Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 )
|
|
|
- Randall Parsons
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 ) homologous recombination arm (middle). The wild-type non-recombinant allele was detected by PCR using a forward primer on E1 and a reverse primer downstream of the right arm homologous recombination genomic sequence (right). (b) Mean ABR measurements (± SEM) from Espn-1 -/- and wild-type mice at six months (each n=9).
2 (c-d) Mean vestibular evoked potential (VsEP) measurements (± standard error) for Espn-1 -/- (n=6) and heterozygous littermates (n=2). No significant differences were observed between genotypes for response peak P1 and P2 latencies (c) or P1 N1 amplitudes (d). VsEP thresholds were -6.5 ± 5.9 db for Espn-1 -/- and -7.5 ± 4.2 db for heterozygous littermates. (e) SEM image of extrastriolar hair bundles of a six-month-old Espn-1 -/- mouse. (f-g) Confocal fluorescence micrographs of stereocilia from vestibular hair cells from the extrastriolar (f) and striolar (g) regions of the utricle from 1-year old Espn-1 -/- mouse. Phalloidin conjugated to AlexaFluor-488 was used to label actin (green); the anti-myo15a antibody (red) shows that MYO15A label persists at stereocilia-tips in both affected and unaffected bundles. (h) Alignment of the first four ankyrin repeats of ESPN-1 and ESPNL. The PB538 antibody epitope is highlighted in red. (i-j) COS7 cells transfected with memerald-espnl (green), and labeled with an anti-espn-1 (i) or anti-espnl (j) antibody to test for selectivity and specificity of antibodies used. Both PB538 (red) and anti-espnl (red) antibody label memerald- ESPNL. (k) The anti-espnl (red) antibody did not label GFP-ESPN-1 (green) expressed in COS7 cells. Scale bars: 5 µm.
3 Supplementary Figure 2. Morphology of the mouse utricle. Figure depicts the hair cells of a single utricle; each dot corresponds to one hair cell. Purple depicts lateral hair cells and light blue medial hair cells; at the boundary between the two zones, the orientation of the hair bundles reverses. The striola is a central region of the utricle with reduced hair cell density and calretinin-positive vestibular nerve calyces surrounding type I hairs; in this figure, the striola is demarked both by dark red arrowheads and by the red dashed line. Hair cells outside the striola are referred to as extrastriolar, and there are medial and lateral populations of these cells. Unlike in other vertebrates, rodent utricles have type I and type II hair cells distributed both in striolar and extrastriolar regions. Figure modified from Figure 2B of Li et al. (2008) J. Neurophysiol. 99, , with permission.
4 Supplementary Figure 3. High-resolution view of MYO3A in the striolar region of the utricle. (a) Survey of utricle at low resolution. (b) Magnification of region indicated by white box in panel (a). Striolar region in small cyan box is magnified 3x in large cyan box; extrastriolar region in small yellow box is magnified 3x in large yellow box. While MYO3A is substantially higher in striolar hair bundles, lower levels of MYO3A can be detected at stereocilia tips in extrastriolar hair cells as well (panel b, lower right yellow-boxed panel). Scale: white box in (a) is 216 µm across, as is full width of (b); cyan and yellow boxes in (b) are 24 µm across.
5 Supplementary Figure 4. High-resolution view of MYO3B in the extrastriolar region of the utricle. Same figure layout as Supplementary Figure 3. While MYO3B is substantially higher in extrastriolar hair bundles, very low levels of MYO3B can be detected at stereocilia tips in extrastriolar hair cells as well (panel b, lower right cyanboxed panel). Scale: white box in (a) is 155 µm across, as is full width of (b); cyan and yellow boxes in (b) are 18 µm across.
6 Supplementary Figure 5. High-resolution view of ESPN in the utricle. Same figure layout as Supplementary Figure 3. Labeling with pan-espn antibody appears similar in striolar and extrastriolar regions. Scale: white box in (a) is 121 µm across, as is (b).
