Photoactivatable mcherry for high-resolution two-color fluorescence microscopy
|
|
|
- Lucy Townsend
- 6 years ago
- Views:
Transcription
1 Photoactivatable mcherry for high-resolution two-color fluorescence microscopy Fedor V Subach 1 3, George H Patterson,3, Suliana Manley, Jennifer M Gillette, Jennifer Lippincott-Schwartz & Vladislav V Verkhusha 1 The reliance of modern microscopy techniques on photoactivatable fluorescent proteins prompted development of mcherry variants that are initially dark but become red fluorescent after violet-light irradiation. Using ensemble and single-molecule characteristics as selection criteria, we developed with excitation/emission maxima at 5/595 nm. Compared to other monomeric red photoactivatable proteins, it has faster maturation, better ph stability, faster photoactivation, higher photoactivation contrast and better photostability. Lack of green fluorescence and single-molecule behavior make monomeric a preferred tag for two-color diffraction-limited photoactivation imaging and for super-resolution techniques such as one- and two-color photoactivated localization microscopy (PALM). We performed PALM imaging using -tagged transferrin receptor expressed alone or with photoactivatable GFP tagged clathrin light chain. Pair correlation and cluster analyses of the resulting PALM images identified r nm clusters of transferrin receptor and clathrin light chain at r5 nm resolution and confirmed the utility of as an intracellular probe. Genetically encoded photoactivatable fluorescent proteins (PAFPs) make up a small category of fluorescent proteins 1, but are beginning to find uses far and above those of normal fluorescent proteins. With initially little or no fluorescence within their associated spectral detection window, photoactivatable proteins can be switched on by irradiation with violet light. Thus they are useful for spatially pulse-labeling subpopulations of molecules in cells in complement to photobleaching applications and can provide other useful features such as a high contrast over background in the photoactivated region and circumvention of fluorescence contributions from newly synthesized, nonactivated PAFPs. PAFPs and photoswitchable dyes also provide probes necessary for high-resolution optical techniques, such as photoactivated localization microscopy (PALM) 3, fluorescence photoactivated localization microscopy (FPALM), stochastic reconstruction microscopy (STORM) 5, PALM with independent running acquisition (PALMIRA) and stroboscopic PALM (SPALM) 7. Of particular interest for PALM is the development of monomeric red PAFPs 8. Several current variants can be activated into a redemitting protein, but these have limitations for both live-cell confocal imaging and for high-resolution localization techniques. The obligate tetrameric state of red Kaede 9, KFP1 (ref. 1) and EosFP 11 frequently causes abnormal localization and function of the tagged proteins. Available monomeric red PAFPs such as Dendra 1, monomeric EosFP (meosfp) or tandem dimeric EosFP (tdeosfp) 11 undergo the photoconversion from a green form to a red form, which complicates two-color photoactivation experiments with green PAGFP 13,PSCFP 1 or Dronpa 15. It is also often difficult to achieve in cells a complete green-to-red photoconversion, which results in a detectable residual amount of the green species of these PAFPs. The available PAmRFP1 (ref. 8) switches from a nonfluorescent to a red fluorescent protein, but lacks the photon yields required for PALM applications. Reversibly photoswitchable fluorescent proteins rscherry and rscherryrev could be used in twocolor experiments with green PAFPs, but have low brightness in their on state, high background fluorescence in their off state and tend to rapidly relax to the dark state after being photoswitched 1. Here we report several irreversibly photoactivatable derivatives of mcherry 17, named PAmCherry proteins, with excellent photoactivation contrast over background, advanced photostability and high single-molecule brightness compatible with PALM imaging. RESULTS