) (Exons E1 and E2 in green) and associated intron encoded
Supplementary Figure 1 RNP purification, components and activity. (a) Intein-mediated RNP production and purification. The Ll.LtrB intron RNA (red) (Exons E1 and E2 in green) and associated intron encoded
Strep-Spin Protein Miniprep Kit Catalog No. P2004, P2005
 INSTRUCTION MANUAL Strep-Spin Protein Miniprep Kit Catalog No. P2004, P2005 Highlights Fast protocol to purify Strep-tagged proteins from cell-free extracts Screen your recombinant colonies directly for
INSTRUCTION MANUAL Strep-Spin Protein Miniprep Kit Catalog No. P2004, P2005 Highlights Fast protocol to purify Strep-tagged proteins from cell-free extracts Screen your recombinant colonies directly for
Supplementary Information
 Supplementary Information Disulfide-Bond Scanning Reveals Assembly State and β-strand Tilt Angle of PFO β-barrel Takehiro K. Sato, Rodney K. Tweten, and Arthur E. Johnson Supplementary Results Supplementary
Supplementary Information Disulfide-Bond Scanning Reveals Assembly State and β-strand Tilt Angle of PFO β-barrel Takehiro K. Sato, Rodney K. Tweten, and Arthur E. Johnson Supplementary Results Supplementary
Structural and functional analysis of Hikeshi, a new nuclear transport receptor of Hsp70
 Supporting information Volume 71 (2015) Supporting information for article: Structural and functional analysis of Hikeshi, a new nuclear transport receptor of Hsp70 Jinsue Song, Shingo Kose, Ai Watanabe,
Supporting information Volume 71 (2015) Supporting information for article: Structural and functional analysis of Hikeshi, a new nuclear transport receptor of Hsp70 Jinsue Song, Shingo Kose, Ai Watanabe,
TECHNICAL BULLETIN. In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits
 In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
Supplementary information, Data S1
 Supplementary information, Data S1 Materials and Methods Reagents Anti-Flag M2 affinity agarose gel was from Sigma; Mono S and Superose 6 were from GE Healthcare; Polyethylenimine (PEI) was from polysciences
Supplementary information, Data S1 Materials and Methods Reagents Anti-Flag M2 affinity agarose gel was from Sigma; Mono S and Superose 6 were from GE Healthcare; Polyethylenimine (PEI) was from polysciences
SUPPLEMENTAL MATERIAL BIOCHEMICAL AND STRUCTURAL STUDIES ON THE M. TUBERCULOSIS O 6 -METHYLGUANINE METHYLTRANSFERASE AND MUTATED VARIANTS
 SUPPLEMENTAL MATERIAL BIOCHEMICAL AND STRUCTURAL STUDIES ON THE M. TUBERCULOSIS O 6 -METHYLGUANINE METHYLTRANSFERASE AND MUTATED VARIANTS Riccardo Miggiano 1, Valentina Casazza 1, Silvia Garavaglia 1,
SUPPLEMENTAL MATERIAL BIOCHEMICAL AND STRUCTURAL STUDIES ON THE M. TUBERCULOSIS O 6 -METHYLGUANINE METHYLTRANSFERASE AND MUTATED VARIANTS Riccardo Miggiano 1, Valentina Casazza 1, Silvia Garavaglia 1,
Protein degradation in a TX-TL cell-free expression system using ClpXP protease
 Technical Report, 8 July 1 http://www.cds.caltech.edu/~murray/papers/sksm1-clpxp.html Protein degradation in a TX-TL cell-free expression system using ClpXP protease AUTHORS: Zachary Z. Sun 1, Jongmin
Technical Report, 8 July 1 http://www.cds.caltech.edu/~murray/papers/sksm1-clpxp.html Protein degradation in a TX-TL cell-free expression system using ClpXP protease AUTHORS: Zachary Z. Sun 1, Jongmin
Site-specific protein cross-linking by peroxidase-catalyzed activation of a tyrosine-containing peptide tag
 Supporting Information Site-specific protein cross-linking by peroxidase-catalyzed activation of a tyrosine-containing peptide tag Kosuke Minamihata 1, Masahiro Goto 1,2, Noriho Kamiya 1,2* 1 Department
Supporting Information Site-specific protein cross-linking by peroxidase-catalyzed activation of a tyrosine-containing peptide tag Kosuke Minamihata 1, Masahiro Goto 1,2, Noriho Kamiya 1,2* 1 Department
The molecular basis of lysine 48 ubiquitin chain synthesis by Ube2K
 Supplementary Information The molecular basis of lysine 48 ubiquitin chain synthesis by Adam J. Middleton, Catherine L. Day* Department of Biochemistry, Otago School of Medical Sciences, University of
Supplementary Information The molecular basis of lysine 48 ubiquitin chain synthesis by Adam J. Middleton, Catherine L. Day* Department of Biochemistry, Otago School of Medical Sciences, University of