7 Supplementary Figure 6. High-resolution view of ESPN-1 in the extrastriolar region of the utricle. Same figure layout as Supplementary Figure 3. While ESPN-1 is substantially higher in extrastriolar hair bundles, lower levels of ESPN-1 can be detected at stereocilia tips in in striolar hair cells as well (panel b, lower right yellowboxed panel). Scale: white box in (a) is 143 µm across, as is full width of (b); cyan and yellow boxes in (b) are 16 µm across.
8 Supplementary Figure 7. High-resolution view of the concentration of ESPNL in the striolar region of the utricle. Same figure layout as Supplementary Figure 3. ESPNL appears to be highly specific for the striola. Scale: white box in (a) is 98 µm across, as is full width of (b); cyan and yellow boxes in (b) are 12 µm across.
9 Supplementary Figure 8. Development of ESPNL immunoreactivity in cochlear and vestibular hair cells. (a) ESPN-1 labeling (green) in P2 cochlear inner hair cells. (b-d) Developmental progression of ab ESPNL labeling (green) in P2-P10 cochlear inner hair cells. (e-g) Developmental progression of BG35961 ESPNL labeling (left, green; right, gray) in P0.5-P20.5 utricles. (h-i) Confocal fluorescence images of stereocilia from vestibular hair cells of rats (P2), expressing ESPNL-mEmerald (green) after gene gun-mediated biolistic transfection, shows ESPNL localization to stereocilia and concentration at stereocilia-tips. (j-k) Supporting cells from rat organ of Corti of rat (P2) expressing ESPNL-mEmerald (green) shows a concentration of ESPNL at actin-based microvilli. Phalloidin conjugated to AlexaFluor-568 was used to label actin (magenta). Scale bars: A-D, H-K, 2 µm; E-G, 100 µm.
10 Supplementary Figure 9. Characterization of Espnl mutations and phenotype, and validation of BG35961 antibody. (a) Sequencing chromatogram for Espnl Δ allele showing breakpoint between exons 1 and 8. (b) PCR genotyping of Espnl Δ allele. Expected sizes for Espnl Δ and wild-type products are indicated on right. (c) Diagrammatic representation of several characterized CRISPR-mediated indel alleles of Espnl. Numbers beside each smaller bar (allele) are the net change in nucleotides in the indicated exon. Red bars are insertions. Only alleles that were distinguished during sequencing runs were illustrated; other genotyping assays provided evidence for mosaic expression of multiple alleles. (d) Validation of BG35961 anti-espnl antibody. Cells were transfected with either memerald-espnl or GFP-ESPN-1, and then were stained with the BG35961 antibody and phalloidin for actin. Note that BG35961 stains the transfected cell in the top row, but not the adjacent untransfected cell. BG35961 stains neither transfected nor untransfected cells in the GFP- ESPN-1 experiment. Each panel is 24.5 µm wide and tall.
11 Supplementary Figure 10. ESPN-1 expression in stereocilia of apical and basal stereocilia from P9 cochleas. (a) Apical. (b) Basal. A and B are 30 µm wide and 9 µm tall. (c) Distribution of signal intensity for 85 (apical) and 161 (basal) stereocilia tips. The apical mean was 2.58 ± 0.05 arbitrary units (n=85); the basal mean was 1.59 ± 0.04 arbitrary units (n=161); applying a Student s t-test, p =
12 Supplementary Figure 11. Structural similarities of ESPN-1 and ESPNL ankyrin-repeat domains. (a) Sequence alignment of mouse ESPN1 and ESPNL N-terminal ankyrin-repeat domain (ARD) colored by sequence identity. Conserved and similar amino acids are highlighted in dark and light gray respectively. Secondary structure is shown schematically above the amino acid sequences, with red cylinders for α-helix and black lines for loops. The ten ARs consisting of two α-helices (helix1 and helix2) separated by loops are numbered. (b) Structural alignment of the ARD of ESPN1 (red) and ESPNL (green), and overlay (right). Homology analysis is described in Supplemental Experimental Procedures.