Development of photoactivatable mcherry variants We analyzed data on color transitions of red fluorescent proteins to the respective nonfluorescent chromoproteins that had been generated by mutagenesis 1 and identified the corresponding crucial amino acid positions on the basis of the mcherry structure 18. Positions spatially close to the chromophore, such as 18, 15, 17 and 3 (numbering is in accordance with GFP alignment; Supplementary Fig. 1 online), appear to be major molecular determinants of color 1,19. We hypothesized that mutagenesis of 1 Department of Anatomy and Structural Biology, and Gruss-Lipper Biophotonics Center, Albert Einstein College of Medicine, 13 Morris Park Ave., Bronx, New York 11, USA. Section on Organelle Biology, Cell Biology and Metabolism Program, National Institute of Child Health and Human Development, National Institutes of Health, 9 Rockville Pike, Bethesda, Maryland 89, USA. 3 These authors contributed equally to this work. Correspondence should be addressed to V.V.V. (vverkhus@aecom.yu.edu). RECEIVED 11 NOVEMBER 8; ACCEPTED DECEMBER 8; PUBLISHED ONLINE 5 JANUARY 9; DOI:1.138/NMETH.198 NATURE METHODS VOL. NO. FEBRUARY 9 153
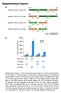
2 a 1 Pre Post b c Abs Ex Em 8 8 mcherry Wavelength (nm) Maturation time (min) Absorbance and fluorescence (a.u.) Relative rate of activation (%) Normalized fluorescence (%) Relative rate Efficiency Time (s) Efficiency of activation (%) d e f 7 8 ph 1 8 Normalized fluorescence (%) 1 8 tdeosfp Photoactivation time (s) g h i Time (s) Normalized fluorescence (%) 1 8 rscherry rscherryrev tdeosfp ph 1 8 tdeosfp Photobleaching time (s) Time (s) Figure 1 Spectral, biochemical and photochemical properties of the purified PAmCherry variants. (a) Absorbance, excitation and emission spectra of before (pre) and after (post) photoactivation with the 399 nm laser line. (b) Maturation kinetics for the indicated proteins at 37 1C. (c) Equilibrium ph dependence for the red fluorescence of the indicated proteins. (d) Rate and efficiency of photoactivation at different physiological ph values, normalized to the fluorescence intensities at the respective ph (data shown in c). (e,f) Photoactivation (e) and photobleaching (f) kinetics for the indicated proteins using arc lamp illumination through oil objective with 39/ nm (e) and 57/3 nm (f) filters. (g i) Cycling illumination of, rscherry and rscherryrev using an arc lamp, 1 oil objective, 57/3 filter for 1 s and one of the following filters: 39/ nm for. s (g), 3/ for.5 s (h) and 8/ for. s (i). The maxima of the fluorescence signals shown in g i were normalized per the relative brightness for each protein indicated in Table 1 to achieve 1%. The ground states of the proteins are indicated with the respective symbols. mcherry at these positions might convert it to a photoactivatable red probe and performed saturating mutagenesis at these positions using the overlap extension approach. We screened the resulting bacterial library of the sitespecific mcherry mutants by fluorescence-activated cell sorting (FACS) and then selected the brightest clones on Petri dishes using a fluorescence stereomicroscope. We identified two weakly photoactivatable mutants with S18L/I15V/Q17P/ I3R and S18L/I15L/Q17A/I3R substitutions compared to mcherry. To enhance their characteristics we subjected these mutants to several rounds of random mutagenesis (Supplementary Methods online). After each round of mutagenesis and sequencing, we purified and analyzed 1 15 candidate mutants in ensemble mode (Supplementary Methods). Then we immobilized 5 1 mutant variants that had the highest product of brightness and photobleaching half-time in ensemble measurements and immobilized them on coverslips for single-molecule imaging using 51 nm excitation and intermittent low-energy pulses of 5 nm light. We imaged the molecules until they became photobleached and then analyzed the data as previously described 3. During the single-molecule screening, we used photon statistics on the localized mcherry mutant proteins as the selection