Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
|
|
|
- Elwin Fleming
- 5 years ago
- Views:
Transcription
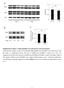
1 Merkestein et al. Supplementary Figure 1 A PLIN WT FTO KO 72 kda FABP4 17 kda HSC70 72 kda B Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation. Western blots of PLIN1 and FABP4 from WT and FTO-KO MEFs 7 days after adipogenic induction. The housekeeping protein HSC70 (heatshock protein 70) is included as a loading control. Replicates are different experiments from the same batch of MEFs.B. Quantitative expression (by densitometry) of PLIN1 and FABP4 protein measured in WT (black) and FTO-KO (green) MEFs 7 days after adipogenic induction. Protein expression was expressed relative to HSC70 and then normalised to WT. Replicates are different experiments from the same batch of MEFs (n=3). Differences were analysed using a multivariate ANOVA with Bonferroni post-hoc testing, * p<0.05. Graph represents mean±sem
2 Merkestein et al. Supplementary Figure 2 A ACTIN FABP4 WT FTO-4 52 kda 42 kda 17 kda B Supplementary Figure 2. Adipogenic protein expression in WT and FTO-4 MEFs after 7 days of adipogenic differentiation. A. Western blots of FABP4 and actin from WT and FTO-4 MEFs, 7 days after adipogenic induction. Actin was included as a loading control. Replicates are different experiments from the same batch of MEFs.B. Protein expression of FABP4 measured in WT (black) and FTO-4 (purple) MEFs 7 days after adipogenic induction. Protein expression was expressed relative to actin and then normalised to WT. Replicates are different experiments from the same batch of MEFs (n=2). Differences were analysed using an independent Student s t-test, * p<0.05. Graph represents mean±sem
3 Merkestein et al. Supplementary Figure 3 Supplementary Figure 3. Confirmation of FTO knockdown in primary adipocytes derived from FTO-4 mice. Quantitative PCR of FTO mrna in FTO-4 MEFs following transfection with control (black) or FTO sirna (green). Replicates (n=3) are different experiments from the same batch of primary adipocytes (derived from one mouse). Statistical significance was analysed with Student s t-test, *** p< Graph represents mean±sem.
4 Merkestein et al. Supplementary Figure 4 A B Supplementary Figure 4. Expression levels of Fto, RPGRIP1L and IRX3 mrnas in gwat and MEFs from WT mice. Quantitative PCR of Fto, RPGRIP1L and IRX3 mrnas measured in gwat (A) and MEFs (B) from WT mice. Data are expressed relative to expression of the housekeeping gene β- actin (ACTB). A, n=4 mice. B, n=3 different experiments using MEFs derived from 3 different mice. Statistical significance was analysed using a oneway ANOVA against RPGRIP1L and IRX3, * p<0.05; **p<0.01. Graph represents mean±sem.
5 A. B. Merkestein et al. Supplementary Figure 5 C. Supplementary Figure 5. Expression levels of Fto, RPGRIP1L and IRX3 mrnas in gwat of WT and FTO-4 mice and WT, FTO-KO and FTO-4 MEFs. Quantitative PCR of Fto, RPGRIP1L and IRX3 mrnas measured (A) in WT (black, n=2) and FTO-4 (purple, n=5) MEFs; (B) WT (black, n=3) and FTO-KO (green, n=3) MEFs; and (C) gwat from WT (black, n=3) and FTO-4 (purple, n=3) mice. N numbers represent biological replicates (MEFs derived from different embryos). Statistical significance was analysed by multivariate ANOVA comparing expression of Fto, RPGRIP1L and IRX3 in WT versus FTO-4 or FTO-KO, * p<0.05. Graphs represent mean±sem.
6 Merkestein et al. Supplementary Figure 6 A. B. Supplementary Figure 6. Adipogenic gene expression in FTO-KO and FTO-4, and their respective WT MEFs, 3 days after induction of adipogenesis. qpcr of FABP4 and PPARγ mrnas from WT (black), FTO-4 (purple) and FTO-KO (green) MEFs 3 days after induction of adipogenic differentiation. n=3 different experiments using MEFs derived from one mouse. Multivariate ANOVA with Bonferroni correction, * p<0.05 against WT. Graphs represent mean±sem.