13 Supplementary Figure 12. Punnett squares indicating expected genotypes arising from three crosses designed to generate myosin-iii double-knockout mice. Right, expected numbers of each genotype and observed numbers.
14 Supplementary Figure 13. MYO3B-ESPNL mimics MYO3A-ESPNL when MYO3A THDII is added to MYO3B. (a) Diagram of MYO3B hybrid protein containing the THDII of MYO3A (mcherry-myo3b+thdii). (b) mcherry- MYO3B+THDII mimics the behavior of MYO3A when co-expressed with ESPNL, co-translocating to filopodia tips in cells expressing relatively low levels of memerald-espnl. (c) The hybrid protein is unable to initiate filopodia in cells expressing high levels of memerald-espn. (d-e) Quantitation of relationship between ESPNL fluorescence level (ESPNL Fl) and filopodia density (d) or length (e). There was an inverse relationship of filopodial number and length with ESPNL fluorescence for MYO3B+THDII (r=0.82 and 0.79). Scale bars: 2 µm.
15 Supplementary Table 1. Two-way ANOVA analysis of filopodia length and number. The data from either MYO3A or MYO3B alone are the reference group. Then, two main effects estimates for +EPSN-1 and +ESPNL effects are added to the reference. Finally, the fourth parameter estimates the interaction of both (+ESPN-1 +ESPNL). MYO3A group, Figure 7o: Estimate Std. Error t value p-value MYO3A+ (reference) < ESPN EPSNL < Interaction <0.001 MYO3B group, Figure 7o: Estimate Std. Error t value p-value MYO3B+ (reference) ESPN EPSNL Interaction MYO3A group, Figure 7p: Estimate Std. Error t value p-value MYO3A+ (reference) < ESPN < EPSNL Interaction MYO3B group, Figure 7p: Estimate Std. Error t value p-value MYO3B+ (reference) ESPN < EPSNL Interaction <0.001
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplementary Figure 1. Serial deletion mutants of BLITz.
 Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte
 Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte Carlo algorithm for one million generations to obtain
Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte Carlo algorithm for one million generations to obtain
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Supplementary Figure 1.
 Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
(A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1
 SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin
 Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin locus. The EcoRI restriction site between exons M1 and M2
Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin locus. The EcoRI restriction site between exons M1 and M2
Supplementary Information
 Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase
 A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase tag insertion at the 3 end using CRISPR-Cas9 genome editing technique Sambuddha Basu 1, 3, Levi Adams 1, 3, Subhrangshu
A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase tag insertion at the 3 end using CRISPR-Cas9 genome editing technique Sambuddha Basu 1, 3, Levi Adams 1, 3, Subhrangshu
SUPPLEMENTARY INFORMATION
 Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supporting Information
 Supporting Information Park et al. 10.1073/pnas.1410555111 5 -TCAAGTCCATCTACATGGCC-3 5 -CAGCTGCCCGGCTACTACTA-3 5 -TGCAGCTGCCCGGCTACTAC-3 5 -AAGCTGGACATCACCTCCCA-3 5 -TGACAGGAACACCTACAAGT-3 5 -AAGGCACCTTTCTGTCTCCA-3
Supporting Information Park et al. 10.1073/pnas.1410555111 5 -TCAAGTCCATCTACATGGCC-3 5 -CAGCTGCCCGGCTACTACTA-3 5 -TGCAGCTGCCCGGCTACTAC-3 5 -AAGCTGGACATCACCTCCCA-3 5 -TGACAGGAACACCTACAAGT-3 5 -AAGGCACCTTTCTGTCTCCA-3
Supplementary Information. Isl2b regulates anterior second heart field development in zebrafish
 Supplementary Information Isl2b regulates anterior second heart field development in zebrafish Hagen R. Witzel 1, Sirisha Cheedipudi 1, Rui Gao 1, Didier Y.R. Stainier 2 and Gergana D. Dobreva 1,3* 1 Origin
Supplementary Information Isl2b regulates anterior second heart field development in zebrafish Hagen R. Witzel 1, Sirisha Cheedipudi 1, Rui Gao 1, Didier Y.R. Stainier 2 and Gergana D. Dobreva 1,3* 1 Origin
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres
 Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres generated from the CD133-positive cells infected with
Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres generated from the CD133-positive cells infected with
Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and
 Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Supplemental Data. Wang et al. (2017). Plant Cell /tpc NIP1;2 NIP7;1
 Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplemental Information. Kinesin 1 Drives Autolysosome Tubulation
 Developmental Cell, Volume 37 Supplemental Information Kinesin 1 Drives Autolysosome Tubulation Wanqing Du, Qian Peter Su, Yang Chen, Yueyao Zhu, Dong Jiang, Yueguang Rong, Senyan Zhang, Yixiao Zhang,
Developmental Cell, Volume 37 Supplemental Information Kinesin 1 Drives Autolysosome Tubulation Wanqing Du, Qian Peter Su, Yang Chen, Yueyao Zhu, Dong Jiang, Yueguang Rong, Senyan Zhang, Yixiao Zhang,
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
CRISPR RNA-guided activation of endogenous human genes
 CRISPR RNA-guided activation of endogenous human genes Morgan L Maeder, Samantha J Linder, Vincent M Cascio, Yanfang Fu, Quan H Ho, J Keith Joung Supplementary Figure 1 Comparison of VEGF activation induced
CRISPR RNA-guided activation of endogenous human genes Morgan L Maeder, Samantha J Linder, Vincent M Cascio, Yanfang Fu, Quan H Ho, J Keith Joung Supplementary Figure 1 Comparison of VEGF activation induced
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
To investigate the heredity of the WFP gene, we selected plants that were homozygous
 Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
embryos. Asterisk represents loss of or reduced expression. Brackets represent
 Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplementary Information
 Supplementary Information Supplementary Figure 1: Synthetic annealed poly(ra):poly(dt) hybrid induces type I interferon response. a) Bone marrow derived macrophages (BMDM) were transfected with or without
Supplementary Information Supplementary Figure 1: Synthetic annealed poly(ra):poly(dt) hybrid induces type I interferon response. a) Bone marrow derived macrophages (BMDM) were transfected with or without
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Information. Plk1/Polo Phosphorylates Sas-4 at the Onset. of Mitosis for an Efficient Recruitment
 Cell Reports, Volume 25 Supplemental Information Plk1/Polo Phosphorylates Sas-4 at the Onset of Mitosis for an Efficient Recruitment of Pericentriolar Material to Centrosomes Anand Ramani, Aruljothi Mariappan,
Cell Reports, Volume 25 Supplemental Information Plk1/Polo Phosphorylates Sas-4 at the Onset of Mitosis for an Efficient Recruitment of Pericentriolar Material to Centrosomes Anand Ramani, Aruljothi Mariappan,
Supplemental Data. Borg et al. Plant Cell (2014) /tpc
 Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
TALEN mediated targeted editing of GM2/GD2-synthase gene modulates anchorage independent growth by reducing anoikis resistance in mouse tumor cells
 Supplementary Information for TALEN mediated targeted editing of GM2/GD2-synthase gene modulates anchorage independent growth by reducing anoikis resistance in mouse tumor cells Barun Mahata 1, Avisek
Supplementary Information for TALEN mediated targeted editing of GM2/GD2-synthase gene modulates anchorage independent growth by reducing anoikis resistance in mouse tumor cells Barun Mahata 1, Avisek
SUPPLEMENTARY INFORMATION
 AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Figure S1 is related to Figure 1B, showing more details of outer segment of
 Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplementary Information
 Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Allele-specific locus binding and genome editing by CRISPR at the
 Supplementary Information Allele-specific locus binding and genome editing by CRISPR at the p6ink4a locus Toshitsugu Fujita, Miyuki Yuno, and Hodaka Fujii Supplementary Figure Legends Supplementary Figure
Supplementary Information Allele-specific locus binding and genome editing by CRISPR at the p6ink4a locus Toshitsugu Fujita, Miyuki Yuno, and Hodaka Fujii Supplementary Figure Legends Supplementary Figure
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Nature Medicine doi: /nm.2548
 Supplementary Table 1: Genotypes of offspring and embryos from matings of Pmm2 WT/F118L mice with Pmm2 WT/R137H mice total events Pmm2 WT/WT Pmm2 WT/R137H Pmm2 WT/F118L Pmm2 R137H/F118L offspring 117 (100%)
Supplementary Table 1: Genotypes of offspring and embryos from matings of Pmm2 WT/F118L mice with Pmm2 WT/R137H mice total events Pmm2 WT/WT Pmm2 WT/R137H Pmm2 WT/F118L Pmm2 R137H/F118L offspring 117 (100%)
Supplementary Figure1: ClustalW comparison between Tll, Dsf and NR2E1.