criteria. We recorded the number of molecules localized within the 1 s integration time for each frame, and the mean and median number of photons for candidate molecules (Supplementary Table 1 online). Finally, we used the mixture of several enhanced variants as a template for the next round of molecular evolution. After four rounds of random mutagenesis, we selected the three photoactivatable variants exhibiting the best performance in both ensemble and single-molecule modes. As compared to parental mcherry, the mutants named, and contained EV/A58T/K9N/L8F/N99K/S18L/ I15V/Q17P/L19V/I3R, M18L/K9N/L8F/A17T/S18L/ I15L/Q17A/L19T/A179T/I3R/RL and M18L/A58T/K9N/ L8F/M137I/E1D/S18L/I15L/Q17A/D178E/I3R substitutions, respectively (Supplementary Fig. 1). Ensemble characterization of the PAmCherry variants Before photoactivation, the purified, and had absorbance maxima at nm but did not fluoresce when excited at these wavelengths. After photoactivation using a 399 nm laser line, new absorbance peaks had maxima at 5 57 nm (Fig. 1a; and PAmCherry 3 exhibited similar spectra). In the photoactivated state,, PAm- Cherry and exhibited red fluorescence with excitation/emission peaks at 5/595 (Fig. 1a), 57/59 and 57/59 nm, respectively. Molar extinction coefficients and quantum yields of the photoactivated PAmCherry proteins were 18,, M 1 cm 1 and..53, respectively (Table 1). After photoactivation, the red forms of the PAmCherry variants were stable with time and did not relax back to the dark state at least for h at 37 1C ortwo weeks at 1C. The protein maturation half-times for the PAm- Cherry variants were fold of that for mcherry, 18 5 min at 37 1C (Fig. 1b). In addition, the PAmCherry variants exhibited monomeric behavior similar to that of mcherry when separated on a native polyacrylamide gel (data not shown). We also compared biochemical and photochemical ensemble characteristics of PAmCherry variants with those of tdeosfp 11.For ensemble experiments we selected tdeosfp over meosfp, which we used for the initial single-molecule screening, because the very low amounts of the latter produced even at low temperatures were insufficient for ensemble measurements. The red fluorescence of the PAmCherry variants exhibited higher ph stability than tdeosfp 15 VOL. NO. FEBRUARY 9 NATURE METHODS
3 Table 1 Ensemble characteristics of the purified PAmCherry variants and of other fluorescent proteins PamCherry tdeosfp Dendra rscherry rscherryrev Before After Before After Before After Before After Before After Ground state After switching Ground state After switching mcherry Absorbance maximum (nm) (571) Extinction coefficient (M 1 cm 1 ),5 18, 1,9,,5 1, 8, 33, (3,) 5, 35, (81,) (8,) a (85,) (8,) a 78, Emission maximum (nm) ND 595 ND 59 ND (58) Quantum yield ND. ND.53 ND (.9) (.) a (.3) (.5) a. Relative brightness (%) ND ND ND (91) 1 81 B.8 (3.7) B8.7 (8.1) B.7 (.13) B8.7 (.1) 87 pk a ND.3 ND. ND. ND (.7)..9 (.) (5.5). Maturation t.5 at 37 1C (min) ND b 9 (179) () (15) Photoactivation t.5 (s).5 c (.) ND ND ND ND Photobleaching t.5 (s) ND () ND ND ND ND (19) Photoactivation contrast (fold), 5, 3, () 3 B.7 (3) d B (3) d ND The tdeosfp 11, Dendra (ref. 1), rscherry 1,rsCherryRev 1 and mcherry 17 characteristics are presented from the respective original papers and/or measured by us using the same conditions as for PAmCherry variants (shown in parentheses). ND, not determined. t.5,half-time. a The post-switching characteristics were determined 1 in the maximally achievable level of fluorescence signal of the on state, using the conditions described in Figure 1h. b The tdeosfp expression in bacteria at 37 1C was too slow to allow for the time-limited production of enough protein for maturation experiments similar to those with PAmCherry variants described in Supplementary Methods. c Photoactivation t.5 for PAGFP is. s using the same conditions. d The maximal contrast observed during the first 17 photocycles using the conditions described in Figure 1h. For photoactivatable proteins, values are given before and after activation, where relevant. with apparent