7 Merkestein et al. Supplementary Figure 7 Supplementary Figure 7. Fto expression during differentiation in MEFs. Fto mrna expression levels measured by qpcr in WT MEFs before (Day 0) and during adipogenic differentiation (16 and 24 hours, 2,3, 5 and 7 days). Data from the 0, 16h and 24h time points consist of data from 2 different batches of MEFs each with 3 technical replicates, the other time points indicate 3 different experiments using MEFs derived from one mouse. Repeated measures ANOVA followed by comparison of all time points to Day 0 with a Dunnett s post-hoc test, and stars indicate significantly different from time point 0, **p<0.01; ***p< Graph represents mean±sem.
8 Calcein AM Emission Ethidium Homodimer Emission Calcein AM Emission Ethidium Homodimer Emission Merkestein et al. Supplementary Figure * WT FTO-KO WT FTO-KO 0 WT FTO-KO WT FTO WT FTO-4 0 WT FTO-4 Supplementary Figure 8. Cell viability assay in WT, FTO-KO and FTO-4 MEFs 24 hours after adipogenic induction. Number of live (left, Calcein EM emission) and dead (right, ethidium homodimer emission) cells 24 hours after adipogenic induction in WT (black), FTO-4 (purple) and FTO-KO (green) MEFs. Data are a minimum of 2 biological replicates, with each 5 technical replicates. Student s t-test, * p<0.05 against WT. Graphs represent mean±sem.
9 C a lc e in A M E m is s io n E th id iu m H o m o d im e r E m is s io n A. FTO-4 sicon Merkestein et al. Supplementary Figure 9 FTO-4 sifto B. C D *** F T O -4 s ic O N F T O -4 s if T O F T O -4 s ic O N F T O -4 s if T O F T O -4 s ic O N F T O -4 s if T O Supplementary Figure 9. BrdU incorporation and cell viability assay 24 hours after adipogenic induction in FTO-4 MEFs treated with FTO sirna. A. BrdU incorporation 24 hours after induction of adipogenic differentiation in MEFs from FTO-4 mice treated with control sirna (left) or FTO sirna (right). Cells were stained for BrdU (green) and nuclei visualised with DAPI (blue), scale bar, 50μM. B. Quantification of BrdU incorporation in FTO-4 MEFs treated with FTO sirna (green) or control sirna (black). Data represent 6 experiments carried out in MEFs derived from one mouse. C, D. Number of live (C, Calcein EM emission) and dead (D, ethidium homodimer emission) cells 24 hours after adipogenic induction in FTO-4 MEFs treated with control sirna (black) and FTO sirna (green). Student s t-test, ***p<0.001, * p<0.05 against FTO-4 sicon. Graphs represent mean±sem.
10 Merkestein et al. Supplementary Figure 10 Supplementary Figure 10. CCND1 expression after induction of adipogenic differentiation in WT MEFs that overexpress Fto. Quantitative PCR of CCND1 mrna 16, 24 and 40 hours after induction of adipogenic differentiation in WT MEFs transfected with a vector expressing full-length FTO (FTO OE, purple) or empty vector (EV, black) as a control. Data represent 3 different experiments using MEFs derived from one mouse. Multivariate ANOVA comparing empty vector to FTO overexpression at each time point, * p<0.05. Graph represents mean±sem.
11 Merkestein et al. Supplementary Figure 11 A B C Supplementary Figure 11. FTO overexpression leads to increased body weight and adiposity. Body weight (A), fat mass (B), and lean mass (C) in FTO-4 mice (n=22, purple) and wildtype (n=20, black) female C57BL/6J mice during chow and high-fat feeding. Animals were maintained on chow from weaning until 17 weeks, at which time they were given a high-fat diet (as indicated by the dotted vertical line) until completion of the study at 28 weeks. Note that the y-axis starts at 10 grams for Supplementary Figures S12A and S12C. Repeated-measures ANOVA, *p<0.05; **p<0.01; ***p<0.001 against WT.Graphs represent means±sem.
12 Merkestein et al. Supplementary Figure 12 4 wks old (weaning) 12 wks old (8 wks HFD) WT 0.078±0.007 mg 0.335±0.05 mg FTO ±0.002 mg 0.413±0.07 mg Supplementary Figure 12. Weights of gwat depots of WT and FTO-4 mice after weaning or following an 8-week HFD. Weights of gwat depots of WT and FTO-4 mice at 4 weeks of age after weaning (WT, n=3; FTO-4, n=5) or at 12 weeks old following an 8-week HFD initiated after weaning (WT, n=4; FTO- 4, n=3). Student s t-test. Data represent means±sem.
