Supplemental Information. Boundary Formation through a Direct. Threshold-Based Readout. of Mobile Small RNA Gradients
|
|
|
- Raymond Wood
- 6 years ago
- Views:
Transcription
1 Developmental Cell, Volume 43 Supplemental Information Boundary Formation through a Direct Threshold-Based Readout of Mobile Small RNA Gradients Damianos S. Skopelitis, Anna H. Benkovics, Aman Y. Husbands, and Marja C.P. Timmermans
2 Supplemental Figure 1. Altering the source and level of mirarf perturbs leaf flatness; Related to Figure 1. (A-I) Tissue specificity of promoters driving artificial mirna expression. (A-C) Representative seedling images of (A) pas2:gus, (B) pmir166a:gus, and (C) pfil:gus reporter lines. (D-I) Transverse sections through young leaf primordia of (D and G) pas2:gus, (E and H) pmir166a:gus and (F and I) pfil:gus reporters in (D-F) wild-type and (G-I) rdr6. The AS2 and MIR166A promoters are active in the adaxial and abaxial leaf epidermis (insets), respectively, while FIL is expressed throughout the abaxial domain. Note that expression patterns in rdr6 are unchanged in comparison to wild-type and that the MIR166A and FIL promoters show activity in the vasculature of mature leaves (arrowheads), but not in the vasculature of young leaf primordia (D-I). Scale bars, 50 µm. (J) Leaf-curling phenotypes seen in T1 seedlings of rdr6 mutants carrying the pas2:mirarf, pmir166a:mirarf, or pfil:mirarf transgenes can be grouped into four classes: (K) severe upwardcurling (dark blue), (L) mild upward-curling (bright blue), (M) flat (light blue) and (N) rdr6-like
3 (lavender). Yellow arrowheads, downward-curling leaves; white arrowheads, upward-curling leaves. (O) Levels of tasiarf (wild-type and rdr6) or mirarf (rdr6 carrying the pas2:mirarf, pmir166a:mirarf, or pfil:mirarf transgenes) as determined by small RNA qrt-pcr. Note the frequency of upward-curled seedlings in (J) positively correlates with the levels of tasiarf/mirarf in (O). tasiarf is not detected (nd) in rdr6 mutants. Expression levels (means ± SE) normalized to wildtype tasiarf expression were calculated based on at least three independent biological replicates.
4 Supplemental Figure 2. Signal intensity in colorometric alkaline phosphatase reactions linearly correlates with probe amount; Related to Figures 1 and 3. (A and B) In situ hybridization reveals endogenous tasiarf forms an abaxially-dissipating concentration gradient in (A) wild-type, and is not detected in (B) rdr6 leaf primordia. (C) Quantification of the colorometric alkaline phosphatase reaction signal shows this to be linearly correlated to probe amount over a wide range of levels provided that the reaction is not saturated. Signal intensities (mean ± SE in arbitrary units) were quantified based on four independent experiments. Note, 30 fmol approximates the estimated total amount of mirnas found in a typical mammalian cell (Bissels et al., 2009).
5 Supplemental Figure 3. Functional complementation and characterization of pphb:phb-yfp and pphb:phb*-yfp lines; Related to Figure 2. (A to C) In contrast to (A) wild-type, (B) phb rev double mutants show defects in meristem maintenance and organ initiation; these defects are completely rescued by (C) the pphb:phb-yfp transgene. (D) Representative PCR genotyping of phb rev, complemented phb rev pphb:phb-yfp, or wild-type seedlings confirms complementation requires the pphb:phb-yfp transgene. Primer sets: 1, phb-6 allele; 2, PHB wild-type; 3, rev-9 allele; 4, REV wild-type; 5, YFP. (E) PHB*-YFP (middle strand) harbors a point mutation that reduces complementarity to wild-type mir166 (top strand) and blocks mir166-directed transcript cleavage. The complementary mutation in mir166* (bottom strand) restores cleavage of PHB*-YFP transcripts. Mutated nucleotides are noted in red. (F and G) Transverse sections show expression of (F) pas2:gus throughout the epidermis of adaxialized pphb:phb*-yfp leaf primordia, whereas (G) pmir166a:gus expression remains polar and restricted to the bottom leaf surface. Scale bars, 50 µm.