pk a values of..3 (Fig. 1c). Both the efficiency and rate of photoactivation of (Fig. 1d for ; and exhibited similar dependences), and the other two PAmCherry variants (data not shown) decreased at acidic ph, suggesting that the PAmCherry photoactivation involves a ph-dependent Glu decarboxylation. We also concluded that photoactivation of PAmCherry variants was still possible at ph 5.5, albeit at B1% of the photoactivation contrast at ph 7., which makes them useful for monitoring lysosomes, the most acidic cellular compartments. Measurements of the photoactivation kinetics by epifluorescence microscopy indicated that the purified became photoactivated more than twofold faster than the other two PAmCherry variants and 1.-fold faster than tdeosfp (Fig. 1e). Under the same conditions, the photoactivated PAmCherry variants photobleached fold slower than tdeosfp (Fig. 1f). The photoactivation contrast achieved with the purified PAmCherry variants was 3, 5,-fold, whereas the increase in the red fluorescence of the purified tdeosfp was -fold (Table 1). As the recently reported mcherry mutants, rscherry and rscherryrev, exhibit reversible photoswitching, we checked this property in using the irradiation conditions used in the original paper 1 but expanded the tests to four different irradiation wavelengths in the range of nm (Fig. 1). When cyclically irradiated using 39/ and 57/3 filter combinations, subpopulations of the sample were sequentially photoactivated with violet light and then photobleached with yellow light (Fig. 1g). As expected for irreversibly photoactivatable proteins, the majority of was photobleached after about 15 light cycles. We did not observe any reversible photoswitching. We observed similar behavior when the 3/ filter was used instead of the 39/ filter (Fig. 1h). did not exhibit photoactivation when irradiated with blue light through a 8/ filter (Fig. 1i). The rscherry and rscherryrev variants were cyclically photoswitched using the same conditions (Fig. 1g i). Note that for data shown in Figure 1g i, the fluorescence signal of was normalized to its maximal fluorescence achievable when activated using a 39/ filter in the absence of the photobleaching yellow light. Evaluation of PAmCherry variants in cells We performed confocal microscopy photoactivation experiments in COS-7 cells using untagged PAmCherry variants co-expressed with enhanced GFP (EGFP) as a transfected cell marker (Supplementary Fig. a c online). Initially the cells displayed little fluorescence in the red channel and after photoactivation showed increases of B1-fold for all PAmCherry variants in the cytoplasm (Supplementary Fig. 3a online). Normalization to the initial signal indicated a lower photoactivation contrast than that obtained for the purified proteins because cellular autofluorescence increased the initial signal used for normalization. To further study in cells, we fused it to five different proteins, expressed the fusions in cells and imaged them by confocal microscopy before and after photoactivation. All five, including histone HB, b-actin and a-tubulin fusion proteins, properly localized when visualized after photoactivation in live cells (Supplementary Fig. online). For PALM assessments, we fused PAm- Cherry variants to two plasma membrane proteins, transferrin receptor (TfR) and a temperature-sensitive version of vesicular stomatitis virus G protein (VSVG) 1, which can be readily imaged using total internal reflection (TIRF) microscopy. The TfR has an intracellular pool in early and recycling endosomes that exchange with the plasma membrane population. TfR binds iron-loaded transferrin for iron uptake, and its cellular distribution, endocytic behavior and diffusion characteristics are well-studied. VSVG is often used in secretory pathway studies, and it is a highly expressed plasma membrane protein when cells are maintained at 3 1C. When expressed in COS-7 cells, TfR-PAmCherry variants (Supplementary Fig. d f) had intracellular pools and plasma membrane populations similar to those of TfR-EGFP, TfR-mCherry (Supplementary Fig. g) and endogenous TfR as previously reported 3. Measurement of transferrin uptake indicated that the NATURE METHODS VOL. NO. FEBRUARY 9 155