13 Original western blot images Supplementary Figure 1a PLIN1 Supplementary Figure 2a ACTIN 95 kda 72 kda 42kDa 52 kda 42 kda 34 kda 28 kda 17 kda 10 kda FABP4 28 kda 17 kda FABP4 HSC70 95 kda 72 kda 42kDa
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector.
 Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1
 Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Supplementary Information
 Supplementary Information Supplementary Figures Supplementary Figure 1. MLK1-4 phosphorylate MEK in the presence of RAF inhibitors. (a) H157 cells were transiently transfected with Flag- or HA-tagged MLK1-4
Supplementary Information Supplementary Figures Supplementary Figure 1. MLK1-4 phosphorylate MEK in the presence of RAF inhibitors. (a) H157 cells were transiently transfected with Flag- or HA-tagged MLK1-4
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates
 Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates vascular calcification (VC). (a) Von Kossa staining shows that TSA potentiated the Pi-induced VC. Scale bar, 100
Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates vascular calcification (VC). (a) Von Kossa staining shows that TSA potentiated the Pi-induced VC. Scale bar, 100
a. Increased active HPSE (50 kda) protein expression upon infection with multiple herpes viruses: bovine
 Fold change vs uninfeced MOI 0.1 MOI 1 MOI 0.1 MOI 1 MOI 0.001 MOI 0.01 Fold change vs uninfected a 6 4 2 50 kda HPSE * ** * * BHV PRV HSV-2 333 ** * 0 - + + - + + - + + b 50 kda HPSE 3 2 ** *** mock inf
Fold change vs uninfeced MOI 0.1 MOI 1 MOI 0.1 MOI 1 MOI 0.001 MOI 0.01 Fold change vs uninfected a 6 4 2 50 kda HPSE * ** * * BHV PRV HSV-2 333 ** * 0 - + + - + + - + + b 50 kda HPSE 3 2 ** *** mock inf
(A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: WT; lower
 Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification.
 Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Supplementary Figure 1. Additional RNAi screen data
 Supplementary Figure 1. Additional RNAi screen data A. Cisplatin induced ATR autophosphorylation. Western blot illustrating ATR and phospho-atr (T1989) in cells exposed to 1 µm cisplatin for 24 hours prior
Supplementary Figure 1. Additional RNAi screen data A. Cisplatin induced ATR autophosphorylation. Western blot illustrating ATR and phospho-atr (T1989) in cells exposed to 1 µm cisplatin for 24 hours prior
Supplementary information. Supplementary Figures
 Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
a KYSE270-CON KYSE270-Id1
 a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Supplementary Table, Figures and Videos
 Supplementary Table, Figures and Videos Table S1. Oligonucleotides used for different approaches. (A) RT-qPCR study. (B) qpcr study after ChIP assay. (C) Probes used for EMSA. Figure S1. Notch activation
Supplementary Table, Figures and Videos Table S1. Oligonucleotides used for different approaches. (A) RT-qPCR study. (B) qpcr study after ChIP assay. (C) Probes used for EMSA. Figure S1. Notch activation
Supplementary Figure 1. GST pull-down analysis of the interaction of GST-cIAP1 (A, B), GSTcIAP1
 Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3164 Supplementary Figure 1 Validation of effective Gnas deletion and epithelial thickness. a, Representative genotyping in mice treated or not with tamoxifen to show Gnas deletion. To
DOI: 10.1038/ncb3164 Supplementary Figure 1 Validation of effective Gnas deletion and epithelial thickness. a, Representative genotyping in mice treated or not with tamoxifen to show Gnas deletion. To
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Gene Forward (5 to 3 ) Reverse (5 to 3 ) Accession # PKA C- TCTGAGGAAATGGGAGAACC CGAGGGTTTTCTTCCTCTCAA NM_011100
 Supplementary Methods: Materials. BRL37344, insulin, 3-isobutyl-1-methylxanthine, dibutyryl camp (Bt2-cAMP) and 8-Bromoadenosine 3,5 -cyclic monophosphate sodium (8-br-cAMP), cilostamide, adenosine deaminase,
Supplementary Methods: Materials. BRL37344, insulin, 3-isobutyl-1-methylxanthine, dibutyryl camp (Bt2-cAMP) and 8-Bromoadenosine 3,5 -cyclic monophosphate sodium (8-br-cAMP), cilostamide, adenosine deaminase,
Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
hnrnp D/AUF1 Rabbit IgG hnrnp M
 Mouse IgG Goat IgG Rabbit IgG Mouse IgG hnrnp F Goat IgG Mouse IgG Kb 6 4 3 2 15 5 Supplementary Figure S1. In vivo binding of TERRA-bound RBPs to target RNAs. Immunoprecipitation (IP) assay using 3 mg
Mouse IgG Goat IgG Rabbit IgG Mouse IgG hnrnp F Goat IgG Mouse IgG Kb 6 4 3 2 15 5 Supplementary Figure S1. In vivo binding of TERRA-bound RBPs to target RNAs. Immunoprecipitation (IP) assay using 3 mg
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplemental Figure 1: Podocyte specific knockdown of Klf4 in Podocin-Cre Klf4 flox/flox mice was confirmed. (A) Primary glomerular epithelial cells
 Supplemental Figure 1: Podocyte specific knockdown of Klf4 in Podocin-Cre Klf4 flox/flox mice was confirmed. (A) Primary glomerular epithelial cells were isolated from 10-week old Podocin-Cre Klf4 flox/flox
Supplemental Figure 1: Podocyte specific knockdown of Klf4 in Podocin-Cre Klf4 flox/flox mice was confirmed. (A) Primary glomerular epithelial cells were isolated from 10-week old Podocin-Cre Klf4 flox/flox
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figures and Legends.