6 Supplemental Figure 4. mirgfp efficiently silences GFP expression; Related to Figure 3. (A and B) GFP fluorescence in (A) p35s:3xnls-gfp seedlings (no mirgfp) is completely eliminated in (B) such seedlings ubiquitously expressing mirgfp (p35s:mirgfp). (C and D) 5 RLM-RACE analysis yields (C) a single GFP cleavage product that (D) initiates at the expected 10 th nucleotide in the mirgfp target site (black arrowhead). (E) Small RNA blot showing mirgfp is 21 nt in size and accumulates specifically in p35s:mirgfp lines. U6 hybridization confirms near even loading of RNA samples. (F) Read counts for mirgfp and GFP-derived secondary sirnas in reads per million (rpm) normalized to the total number of mapped nt small RNA reads in libraries constructed from p35s:3xnls-gfp (no mirgfp) and p35s:3xnls-gfp p35s:mirgfp lines. The absence of GFP-derived secondary sirnas confirms the lack of transitivity in these lines.
7 Supplemental Figure 5. Target readout is affected by mirgfp levels at the source; Related to Figures 3 and 4. (A-C) Sequential optical sections of a pas2:mirgfp seedling leaf show cells in the presumptive spongy parenchyma and abaxial epidermal layers all retain fluorescence. Nuclei not visible in one section (e.g. arrowheads) show fluorescence in adjacent focal planes. Fluorescence is never detected in presumptive palisade parenchyma and adaxial epidermal cells, regardless of the focal plane. (D) Fluorescence signal intensity (mean ± SE in arbitrary units) quantified from multiple nuclei per cell layer (see Materials and Methods) shows GFP fluorescence is completely silenced in the two adaxialmost cell layers in pas2:mirgfp and the two abaxial-most cell layers in pmir166a:mirgfp primordia. GFP fluorescence intensity in the remaining cell layers is reduced and less variable, showing a lower coefficient of variation (CV: standard deviation divided by mean) than the p35s:3xnls-gfp (no mirgfp) control. (E-G) Sequential optical sections through a 24 hours estradiol-induced pas2>>mirgfp seedling leaf show a lack of GFP fluorescence in adaxial epidermal cells, regardless of the focal plane. Cells in the
8 remaining layers all retain fluorescence with nuclei not visible in one section (e.g. arrowheads) showing fluorescence in adjacent focal planes. (H-J) Independent leaf sections of estradiol-induced pas2>>mirgfp seedlings 72 hours post-induction demonstrating GFP fluorescence on the abaxial side has a stochastic distribution. Ep, epidermis; Pp, presumptive palisade parenchyma; Sp, presumptive spongy parenchyma.
9 Supplemental Figure 6. Characterization of the estradiol-inducible mirgfp system; Related to Figure 4. (A) Schematic of the epidermal-specific, estradiol-inducible patml1>>3xnls-gfp construct. (B-E) GFP fluorescence progressively increases in patml1>>gfp seedlings induced with 20 µm estradiol for (B) 0, (C) 6, (D) 12, and (E) 24 hours. (F-H) Transverse sections of estradiol-induced patml1>>3xnls-gfp seedlings (F) 0, (G) 12, and (H) 24 hours post-induction show GFP fluorescence, and hence OlexA accumulation, is cell-autonomous. (I) Schematic of the adaxial epidermal-specific, estradiol-inducible pas2>>mirgfp construct. (J-M) GFP fluorescence progressively decreases in pas2>>mirgfp seedlings upon (J) 0, (K) 24, (L) 48, and (M) 72 hours of induction with 20 µm estradiol. Seedlings after 48h of induction resemble (N) stably-silenced pas2:mirgfp seedlings. (O) Small RNA qrt-pcr shows a time-dependent increase in mirgfp levels upon induction with 20 µm estradiol that negatively correlates with GFP fluorescence (K to L). mirgfp levels (means ± SE) normalized to U6 were calculated based on at least three independent biological replicates, and plotted
10 relative to mean mirgfp levels 24 hpi. n.d., not detected; no mirgfp, input RNA from p35s:3xnls- GFP seedlings; no RT, input RNA from 72 hpi pas2>>mirgfp seedlings.
11 Supplemental Figure 7. Aligning the mir166 and tasiarf gradients compromises the adaxialabaxial boundary and robust flat leaf architecture; Related to Figure 5. (A and B) Images of 28-dayold plants showing that unlike (A) wild-type, (B) pmir166a:mirarf leaves are not flat, presenting substantial downward-curling (yellow arrowheads) and upward-curling (blue arrowhead) phenotypes. (C and D) Dissected (C) wild-type and (D) pmir166a:mirarf mature leaves reveal the latter develop ectopic outgrowths on their abaxial surface (white arrowheads). (E) Transverse sections of pmir166a:mirarf primordia show misexpression of the pas2:gus adaxial epidermal reporter in random cells of the abaxial epidermis (black arrowheads). (F) Leaves of pphb:phb*-yfp
12 pas2:mir166* seedlings display a range of phenotypes including downward-curling (yellow arrowheads), upward-curling (blue arrowheads), and radial leaf morphologies (black arrowhead). (G) Transverse sections of primordia from a single pphb:phb*-yfp pas2:mir166* seedling show a variable misexpression of the pas2:gus reporter demonstrating adaxial-abaxial polarity and flat leaf production are no longer robustly achieved.