 P-Box Dsf -----------------MG-TAG--DRLLD-IPCKVCGDRSSGKHYGIYSCDGCSGFFKR 39 NR2E1 -----------------MSKPAGSTSRILD-IPCKVCGDRSSGKHYGVYACDGCSGFFKR 42 Tll MQSSEGSPDMMDQKYNSVRLSPAASSRILYHVPCKVCRDHSSGKHYGIYACDGCAGFFKR
P-Box Dsf -----------------MG-TAG--DRLLD-IPCKVCGDRSSGKHYGIYSCDGCSGFFKR 39 NR2E1 -----------------MSKPAGSTSRILD-IPCKVCGDRSSGKHYGVYACDGCSGFFKR 42 Tll MQSSEGSPDMMDQKYNSVRLSPAASSRILYHVPCKVCRDHSSGKHYGIYACDGCAGFFKR
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 Sox2 localizes in neutrophils of mouse spleen. (a) Microscopy analysis of Sox2 and MPO in wild-type mouse spleen. Nucl, nucleus. (b) Immunohistochemistry staining of Sox2 and MPO
Supplementary Figure 1 Sox2 localizes in neutrophils of mouse spleen. (a) Microscopy analysis of Sox2 and MPO in wild-type mouse spleen. Nucl, nucleus. (b) Immunohistochemistry staining of Sox2 and MPO
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing
 Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
A Low salt diet. C Low salt diet + mf4-31c1 3. D High salt diet + mf4-31c1 3. B High salt diet
 A Low salt diet GV [AU]:. [mmhg]: 0..... 09 9 7 B High salt diet GV [AU]:. [mmhg]: 7 8...0. 7.8 8.. 0 8 7 C Low salt diet + mf-c GV [AU]:. [mmhg]:.0.9 8.7.7. 7 8 0 D High salt diet + mf-c E Lymph capillary
A Low salt diet GV [AU]:. [mmhg]: 0..... 09 9 7 B High salt diet GV [AU]:. [mmhg]: 7 8...0. 7.8 8.. 0 8 7 C Low salt diet + mf-c GV [AU]:. [mmhg]:.0.9 8.7.7. 7 8 0 D High salt diet + mf-c E Lymph capillary
Nature Methods: doi: /nmeth Supplementary Figure 1. Construction of a sensitive TetR mediated auxotrophic off-switch.
 Supplementary Figure 1 Construction of a sensitive TetR mediated auxotrophic off-switch. A Production of the Tet repressor in yeast when conjugated to either the LexA4 or LexA8 promoter DNA binding sequences.
Supplementary Figure 1 Construction of a sensitive TetR mediated auxotrophic off-switch. A Production of the Tet repressor in yeast when conjugated to either the LexA4 or LexA8 promoter DNA binding sequences.