 Supplementary Figures and Legends. Supplementary Figure 1: Impact of injury on Rb1 and PPARϒ expression. Following ipsilateral axotomy injury, adult DRG expression of Rb1 mrna declined (*p
Supplementary Figures and Legends. Supplementary Figure 1: Impact of injury on Rb1 and PPARϒ expression. Following ipsilateral axotomy injury, adult DRG expression of Rb1 mrna declined (*p
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
WT Day 90 after injections
 Supplementary Figure 1 a Day 1 after injections Day 9 after injections Klf5 +/- Day 1 after injections Klf5 +/- Day 9 after injections BLM PBS b Day 1 after injections Dermal thickness (μm) 3 1 Day 9 after
Supplementary Figure 1 a Day 1 after injections Day 9 after injections Klf5 +/- Day 1 after injections Klf5 +/- Day 9 after injections BLM PBS b Day 1 after injections Dermal thickness (μm) 3 1 Day 9 after
Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr
 Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in
 Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
TRIM31 is recruited to mitochondria after infection with SeV.
 Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Flowcytometry-based purity analysis of peritoneal macrophage culture.
 Liao et al., KLF4 regulates macrophage polarization Revision of Manuscript 45444-RG- Supplementary Figure Legends Figure S Flowcytometry-based purity analysis of peritoneal macrophage culture. Thioglycollate
Liao et al., KLF4 regulates macrophage polarization Revision of Manuscript 45444-RG- Supplementary Figure Legends Figure S Flowcytometry-based purity analysis of peritoneal macrophage culture. Thioglycollate
Supplementary Information
 Supplementary Information Supplementary Figure S1 (a) P-cRAF colocalizes with LC3 puncta. Immunofluorescence (IF) depicting colocalization of P-cRAF (green) and LC3 puncta (red) in NIH/3T3 cells treated
Supplementary Information Supplementary Figure S1 (a) P-cRAF colocalizes with LC3 puncta. Immunofluorescence (IF) depicting colocalization of P-cRAF (green) and LC3 puncta (red) in NIH/3T3 cells treated
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State
 Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Li et al., Supplemental Figures
 Li et al., Supplemental Figures Fig. S1. Suppressing TGM2 expression with TGM2 sirnas inhibits migration and invasion in A549-TR cells. A, A549-TR cells transfected with negative control sirna (NC sirna)
Li et al., Supplemental Figures Fig. S1. Suppressing TGM2 expression with TGM2 sirnas inhibits migration and invasion in A549-TR cells. A, A549-TR cells transfected with negative control sirna (NC sirna)
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
Supplementary Figure 1. Nur77 and leptin-controlled obesity. (A) (B) (C)
 Supplementary Figure 1. Nur77 and leptin-controlled obesity. (A) Effect of leptin on body weight and food intake between WT and KO mice at the age of 12 weeks (n=7). Mice were i.c.v. injected with saline
Supplementary Figure 1. Nur77 and leptin-controlled obesity. (A) Effect of leptin on body weight and food intake between WT and KO mice at the age of 12 weeks (n=7). Mice were i.c.v. injected with saline
FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs
 FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs amount of standard analysed. Each point represents the
FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs amount of standard analysed. Each point represents the
Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein 22 mutant that causes type 1A Charcot-Marie-Tooth disease
 Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein mutant that causes type 1A Charcot-Marie-Tooth disease Taichi Hara, Yukiko Hashimoto, Tomoko Akuzawa, Rika Hirai,
Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein mutant that causes type 1A Charcot-Marie-Tooth disease Taichi Hara, Yukiko Hashimoto, Tomoko Akuzawa, Rika Hirai,