Pattern Formation via Small RNA Mobility SUPPLEMENTAL FIGURE 1 SUPPLEMENTAL INFORMATION. Daniel H. Chitwood et al.
 SUPPLEMENTAL INFORMATION Pattern Formation via Small RNA Mobility Daniel H. Chitwood et al. SUPPLEMENTAL FIGURE 1 Supplemental Figure 1. The ARF3 promoter drives expression throughout leaves. (A, B) Expression
SUPPLEMENTAL INFORMATION Pattern Formation via Small RNA Mobility Daniel H. Chitwood et al. SUPPLEMENTAL FIGURE 1 Supplemental Figure 1. The ARF3 promoter drives expression throughout leaves. (A, B) Expression
B. Transgenic plants with strong phenotype (%)
 A. TCTAGTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT AAATATGGTCTAAAGAAGAAGAATATGGTCTAAAGAAGAAGAATATGG 2XP35S STTM165 5 GGGGGATGAAGctaCCTGGTCCGA3 3 CCCCCUACUUC---GGACCAGGCU5 mir165 HindIII mir165 96 nt GTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT
A. TCTAGTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT AAATATGGTCTAAAGAAGAAGAATATGGTCTAAAGAAGAAGAATATGG 2XP35S STTM165 5 GGGGGATGAAGctaCCTGGTCCGA3 3 CCCCCUACUUC---GGACCAGGCU5 mir165 HindIII mir165 96 nt GTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT
Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte
 Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte Carlo algorithm for one million generations to obtain
Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte Carlo algorithm for one million generations to obtain
sides of the aleurone (Al) but it is excluded from the basal endosperm transfer layer
 Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
SUPPLEMENTARY INFORMATION
 a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
SUPPLEMENTARY INFORMATION
 AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
Supplementary Fig. 1. Microscopic image of cotyledon adaxial epidermis of 3-dayold Col, epf2-1, ros1-4 and rdd. Scale bar, 100 m.
 Supplementary Fig. 1. Microscopic image of cotyledon adaxial epidermis of 3-dayold Col, epf2-1, ros1-4 and rdd. Scale bar, 100 m. Supplementary Fig. 2. The small-cell-cluster phenotype of different ros1
Supplementary Fig. 1. Microscopic image of cotyledon adaxial epidermis of 3-dayold Col, epf2-1, ros1-4 and rdd. Scale bar, 100 m. Supplementary Fig. 2. The small-cell-cluster phenotype of different ros1
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Poly-A signals. amirna. pentr/d-topo for Gateway cloning
 popon inducible system A Poly-A signals CaMV 35S minimal promoter TMV omega translational enhancer HYG KAN RR 35S 35S LhGR GUS Ω pop6 Ω Gene construct B - Dex + Dex CACC prs300 (mir319a) CACC amirna pentr/d-topo
popon inducible system A Poly-A signals CaMV 35S minimal promoter TMV omega translational enhancer HYG KAN RR 35S 35S LhGR GUS Ω pop6 Ω Gene construct B - Dex + Dex CACC prs300 (mir319a) CACC amirna pentr/d-topo
Supplemental data. Zhao et al. (2009). The Wuschel-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice.
 Supplemental data. Zhao et al. (2009). The Wuschel-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. A B Supplemental Figure 1. Expression of WOX11p-GUS WOX11-GFP
Supplemental data. Zhao et al. (2009). The Wuschel-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. A B Supplemental Figure 1. Expression of WOX11p-GUS WOX11-GFP
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for
 Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Supplementary Information. c d e
 Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
Supplementary Data Supplementary Figures
 Supplementary Data Supplementary Figures Supplementary Figure 1. Pi04314 is expressed during infection, each GFP-Pi04314 fusion is stable and myr GFP-Pi04314 is removed from the nucleus while NLS GFP-Pi04314
Supplementary Data Supplementary Figures Supplementary Figure 1. Pi04314 is expressed during infection, each GFP-Pi04314 fusion is stable and myr GFP-Pi04314 is removed from the nucleus while NLS GFP-Pi04314
Supplementary Information
 Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
Supplemental Materials
 Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microrna mir166g and its AtHD-ZIP target genes
 Research article 3657 Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microrna mir166g and its AtHD-ZIP target genes Leor Williams 1, Stephen P. Grigg 1, *, Mingtang Xie
Research article 3657 Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microrna mir166g and its AtHD-ZIP target genes Leor Williams 1, Stephen P. Grigg 1, *, Mingtang Xie
Chinese Academy of Sciences, Datun Road, Chaoyang District, Beijing, 10101, China
 SUPPLEMENTARY INFORMATION Title: Bi-directional processing of pri-mirnas with branched terminal loops by Arabidopsis Dicer-like1 Hongliang Zhu 1,2,3, Yuyi Zhou 1,2,4, Claudia Castillo-González 1,2, Amber
SUPPLEMENTARY INFORMATION Title: Bi-directional processing of pri-mirnas with branched terminal loops by Arabidopsis Dicer-like1 Hongliang Zhu 1,2,3, Yuyi Zhou 1,2,4, Claudia Castillo-González 1,2, Amber
that two types of cis-acting elements control the abaxial epidermis-specific
 FEBS Letters 583 (2009) 3711 3717 journal homepage: www.febsletters.org Two types of cis-acting elements control the abaxial epidermis-specific transcription of the MIR165a and MIR166a genes Xiaozhen Yao,
FEBS Letters 583 (2009) 3711 3717 journal homepage: www.febsletters.org Two types of cis-acting elements control the abaxial epidermis-specific transcription of the MIR165a and MIR166a genes Xiaozhen Yao,
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplemental Data. Wang et al. (2017). Plant Cell /tpc NIP1;2 NIP7;1
 Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination.
 Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
SUPPLEMENTARY INFORMATION
 Materials and Methods Transgenic Plant Materials and DNA Constructs. VEX1::H2B-GFP, ACA3::H2B-GFP, KRP6::H2B-GFP, KRP6::mock21ts-GFP, KRP6::TE21ts- GFP and KRP6::miR161ts-GFP constructs were generated
Materials and Methods Transgenic Plant Materials and DNA Constructs. VEX1::H2B-GFP, ACA3::H2B-GFP, KRP6::H2B-GFP, KRP6::mock21ts-GFP, KRP6::TE21ts- GFP and KRP6::miR161ts-GFP constructs were generated
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Figure S1a. The modular dcas9 fusion system works efficiently to suppress and activate endogenous gene expression in C. elegans
 Figure S1a. The modular dcas9 fusion system works efficiently to suppress and activate endogenous gene expression in C. elegans A, a normal wild-type hermaphrodite N2 is shown. B, a dpy-5-supressed F1
Figure S1a. The modular dcas9 fusion system works efficiently to suppress and activate endogenous gene expression in C. elegans A, a normal wild-type hermaphrodite N2 is shown. B, a dpy-5-supressed F1
Supplementary Information
 Supplementary Information COP1 E3 ligase protects HYL1 to retain microrna biogenesis Seok Keun Cho 1, Samir Ben Chaabane 1, Pratik Shah 1, Christian Peter Poulsen 1, and Seong Wook Yang 1 * 1 Laboratory
Supplementary Information COP1 E3 ligase protects HYL1 to retain microrna biogenesis Seok Keun Cho 1, Samir Ben Chaabane 1, Pratik Shah 1, Christian Peter Poulsen 1, and Seong Wook Yang 1 * 1 Laboratory
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 )
 Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
(A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1
 SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
sirnas compete with mirnas for methylation by HEN1 in Arabidopsis (Supplementary Material)
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Bin Yu Publications Papers in the Biological Sciences 2010 sirnas compete with mirnas for methylation by HEN1 in Arabidopsis
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Bin Yu Publications Papers in the Biological Sciences 2010 sirnas compete with mirnas for methylation by HEN1 in Arabidopsis
Fig. S1. TPL and TPL N176H protein interactions. (A) Semi-in vivo pull-down assays using recombinant GST N-TPL and GST N-TPL N176H fusions and
 Fig. S1. TPL and TPL N176H protein interactions. (A) Semi-in vivo pull-down assays using recombinant GST N-TPL and GST N-TPL N176H fusions and transgenic Arabidopsis TPL-HA lysates. Immunoblotting of input
Fig. S1. TPL and TPL N176H protein interactions. (A) Semi-in vivo pull-down assays using recombinant GST N-TPL and GST N-TPL N176H fusions and transgenic Arabidopsis TPL-HA lysates. Immunoblotting of input
Supplementary information
 Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
6/256 1/256 0/256 1/256 2/256 7/256 10/256. At3g06290 (SAC3B)
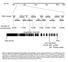 Chr.III M 5M 1M 15M 2M 23M BAC clones F22F7 F1A16 F24F17 F24P17 T8E24 F17A9 F21O3 F17A17 F18C1 F2O1 F28L1 F5E6 F3E22 T1B9 MLP3 Number of recombinants 6/256 1/256 /256 1/256 2/256 7/256 1/256 At3g629 (SAC3B)
Chr.III M 5M 1M 15M 2M 23M BAC clones F22F7 F1A16 F24F17 F24P17 T8E24 F17A9 F21O3 F17A17 F18C1 F2O1 F28L1 F5E6 F3E22 T1B9 MLP3 Number of recombinants 6/256 1/256 /256 1/256 2/256 7/256 1/256 At3g629 (SAC3B)
Aminoacid change in chromophore. PIN3::PIN3-GFP GFP S65 (no change) (5.9) (Kneen et al., 1998)
 Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Supplemental Figure legends Figure S1. (A) (B) (C) (D) Figure S2. Figure S3. (A-E) Figure S4. Figure S5. (A, C, E, G, I) (B, D, F, H, Figure S6.
 Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Data. Borg et al. Plant Cell (2014) /tpc
 Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplemental Data. Tilbrook et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. Protein alignment of with. Identical aligned residues highlighted in black and similar and non-similar residues highlighted in grey and white, respectively. Position of Trp residues
Supplemental Figure 1. Protein alignment of with. Identical aligned residues highlighted in black and similar and non-similar residues highlighted in grey and white, respectively. Position of Trp residues
SUPPLEMENTARY INFORMATION
 Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Sperm cells are passive cargo of the pollen tube in plant fertilization
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplementary Information. A superfolding Spinach2 reveals the dynamic nature of. trinucleotide repeat RNA
 Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15
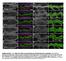 Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Supplemental Data. Guo et al. (2015). Plant Cell /tpc
 Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
kda
 Relative VENUS/RFP fluorescence Relative fluorescence a min 2 min 4 min 6 min 8 min min Jas9-VENUS d kda 8 3 75 63 48 Jas9-VENUS Col- - + - + COR µm Jas9-VENUS 35 28 H2B-RFP 7 b c Overlay,2,8,6,4,2,6,4,2,8,6,4,2
Relative VENUS/RFP fluorescence Relative fluorescence a min 2 min 4 min 6 min 8 min min Jas9-VENUS d kda 8 3 75 63 48 Jas9-VENUS Col- - + - + COR µm Jas9-VENUS 35 28 H2B-RFP 7 b c Overlay,2,8,6,4,2,6,4,2,8,6,4,2
Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis
 Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis seedlings expressing ERF109pro-GUS. The GUS staining
Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis seedlings expressing ERF109pro-GUS. The GUS staining
Supplemental Data. Steiner et al. Plant Cell. (2012) /tpc
 Supplemental Figure 1. SPY does not interact with free GST. Invitro pull-down assay using E. coli-expressed MBP-SPY and GST, GST-TCP14 and GST-TCP15. MBP-SPY was used as bait and incubated with equal amount
Supplemental Figure 1. SPY does not interact with free GST. Invitro pull-down assay using E. coli-expressed MBP-SPY and GST, GST-TCP14 and GST-TCP15. MBP-SPY was used as bait and incubated with equal amount
SUPPLEMENTARY INFORMATION
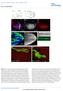 DOI: 10.1038/ncb2806 Supplemental Material: Figure S1 Wnt4 PCP phenotypes are not mediated through canonical Wnt signaling. (a-a ) Illustration of wg, dwnt4 and dwnt6 expression patterns in wing (a) and
DOI: 10.1038/ncb2806 Supplemental Material: Figure S1 Wnt4 PCP phenotypes are not mediated through canonical Wnt signaling. (a-a ) Illustration of wg, dwnt4 and dwnt6 expression patterns in wing (a) and
Supplemental Data. mir156-regulated SPL Transcription. Factors Define an Endogenous Flowering. Pathway in Arabidopsis thaliana
 Cell, Volume 138 Supplemental Data mir156-regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana Jia-Wei Wang, Benjamin Czech, and Detlef Weigel Table S1. Interaction
Cell, Volume 138 Supplemental Data mir156-regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana Jia-Wei Wang, Benjamin Czech, and Detlef Weigel Table S1. Interaction
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues.
 Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Supplemental Figure 1
 12 1 8 Embryo 6 5 4 Silk 6 4 2 3 2 1 1 15 21 24 3 35 4 45 days after pollination 2 6 12 24 72 hours after pollination 35 6 3 25 2 15 1 5 Endosperm 8 12 21 22 3 35 4 45 days after pollination 5 4 3 2 1
12 1 8 Embryo 6 5 4 Silk 6 4 2 3 2 1 1 15 21 24 3 35 4 45 days after pollination 2 6 12 24 72 hours after pollination 35 6 3 25 2 15 1 5 Endosperm 8 12 21 22 3 35 4 45 days after pollination 5 4 3 2 1
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Kunduri et al., http://www.jcb.org/cgi/content/full/jcb.201405020/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Subcellular localization and interactions of PAPLA1.
Supplemental material Kunduri et al., http://www.jcb.org/cgi/content/full/jcb.201405020/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Subcellular localization and interactions of PAPLA1.