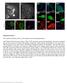
4 a d TIRF PALM Cluster analysis b c e f Figure Plasma membrane distribution of TfR observed using PALM. (a) Cells expressing TfR- were subjected to low-level 5-nm laser photoactivation, and we simultaneously collected 51-nm-laser-light excited single-molecule fluorescence. Data were collected at 1 frames per second for 15, frames. The fluorescence collected during the 15, frames is shown in a and represents a diffraction-limited TIRF microscopy image. (b) The position and uncertainty of the molecules plotted as Gaussiannormalized spots to form a PALM image. (c) Cluster analysis was performed on the molecules localized over the entire PALM image. The threshold used for clusters required the local molecular density to be fivefold that of the average molecular density. Individual clusters are plotted in different colors with nonclustered molecules represented in black. (d f) Magnified views of the boxed region in a are shown as TIRF microscopy (d), PALM (e) and cluster analysis images (f). PALM data in these images were limited to molecules that localized to o5 nm uncertainty. Scale bars, 5 mm (a,b) and 1 mm (d,e). chimeras are functional and capable of binding and internalizing transferrin (data not shown). We performed photoactivation kinetics measurements using the more highly expressed VSVG- fusion and observed an B1-fold increase in fluorescence at the plasma membrane (Supplementary Fig. 3b). Similar to the results with purified proteins (Fig. 1e), in cells activated with less irradiation when compared to the other variants (Supplementary Fig. 3a), which is an important property for live-cell photoactivation experiments. Evaluation of as a PALM marker in cells Initial PALM experiments with TfR-PAmCherry variants in COS-7 cells allowed single-molecule characterization of the PAmCherry variants. The number of photons collected from single molecules revealed that these new variants should work well in cellular imaging experiments (Supplementary Table 1). The slight difference in detected photon yields between cells and purified protein is not surprising, as the proteins are subject to different environments by being attached to TfR and fixed before imaging. In subsequent studies we used because of its brightness compared to, its photostability compared with and its easier photoactivation compared to both other proteins (Table 1 and Supplementary Fig. 3). We directly compared (Fig. ) with rscherry and rscherryrev, as well as with tdeosfp, which has been used and characterized in previous PALM studies 3,,5. The fluorescence from all frames collected in a PALM experiment with TfR- produced the equivalent of a diffractionlimited TIRF microscopy image (Fig. a). Comparison of the TIRF microscopy image with the PALM image showed the greater detail in the more highly resolved (o5 nm molecular organization uncertainty (s)) PALM image (Fig. b). A highmagnification TIRF microscopy image showed many bright and dim spots, which corresponded in the PALM image to high or low densities of molecules (Fig. d). We performed cluster analysis 5 to highlight the dense structures, which revealed that many of these heterogeneities correspond to B1 nm clusters of TfR- (Fig. c,f). We performed PALM experiments with TfR-rsCherry- and TfRrsCherryRev-expressing cells using a modified protocol. The high background from both fluorescent proteins required preirradiations with 88 nm for rscherry and 51 nm for rscherryrev to switch them off from the ground states. The residual off-state fluorescence for both proteins still presented a problem for singlemolecule localization and required an increase in the preactivation irradiation. In addition, experiments with rscherry were complicated by the reversal of its light sensitivity (Fig. 1g i). Comparison of the TfR- with TfR-rsCherry and TfR-rsCherryRev molecules showed that had substantially better key PALM properties, such