SUPPLEMENTARY INFORMATION. Small molecule activation of the TRAIL receptor DR5 in human cancer cells
 SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by. Regulating Occludin
 Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by Regulating Occludin Chaithanya Chelakkot 1,ǂ, Jaewang Ghim 2,3,ǂ, Nirmal Rajasekaran 4, Jong-Sun Choi 5, Jung-Hwan
Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by Regulating Occludin Chaithanya Chelakkot 1,ǂ, Jaewang Ghim 2,3,ǂ, Nirmal Rajasekaran 4, Jong-Sun Choi 5, Jung-Hwan
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/428/ra5/1 Supplementary Materials for ameliorates liver steatosis by decreasing stearoyloa desaturase 1 (S1) abundance and altering hepatic lipid metabolism
www.sciencesignaling.org/cgi/content/full/9/428/ra5/1 Supplementary Materials for ameliorates liver steatosis by decreasing stearoyloa desaturase 1 (S1) abundance and altering hepatic lipid metabolism
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas.
 Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
neutrophils (CD11b+, the total Statistical multiple
 Supplementary Figure 1. (A) Wild type and Vim / mice were treated with (24mg/kg LPS); lungs were harvested at 48h and enzymaticallyy digested. CD45+ cells were excluded from the total cell population and
Supplementary Figure 1. (A) Wild type and Vim / mice were treated with (24mg/kg LPS); lungs were harvested at 48h and enzymaticallyy digested. CD45+ cells were excluded from the total cell population and
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various
 Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/282/ra53/dc1 Supplementary Materials for Estrogen Alters the Splicing of Type 1 Corticotropin-Releasing Hormone Receptor in Breast Cancer Cells Suchita Lal,
www.sciencesignaling.org/cgi/content/full/6/282/ra53/dc1 Supplementary Materials for Estrogen Alters the Splicing of Type 1 Corticotropin-Releasing Hormone Receptor in Breast Cancer Cells Suchita Lal,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/496/eaam6291/dc1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells
www.sciencesignaling.org/cgi/content/full/10/496/eaam6291/dc1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells
Supplementary Methods
 Supplementary Methods Microarray Data Analysis Gene expression data were obtained by hybridising a total of 24 samples from 6 experimental groups (n=4 per group) to Illumina HumanHT-12 Expression BeadChips.
Supplementary Methods Microarray Data Analysis Gene expression data were obtained by hybridising a total of 24 samples from 6 experimental groups (n=4 per group) to Illumina HumanHT-12 Expression BeadChips.
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
SUPPLEMENTARY DATA. Supplementary Table 1. Sequences of qpcr primers.
 Supplementary Table 1. Sequences of qpcr primers. Lpl Fatp1 (Slc27a1) Fatp4 (Slc27a4) Cd36 Acsl1 Gpam Agpat2 Dgat1 Dgat2 Plin1 Atgl Hsl Mgll Lpin1 Got2 Cav1 Cav2 Fitm2 Nr1h2 Nr1h3 Acsl4 Acsl5 Agpat6 Agpat9
Supplementary Table 1. Sequences of qpcr primers. Lpl Fatp1 (Slc27a1) Fatp4 (Slc27a4) Cd36 Acsl1 Gpam Agpat2 Dgat1 Dgat2 Plin1 Atgl Hsl Mgll Lpin1 Got2 Cav1 Cav2 Fitm2 Nr1h2 Nr1h3 Acsl4 Acsl5 Agpat6 Agpat9
Supplementary Figure 1
 Supplementary Figure 1 Virus infection induces RNF128 expression. (a,b) RT-PCR analysis of Rnf128 (RNF128) mrna expression in mouse peritoneal macrophages (a) and THP-1 cells (b) upon stimulation with
Supplementary Figure 1 Virus infection induces RNF128 expression. (a,b) RT-PCR analysis of Rnf128 (RNF128) mrna expression in mouse peritoneal macrophages (a) and THP-1 cells (b) upon stimulation with
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Figures and supplementary figure legends
 Supplementary Figures and supplementary figure legends Figure S1. Effect of different percentage of FGF signaling knockdown on TGF signaling and EndMT marker gene expression. HUVECs were subjected to different