Supplemental Figure S1. Nucleotide and deduced amino acid sequences of pepper CaHSP70a (Capsicum annuum heat shock protein 70a) cdna.
 Supplemental Figure S1. Nucleotide and deduced amino acid sequences of pepper (Capsicum annuum heat shock protein 70a) cdna. Translation initiation codon is shown in bold typeface; termination codon is
Supplemental Figure S1. Nucleotide and deduced amino acid sequences of pepper (Capsicum annuum heat shock protein 70a) cdna. Translation initiation codon is shown in bold typeface; termination codon is
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
fga phenotypes were also analyzed in the tracheal system, the organ responsible for
 SUPPLEMENTARY TEXT Tracheal defects fga phenotypes were also analyzed in the tracheal system, the organ responsible for oxygen delivery in the fly. In fga homozygous mutant alleles, we observed defects
SUPPLEMENTARY TEXT Tracheal defects fga phenotypes were also analyzed in the tracheal system, the organ responsible for oxygen delivery in the fly. In fga homozygous mutant alleles, we observed defects
A Repressor Complex Governs the Integration of
 Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
To investigate the heredity of the WFP gene, we selected plants that were homozygous
 Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
A Survey of Genetic Methods
 IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
Effective Small RNA Destruction by the Expression of a Short Tandem Target Mimic in Arabidopsis C W
 The Plant Cell, Vol. 24: 415 427, February 2012, www.plantcell.org ã 2012 American Society of Plant Biologists. All rights reserved. RESEARCH ARTICLES Effective Small RNA Destruction by the Expression
The Plant Cell, Vol. 24: 415 427, February 2012, www.plantcell.org ã 2012 American Society of Plant Biologists. All rights reserved. RESEARCH ARTICLES Effective Small RNA Destruction by the Expression
Effective Small RNA Destruction by the Expression of a Short Tandem Target Mimic in Arabidopsis C W
 The Plant Cell, Vol. 24: 415 427, February 2012, www.plantcell.org ã 2012 American Society of Plant Biologists. All rights reserved. RESEARCH ARTICLES Effective Small RNA Destruction by the Expression
The Plant Cell, Vol. 24: 415 427, February 2012, www.plantcell.org ã 2012 American Society of Plant Biologists. All rights reserved. RESEARCH ARTICLES Effective Small RNA Destruction by the Expression
VIFMGAD IFFRDGVS IVPPAYYAHLAAY
 Supplemental Data. Carbonell et al. (0). Plant Cell 0.0/tpc..0999 A C D E At AGO () At AGO () At AGO7 () At AGO0 () AGO:AGO Or S:AGO AGO:AGO Or S:xAGO AGO7:AGO7 S: AGO0 TIIFGAD IFYRDGVS IVPPAYYAHLAAF S:AGO
Supplemental Data. Carbonell et al. (0). Plant Cell 0.0/tpc..0999 A C D E At AGO () At AGO () At AGO7 () At AGO0 () AGO:AGO Or S:AGO AGO:AGO Or S:xAGO AGO7:AGO7 S: AGO0 TIIFGAD IFYRDGVS IVPPAYYAHLAAF S:AGO
Identification and Characterization of a Plastidic Adenine. Nucleotide Uniporter (OsBT1-3) Required for Chloroplast
 Identification and Characterization of a Plastidic Adenine Nucleotide Uniporter (OsBT1-3) Required for Chloroplast Development in the Early Leaf Stage of Rice Authors: Daoheng Hu 1, Yang Li 1, Wenbin Jin
Identification and Characterization of a Plastidic Adenine Nucleotide Uniporter (OsBT1-3) Required for Chloroplast Development in the Early Leaf Stage of Rice Authors: Daoheng Hu 1, Yang Li 1, Wenbin Jin
Supplemental Data. Meng et al. (2011). Plant Cell /tpc B73 CML311 CML436. Gaspé Flint
 Gaspé Flint B73 CML311 CML436 A B C D 10 th leaf 10 th leaf 10 th leaf Supplemental Figure 1. Whole plant images of the four varieties used in this study (A) Extreme early flowering temperate line Gaspé
Gaspé Flint B73 CML311 CML436 A B C D 10 th leaf 10 th leaf 10 th leaf Supplemental Figure 1. Whole plant images of the four varieties used in this study (A) Extreme early flowering temperate line Gaspé
Nature Genetics: doi: /ng Supplementary Figure 1. High-confidence PRC2 targets and candidate PREs.