Optimizing Cas9 and sgrna concentrations for increasing mutation efficiency. Nature Methods: doi: /nmeth.3360
 Supplementary Figure 1 Optimizing Cas9 and sgrna concentrations for increasing mutation efficiency. (a) Wild-type, 2-dpf slc24a5 b1 embryos and a series of slc24a5 CRISPR-injected embryos; anterior is
Supplementary Figure 1 Optimizing Cas9 and sgrna concentrations for increasing mutation efficiency. (a) Wild-type, 2-dpf slc24a5 b1 embryos and a series of slc24a5 CRISPR-injected embryos; anterior is
SUPPLEMENTARY INFORMATION
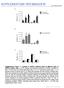 SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1
 Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Negative selection analysis of sgrnas targeting all Brd4 exons comparing day 2 to day 10 time points. Systematic evaluation of 64 Brd4 sgrnas in negative selection experiments, targeting
Supplementary Figure 1 Negative selection analysis of sgrnas targeting all Brd4 exons comparing day 2 to day 10 time points. Systematic evaluation of 64 Brd4 sgrnas in negative selection experiments, targeting
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Supplementary Materials for
 www.advances.sciencemag.org/cgi/content/full/1/7/e1500454/dc1 Supplementary Materials for CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation Xin Han, Zongbin Liu, Myeong chan Jo,
www.advances.sciencemag.org/cgi/content/full/1/7/e1500454/dc1 Supplementary Materials for CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation Xin Han, Zongbin Liu, Myeong chan Jo,
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES
 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Generation of inducible BICD2 knock-out mice. A) The mouse BICD2 locus and gene targeting constructs. To generate an inducible Bicd2
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Generation of inducible BICD2 knock-out mice. A) The mouse BICD2 locus and gene targeting constructs. To generate an inducible Bicd2
Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana.
 SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Map-Based Cloning of Qualitative Plant Genes
 Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
MeCP2. MeCP2/α-tubulin. GFP mir1-1 mir132
 Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Supplemental Figure 1.
 Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplement Figure 1. Plin5 Plin2 Plin1. KDEL-DSRed. Plin-YFP. Merge
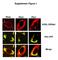 Supplement Figure 1 Plin5 Plin2 Plin1 KDEL-DSRed Plin-YFP Merge Supplement Figure 2 A. Plin5-Ab MitoTracker Merge AML12 B. Plin5-YFP Cytochrome c-cfp merge Supplement Figure 3 Ad.GFP Ad.Plin5 Supplement
Supplement Figure 1 Plin5 Plin2 Plin1 KDEL-DSRed Plin-YFP Merge Supplement Figure 2 A. Plin5-Ab MitoTracker Merge AML12 B. Plin5-YFP Cytochrome c-cfp merge Supplement Figure 3 Ad.GFP Ad.Plin5 Supplement
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast. Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice
 A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
Nature Immunology: doi: /ni.3101
 Supplementary Figure 1 Generation of Plvap / mice. (a) Schematic presentation of the targeting construct as well as the wild-type and targeted Plvap alleles]. PuroR, puromycin resistance. The location
Supplementary Figure 1 Generation of Plvap / mice. (a) Schematic presentation of the targeting construct as well as the wild-type and targeted Plvap alleles]. PuroR, puromycin resistance. The location
sides of the aleurone (Al) but it is excluded from the basal endosperm transfer layer
 Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Supplementary Information
 Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Chapter 14 Regulation of Transcription
 Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Supplemental Data. Seo et al. (2014). Plant Cell /tpc
 Supplemental Figure 1. Protein alignment of ABD1 from other model organisms. The alignment was performed with H. sapiens DCAF8, M. musculus DCAF8 and O. sativa Os10g0544500. The WD40 domains are underlined.
Supplemental Figure 1. Protein alignment of ABD1 from other model organisms. The alignment was performed with H. sapiens DCAF8, M. musculus DCAF8 and O. sativa Os10g0544500. The WD40 domains are underlined.
over time using live cell microscopy. The time post infection is indicated in the lower left corner.