Supplementary Figures and supplementary figure legends Figure S1. Effect of different percentage of FGF signaling knockdown on TGF signaling and EndMT marker gene expression. HUVECs were subjected to different
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplemental Table 1: Sequences of real time PCR primers. Primers were intronspanning
 Symbol Accession Number Sense-primer (5-3 ) Antisense-primer (5-3 ) T a C ACTB NM_001101.3 CCAGAGGCGTACAGGGATAG CCAACCGCGAGAAGATGA 57 HSD3B2 NM_000198.3 CTTGGACAAGGCCTTCAGAC TCAAGTACAGTCAGCTTGGTCCT 60
Symbol Accession Number Sense-primer (5-3 ) Antisense-primer (5-3 ) T a C ACTB NM_001101.3 CCAGAGGCGTACAGGGATAG CCAACCGCGAGAAGATGA 57 HSD3B2 NM_000198.3 CTTGGACAAGGCCTTCAGAC TCAAGTACAGTCAGCTTGGTCCT 60
Supplemental Information. The TRAIL-Induced Cancer Secretome. Promotes a Tumor-Supportive Immune. Microenvironment via CCR2
 Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/5/233/ra50/dc1 Supplementary Materials for Epidermal Growth Factor Receptor Is Essential for Toll-Like Receptor 3 Signaling Michifumi Yamashita, Saurabh Chattopadhyay,
www.sciencesignaling.org/cgi/content/full/5/233/ra50/dc1 Supplementary Materials for Epidermal Growth Factor Receptor Is Essential for Toll-Like Receptor 3 Signaling Michifumi Yamashita, Saurabh Chattopadhyay,
Nature Medicine: doi: /nm.4169
 Supplementary Fig.1. EC-specific deletion of Ccm3 by Cdh5-CreERT2. a. mt/mg reporter mice were bred with Cdh5CreERT2 deleter mice followed by tamoxifen feeding from P1 to P3. mg expression was specifically
Supplementary Fig.1. EC-specific deletion of Ccm3 by Cdh5-CreERT2. a. mt/mg reporter mice were bred with Cdh5CreERT2 deleter mice followed by tamoxifen feeding from P1 to P3. mg expression was specifically
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines
 Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Supplementary Figure 1. ERK signaling is not activated at early hypertension. a, Western blot analysis for the level of phospho-erk (perk) and total
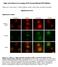
 Supplementary Figure 1. ERK signaling is not activated at early hypertension. a, Western blot analysis for the level of phospho-erk (perk) and total ERK in the aortic tissue from the saline- or AngII-infused
Supplementary Figure 1. ERK signaling is not activated at early hypertension. a, Western blot analysis for the level of phospho-erk (perk) and total ERK in the aortic tissue from the saline- or AngII-infused
Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice
 Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice Supplementary Material Supplementary Methods Materials Spironolactone, aldosterone and β-glycerophosphate were
Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice Supplementary Material Supplementary Methods Materials Spironolactone, aldosterone and β-glycerophosphate were
Supplementary Fig. 1. Multiple five micron sections of liver tissues of rats treated
 Supplementary Figure Legends Supplementary Fig. 1. Multiple five micron sections of liver tissues of rats treated with either vehicle (left; n=3) or CCl 4 (right; n=3) were co-immunostained for NRP-1 (green)
Supplementary Figure Legends Supplementary Fig. 1. Multiple five micron sections of liver tissues of rats treated with either vehicle (left; n=3) or CCl 4 (right; n=3) were co-immunostained for NRP-1 (green)
Supplementary Figure 1. Generation of B2M -/- ESCs. Nature Biotechnology: doi: /nbt.3860
 Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
Resveratrol inhibits epithelial-mesenchymal transition of retinal. pigment epithelium and development of proliferative vitreoretinopathy
 Resveratrol inhibits epithelial-mesenchymal transition of retinal pigment epithelium and development of proliferative vitreoretinopathy Keijiro Ishikawa, 1,2 Shikun He, 2, 3 Hiroto Terasaki, 1 Hossein