 Supplementary Figure 1 High-confidence PRC2 targets and candidate PREs. (a) Flowchart for identification of candidate Arabidopsis PREs. We identified 1504 genomic regions marked by at least 3 of the following:
Supplementary Figure 1 High-confidence PRC2 targets and candidate PREs. (a) Flowchart for identification of candidate Arabidopsis PREs. We identified 1504 genomic regions marked by at least 3 of the following:
ABI3 Controls Embryo De-greening Through Mendel's I locus
 Supporting Online Material for ABI3 Controls Embryo De-greening Through Mendel's I locus Frédéric Delmas a,b,c, Subramanian Sankaranarayanan d, Srijani Deb d, Ellen Widdup d, Céline Bournonville b,c,norbert
Supporting Online Material for ABI3 Controls Embryo De-greening Through Mendel's I locus Frédéric Delmas a,b,c, Subramanian Sankaranarayanan d, Srijani Deb d, Ellen Widdup d, Céline Bournonville b,c,norbert
Technical Review. Real time PCR
 Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Intron distance to transcription start (nt)*
 Schwab et al. SOM: Enhanced microrna accumulation through stemloop-adjacent introns 1 Supplemental Table 1. Characteristics of introns used in this study Intron name Length intron (nt) Length cloned fragment
Schwab et al. SOM: Enhanced microrna accumulation through stemloop-adjacent introns 1 Supplemental Table 1. Characteristics of introns used in this study Intron name Length intron (nt) Length cloned fragment
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Nature Structural and Molecular Biology: doi: /nsmb.2847
 Supplementary Figure 1 Specificity coefficients and generality of the TRM code. (A) The ranking of TRM specificity is shown based on the percent enrichment of the dominant base at position +2 multiplied
Supplementary Figure 1 Specificity coefficients and generality of the TRM code. (A) The ranking of TRM specificity is shown based on the percent enrichment of the dominant base at position +2 multiplied
Supplemental Materials
 Supplemental Materials Supplemental Figure S. Phenotypic assessment of alb4 mutant plants under different stress conditions. (A) High-light stress and drought stress. Wild-type (WT) and alb4 mutant plants
Supplemental Materials Supplemental Figure S. Phenotypic assessment of alb4 mutant plants under different stress conditions. (A) High-light stress and drought stress. Wild-type (WT) and alb4 mutant plants
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Supplemental Figure 1. Rosette Leaf Morphology of Single, Double and Triple Mutants of eid3, phya-201 and phyb-5. Photographs of Ler wild type,
 Supplemental Figure 1. Rosette Leaf Morphology of Single, Double and Triple Mutants of eid3, phya-201 and phyb-5. Photographs of Ler wild type, phya-201, phyb-5, phya-201 phyb-5, eid3, phya-201 eid3, phyb-5
Supplemental Figure 1. Rosette Leaf Morphology of Single, Double and Triple Mutants of eid3, phya-201 and phyb-5. Photographs of Ler wild type, phya-201, phyb-5, phya-201 phyb-5, eid3, phya-201 eid3, phyb-5
Nature Genetics: doi: /ng.3556 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE. Supplementary Figure 1
 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
Targeted modification of gene function exploiting homology directed repair of TALENmediated double strand breaks in barley
 Targeted modification of gene function exploiting homology directed repair of TALENmediated double strand breaks in barley Nagaveni Budhagatapalli a, Twan Rutten b, Maia Gurushidze a, Jochen Kumlehn a,
Targeted modification of gene function exploiting homology directed repair of TALENmediated double strand breaks in barley Nagaveni Budhagatapalli a, Twan Rutten b, Maia Gurushidze a, Jochen Kumlehn a,
WiscDsLox485 ATG < > //----- E1 E2 E3 E4 E bp. Col-0 arr7 ARR7 ACTIN7. s of mrna/ng total RNA (x10 3 ) ARR7.