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Supplementary Movie 1 Description: Fusion of NBs. BSR cells were infected
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Supplementary Movie 1 Description: Fusion of NBs. BSR cells were infected
LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation
 LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation Mi Young Park, 1# Raki Sudan, 1# Neetu Srivastava, 1 Sudha Neelam, 1 Christie Youngs, 1 Jia- Wang Wang, 4 Robert W. Engelman, 5,6,7
LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation Mi Young Park, 1# Raki Sudan, 1# Neetu Srivastava, 1 Sudha Neelam, 1 Christie Youngs, 1 Jia- Wang Wang, 4 Robert W. Engelman, 5,6,7
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure S1. Generation of a synaptobrevin2-mrfp knock-in mouse. (a) Targeting strategy of Syb2-mRFP knock-in mouse leaving the synaptobrevin2 gene locus intact except
SUPPLEMENTARY FIGURES Supplementary Figure S1. Generation of a synaptobrevin2-mrfp knock-in mouse. (a) Targeting strategy of Syb2-mRFP knock-in mouse leaving the synaptobrevin2 gene locus intact except
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl
 Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Erhard et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. c1-hbr allele structure. Diagram of the c1-hbr allele found in stocks segregating 1:1 for rpd1-1 and rpd1-2 homozygous mutants showing the presence of a 363 base pair (bp) Heartbreaker
Supplemental Figure 1. c1-hbr allele structure. Diagram of the c1-hbr allele found in stocks segregating 1:1 for rpd1-1 and rpd1-2 homozygous mutants showing the presence of a 363 base pair (bp) Heartbreaker
High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas
 Supplementary information High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas Kei Endo *, Karin Hayashi, Hirohide Saito * Contents: 1. Supplementary
Supplementary information High-resolution Identification and Separation of Living Cell Types by Multiple microrna-responsive Synthetic mrnas Kei Endo *, Karin Hayashi, Hirohide Saito * Contents: 1. Supplementary
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplemental Information. Boundary Formation through a Direct. Threshold-Based Readout. of Mobile Small RNA Gradients
 Developmental Cell, Volume 43 Supplemental Information Boundary Formation through a Direct Threshold-Based Readout of Mobile Small RNA Gradients Damianos S. Skopelitis, Anna H. Benkovics, Aman Y. Husbands,
Developmental Cell, Volume 43 Supplemental Information Boundary Formation through a Direct Threshold-Based Readout of Mobile Small RNA Gradients Damianos S. Skopelitis, Anna H. Benkovics, Aman Y. Husbands,
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15
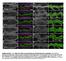 Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues.
 Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Nature Structural & Molecular Biology: doi: /nsmb.3018
 Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss
 SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
Quantitative analysis of recombination in YFP and CFP gene of FRET biosensor induced by lentiviral or retroviral gene transfer.
 Supplementary Information: Quantitative analysis of recombination in and gene of FRET biosensor induced by lentiviral or retroviral gene transfer. Akira T. Komatsubara, Michiyuki Matsuda,, and Kazuhiro
Supplementary Information: Quantitative analysis of recombination in and gene of FRET biosensor induced by lentiviral or retroviral gene transfer. Akira T. Komatsubara, Michiyuki Matsuda,, and Kazuhiro
Gene Therapy for MYO7A USH1B. Shannon Boye, Ph.D. Assistant Professor Department of Ophthalmology University of Florida
 Gene Therapy for MYO7A USH1B Shannon Boye, Ph.D. Assistant Professor Department of Ophthalmology University of Florida Hearing Vision Vestibular Function (balance) Type 1 Type 2 Type 3 Profound deafness
Gene Therapy for MYO7A USH1B Shannon Boye, Ph.D. Assistant Professor Department of Ophthalmology University of Florida Hearing Vision Vestibular Function (balance) Type 1 Type 2 Type 3 Profound deafness
T H E J O U R N A L O F C E L L B I O L O G Y
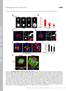 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
Background for Schizophrenia Research Article. Sekar A, et al., Schizophrenia risk from complex variation of complement component 4, Nature, 2016
 Background for Schizophrenia Research Article Sekar A, et al., Schizophrenia risk from complex variation of complement component 4, Nature, 2016 Topics The Major Histocompatibility Complex (MHC) region
Background for Schizophrenia Research Article Sekar A, et al., Schizophrenia risk from complex variation of complement component 4, Nature, 2016 Topics The Major Histocompatibility Complex (MHC) region
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