Resveratrol inhibits epithelial-mesenchymal transition of retinal pigment epithelium and development of proliferative vitreoretinopathy Keijiro Ishikawa, 1,2 Shikun He, 2, 3 Hiroto Terasaki, 1 Hossein
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Supplementary Figure S1: (A) Schematic representation of the Jarid2 Flox/Flox
 Supplementary Figure S1: (A) Schematic representation of the Flox/Flox locus and the strategy used to visualize the wt and the Flox/Flox mrna by PCR from cdna. An example of the genotyping strategy is
Supplementary Figure S1: (A) Schematic representation of the Flox/Flox locus and the strategy used to visualize the wt and the Flox/Flox mrna by PCR from cdna. An example of the genotyping strategy is
SUPPLEMENTARY INFORMATION
 doi:1.138/nature9464 C 3 K 3 -d 8 C 3 PK-d 7 MK-4-d 12 C 3 C 3 MK-4-d 7 C 3 Supplementary Figure 1 Chemical structures of PK-d 7, MK-4-d 12, K 3 -d 8 and MK-4-d 7. WWW NATURE.CM/NATURE 1 a Relative ratio
doi:1.138/nature9464 C 3 K 3 -d 8 C 3 PK-d 7 MK-4-d 12 C 3 C 3 MK-4-d 7 C 3 Supplementary Figure 1 Chemical structures of PK-d 7, MK-4-d 12, K 3 -d 8 and MK-4-d 7. WWW NATURE.CM/NATURE 1 a Relative ratio
SUPPLEMENTARY INFORMATION
 Figure S1 The effect of T198A mutation on p27 stability. a, Hoechst 33342 staining for nuclei (see Fig 1d). Scale bar, 100 μm. b, Densitometric analysis of wild type and mutant p27 protein levels represented
Figure S1 The effect of T198A mutation on p27 stability. a, Hoechst 33342 staining for nuclei (see Fig 1d). Scale bar, 100 μm. b, Densitometric analysis of wild type and mutant p27 protein levels represented
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Analyses of ECTRs by C-circle and T-circle assays.
 Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
Utility of the dual-specificity protein kinase TTK as a therapeutic target
 Utility of the dual-specificity protein kinase TTK as a therapeutic target for intrahepatic spread of liver cancer Ruoyu Miao, 1,2* Yan Wu, 2* Haohai Zhang, 1 Huandi Zhou, 3 Xiaofeng Sun, 2 Eva Csizmadia,
Utility of the dual-specificity protein kinase TTK as a therapeutic target for intrahepatic spread of liver cancer Ruoyu Miao, 1,2* Yan Wu, 2* Haohai Zhang, 1 Huandi Zhou, 3 Xiaofeng Sun, 2 Eva Csizmadia,
Supplementary Information
 Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Supplemental Table 1 Primers used in study. Human. Mouse
 Supplemental Table 1 Primers used in study Human Forward primer region(5-3 ) Reverse primer region(5-3 ) RT-PCR GAPDH gagtcaacggatttggtcgt ttgattttggagggatctcg Raftlin atgggttgcggattgaacaagttaga ctgaggtataacaccaacgaatttcaggc
Supplemental Table 1 Primers used in study Human Forward primer region(5-3 ) Reverse primer region(5-3 ) RT-PCR GAPDH gagtcaacggatttggtcgt ttgattttggagggatctcg Raftlin atgggttgcggattgaacaagttaga ctgaggtataacaccaacgaatttcaggc
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA
 Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
The p53-puma axis suppresses ipsc generation
 The p53-puma axis suppresses ipsc generation Yanxin Li, Haizhong Feng, Haihui Gu, Dale W. Lewis, Youzhong Yuan, Lei Zhang, Hui Yu, Peng Zhang, Haizi Cheng, Weimin Miao, Weiping Yuan, Shi-Yuan Cheng, Susanne
The p53-puma axis suppresses ipsc generation Yanxin Li, Haizhong Feng, Haihui Gu, Dale W. Lewis, Youzhong Yuan, Lei Zhang, Hui Yu, Peng Zhang, Haizi Cheng, Weimin Miao, Weiping Yuan, Shi-Yuan Cheng, Susanne
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
DRG Pituitary Cerebral Cortex
 Liver Spinal cord Pons Atg5 -/- Atg5 +/+ DRG Pituitary Cerebral Cortex WT KO Supplementary Figure S1 Ubiquitin-positive IBs accumulate in Atg5 -/- tissues. Atg5 -/- neonatal tissues were fixed and decalcified.
Liver Spinal cord Pons Atg5 -/- Atg5 +/+ DRG Pituitary Cerebral Cortex WT KO Supplementary Figure S1 Ubiquitin-positive IBs accumulate in Atg5 -/- tissues. Atg5 -/- neonatal tissues were fixed and decalcified.