 A WiscDsLox8 ATG < > -9 +0 > ---------//----- E E E E E UTR +9 UTR +0 > 00bp B S D ol-0 arr7 ol-0 arr7 ARR7 ATIN7 D s of mrna/ng total RNA opie 0. ol-0 (x0 ) arr7 ARR7 Supplemental Fig.. Genotyping and
A WiscDsLox8 ATG < > -9 +0 > ---------//----- E E E E E UTR +9 UTR +0 > 00bp B S D ol-0 arr7 ol-0 arr7 ARR7 ATIN7 D s of mrna/ng total RNA opie 0. ol-0 (x0 ) arr7 ARR7 Supplemental Fig.. Genotyping and
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Supplementary Figure 1 Telomerase RNA fragments used in single-molecule FRET experiments. A pseudoknot fragment (nts ) labeled at position U42
 Supplementary Figure 1 Telomerase RNA fragments used in single-molecule FRET experiments. A pseudoknot fragment (nts 32-195) labeled at position U42 with Cy3 (green circle) was constructed by a two piece
Supplementary Figure 1 Telomerase RNA fragments used in single-molecule FRET experiments. A pseudoknot fragment (nts 32-195) labeled at position U42 with Cy3 (green circle) was constructed by a two piece
Chapter 20 Biotechnology
 Chapter 20 Biotechnology Manipulation of DNA In 2007, the first entire human genome had been sequenced. The ability to sequence an organisms genomes were made possible by advances in biotechnology, (the
Chapter 20 Biotechnology Manipulation of DNA In 2007, the first entire human genome had been sequenced. The ability to sequence an organisms genomes were made possible by advances in biotechnology, (the
Supplementary Information
 Supplementary Information ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis Supplemental Figures a 8 6 4 2 Absolute Expression b Absolute Expression
Supplementary Information ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis Supplemental Figures a 8 6 4 2 Absolute Expression b Absolute Expression
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplemental Data. Benstein et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Supplemental Figure 1. Purification of the heterologously expressed PGDH1, PGDH2 and PGDH3 enzymes by Ni-NTA affinity chromatography. Protein extracts (2 µl) of different fractions (lane 1 = total extract,
Supplementary Information. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato
 Supplementary Information The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato Uri Krieger 1, Zachary B. Lippman 2 *, and Dani Zamir 1 * 1. The Hebrew University of Jerusalem Faculty
Supplementary Information The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato Uri Krieger 1, Zachary B. Lippman 2 *, and Dani Zamir 1 * 1. The Hebrew University of Jerusalem Faculty
Supplemental Data. Huo et al. (2013). Plant Cell /tpc
 Supplemental Data. Huo et al. (2013). Plant Cell 10.1105/tpc.112.108902 1 Supplemental Figure 1. Alignment of NCED4 Amino Acid Sequences from Different Lettuce Varieties. NCED4 amino acid sequences are
Supplemental Data. Huo et al. (2013). Plant Cell 10.1105/tpc.112.108902 1 Supplemental Figure 1. Alignment of NCED4 Amino Acid Sequences from Different Lettuce Varieties. NCED4 amino acid sequences are
Endogenous and Synthetic MicroRNAs Stimulate Simultaneous, Efficient, and Localized Regulation of Multiple Targets in Diverse Species W
 The Plant Cell, Vol. 18, 1134 1151, May 2006, www.plantcell.org ª 2006 American Society of Plant Biologists Endogenous and Synthetic MicroRNAs Stimulate Simultaneous, Efficient, and Localized Regulation
The Plant Cell, Vol. 18, 1134 1151, May 2006, www.plantcell.org ª 2006 American Society of Plant Biologists Endogenous and Synthetic MicroRNAs Stimulate Simultaneous, Efficient, and Localized Regulation
Plant virus mediated induction of mir168 is associated with repression of ARGONAUTE1 accumulation
 Manuscript EMBO-2010-73919 Plant virus mediated induction of mir168 is associated with repression of ARGONAUTE1 accumulation Éva Várallyay, Anna Válóczi, Ákos Ágyi, József Burgyán and Zoltan Havelda Corresponding
Manuscript EMBO-2010-73919 Plant virus mediated induction of mir168 is associated with repression of ARGONAUTE1 accumulation Éva Várallyay, Anna Válóczi, Ákos Ágyi, József Burgyán and Zoltan Havelda Corresponding
The early extra petals1 Mutant Uncovers a Role for MicroRNA mir164c in Regulating Petal Number in Arabidopsis
 Current Biology, Vol. 15, 303 315, February 22, 2005, 2005 Elsevier Ltd All rights reserved. DOI 10.1016/j.cub.2005.02.017 The early extra petals1 Mutant Uncovers a Role for MicroRNA mir164c in Regulating
Current Biology, Vol. 15, 303 315, February 22, 2005, 2005 Elsevier Ltd All rights reserved. DOI 10.1016/j.cub.2005.02.017 The early extra petals1 Mutant Uncovers a Role for MicroRNA mir164c in Regulating
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
C24. ros1-1. ros1-1 rdm18-1. ros1-1 rdm18-2. ros1-1 nrpe1
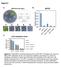 Figure S1 Methylation level 1 0.8 0.6 0.4 0.2 0 p35s methylation levels 24 ros1-1 ros1-1 rdm18-1 ros1-1 rdm18-2 ros1-1 nrpe1 mg mhg mhh Figure S1. RM18/PKL promotes silencing at the p35s-npt II transgene
Figure S1 Methylation level 1 0.8 0.6 0.4 0.2 0 p35s methylation levels 24 ros1-1 ros1-1 rdm18-1 ros1-1 rdm18-2 ros1-1 nrpe1 mg mhg mhh Figure S1. RM18/PKL promotes silencing at the p35s-npt II transgene
Two classes of silencing RNAs move between Caenorhabditis elegans tissues.
 Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