Supplementary Figure 1
 Supplementary Figure 1 PTEN promotes virus-induced expression of IFNB1 and its downstream genes. (a) Quantitative RT-PCR analysis of IFNB1 mrna (left) and ELISA of IFN-β (right) in HEK 293 cells (2 10
Supplementary Figure 1 PTEN promotes virus-induced expression of IFNB1 and its downstream genes. (a) Quantitative RT-PCR analysis of IFNB1 mrna (left) and ELISA of IFN-β (right) in HEK 293 cells (2 10
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
 Figure S1. Generation of HspA4 -/- mice. (A) Structures of the wild-type (Wt) HspA4 -/- allele, targeting vector and targeted allele are shown together with the relevant restriction sites. The filled rectangles
Figure S1. Generation of HspA4 -/- mice. (A) Structures of the wild-type (Wt) HspA4 -/- allele, targeting vector and targeted allele are shown together with the relevant restriction sites. The filled rectangles
Supplementary Figure1: ClustalW comparison between Tll, Dsf and NR2E1.
 P-Box Dsf -----------------MG-TAG--DRLLD-IPCKVCGDRSSGKHYGIYSCDGCSGFFKR 39 NR2E1 -----------------MSKPAGSTSRILD-IPCKVCGDRSSGKHYGVYACDGCSGFFKR 42 Tll MQSSEGSPDMMDQKYNSVRLSPAASSRILYHVPCKVCRDHSSGKHYGIYACDGCAGFFKR
P-Box Dsf -----------------MG-TAG--DRLLD-IPCKVCGDRSSGKHYGIYSCDGCSGFFKR 39 NR2E1 -----------------MSKPAGSTSRILD-IPCKVCGDRSSGKHYGVYACDGCSGFFKR 42 Tll MQSSEGSPDMMDQKYNSVRLSPAASSRILYHVPCKVCRDHSSGKHYGIYACDGCAGFFKR
Single cell resolution in vivo imaging of DNA damage following PARP inhibition. Supplementary Data
 Single cell resolution in vivo imaging of DNA damage following PARP inhibition Katherine S. Yang, Rainer H. Kohler, Matthieu Landon, Randy Giedt, and Ralph Weissleder Supplementary Data Supplementary Figures
Single cell resolution in vivo imaging of DNA damage following PARP inhibition Katherine S. Yang, Rainer H. Kohler, Matthieu Landon, Randy Giedt, and Ralph Weissleder Supplementary Data Supplementary Figures
SUPPLEMENTAL FIGURES AND TABLES
 SUPPLEMENTAL FIGURES AND TABLES A B Flag-ALDH1A1 IP: α-ac HEK293T WT 91R 128R 252Q 367R 41/ 419R 435R 495R 412R C Flag-ALDH1A1 NAM IP: HEK293T + + - + D NAM - + + E Relative ALDH1A1 activity 1..8.6.4.2
SUPPLEMENTAL FIGURES AND TABLES A B Flag-ALDH1A1 IP: α-ac HEK293T WT 91R 128R 252Q 367R 41/ 419R 435R 495R 412R C Flag-ALDH1A1 NAM IP: HEK293T + + - + D NAM - + + E Relative ALDH1A1 activity 1..8.6.4.2
Nature Immunology: doi: /ni Supplementary Figure 1. Control experiments for Figure 1.
 Supplementary Figure 1 Control experiments for Figure 1. (a) Localization of SETX in untreated or PR8ΔNS1 virus infected A549 cells (4hours). Nuclear (DAPI) and Tubulin staining are shown. SETX antibody
Supplementary Figure 1 Control experiments for Figure 1. (a) Localization of SETX in untreated or PR8ΔNS1 virus infected A549 cells (4hours). Nuclear (DAPI) and Tubulin staining are shown. SETX antibody
Nature Biotechnology: doi: /nbt Supplementary Figure 1. In vitro validation of OTC sgrnas and donor template.
 Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
FIGURE S1. Representative images to illustrate RAD51 foci induction in FANCD2 wild-type (wt) and
 Supplementary Figures FIGURE S1. Representative images to illustrate RAD51 foci induction in FANCD2 wild-type (wt) and mutant (mut) cells. Higher magnification is shown in Figure 1A. Arrows indicate cisplatin-induced
Supplementary Figures FIGURE S1. Representative images to illustrate RAD51 foci induction in FANCD2 wild-type (wt) and mutant (mut) cells. Higher magnification is shown in Figure 1A. Arrows indicate cisplatin-induced