as mean number of photons, mean s values and duration of the molecular fluorescence, than rscherryrev and rscherry (Supplementary Fig. 5 Figure 3 Comparison of and tdeosfp fusions in fixed cells. (a) Distribution of the number of photons collected from molecules in cells expressing TfR- (mean, 7; median, 13) and TfR-tdEosFP (mean, 9; median, 39). (b) Distribution of s in cells expressing TfR- (mean, 15.18; median, 1.99) and TfR-tdEosFP (mean, 1.75; median, 1.9). (c) Distribution of the duration of the molecular fluorescence (.1-s frames) in cells expressing TfR- (mean,.3; median, 1) and TfR-tdEosFP (mean, 3.; median, 1). Two-tailed Wilcoxon-Mann-Whitney tests indicated differences between the TfR- and TfR-tdEosFP data medians for all three distributions (P o.1, n ¼ 757,18 from five TfR- expressing cells and n ¼ 5,9 from five TfR-tdEosFP expressing cells). (d) Distribution of the number of photons collected from molecules in cells expressing VSVG- (mean, 57; median, 38) and VSVG-tdEosFP (mean, 1,57; median, ). (e) Distribution of s in cells expressing VSVG- (mean,.9; median, 19.8) and VSVG-tdEosFP (mean, 19.91; median, 19.). (f) Distribution of the duration of the molecular fluorescence (.1-s frames) in cells expressing VSVG- (mean, 1.8; median, 1) a b c 1 1 TfR-tdEosFP TfR ,, Photons and VSVG-tdEosFP (mean,.37; median, 1). Two-tailed Wilcoxon-Mann-Whitney tests indicated differences between the VSVG- and VSVG-tdEosFP data medians for all three distributions (P o.1, n ¼ 377,1 from three VSVG- cells and n ¼ 78, from three VSVG-tdEosFP cells). VSVG-tdEosFP VSVG σ (nm) d e f ,, Photons σ (nm) Frames Frames 15 VOL. NO. FEBRUARY 9 NATURE METHODS
5 online). Notably, the density of molecules in the TfR-rsCherry and TfR-rsCherryRev PALM images was substantially lower than that for TfR-. This observation may explain why the rscherry and rscherryrev produce moderate numbers of photons from single molecules (Supplementary Fig. 5) but have very low brightness in the ensemble (Table 1). The simplest explanation is that there is a large subpopulation of the rscherry and rscherryrev molecules, which can form the chromophore, but remain nonfluorescent, that is, in a chromo state 1, and cannot be photoactivated. We also compared with a more thoroughly characterized PALM probe, tdeosfp 3,,5. TfR-tdEosFP did not show the normal punctate intracellular and plasma membrane TfR distributions but was located in the cytosol with several large punctate signals throughout the cytoplasm (Supplementary Fig. online). Nevertheless, sufficient TfR-tdEosFP molecules were within the TIRF microscopy excitation range for comparing properties important for PALM with those of TfR- (Fig. 3). In cells imaged and analyzed under the same conditions the number of photons per molecule had slightly different distributions (Fig. 3a) withb9 and B7 mean number of photons collected for tdeosfp and PAm- Cherry1, respectively. Despite the slight shift toward more photons per molecule in the distribution of, tdeosfp has the higher mean due to a population of molecules that produce up to 3, photons. s distributions were centered around B15 1 nm uncertainty (Fig. 3b) indicating that and tdeosfp were comparable in fixed cell PALM experiments. Analysis of the duration of fluorescence from each molecule revealed that the tdeosfp molecules tend Figure Distributions of TfR and CLC by twocolor PALM. (a i) COS-7 cells expressing TfR- and PAGFP-CLC were subjected to low-level 5-nm laser photoactivation while we alternately collected 51-nm-light excited (a c) and 88-nm-light excited PAGFP (d f) single-molecule fluorescence. TIRF microscopy images of fluorescence (a), PAGFP fluorescence (d) and the merge (g) are shown. Position and uncertainty for TfR- (b,c) and PAGFP-CLC (e,f) are plotted as Gaussian-normalized spots. Magnified views of the boxed area in b are shown in c, f and i. Merge images (g i) show the relative distributions of TfR- (red) and PAGFP-CLC (green). (j) Pair correlation analysis (top) and normalized results (bottom) for the presence and size of TfR- and PAGFP-CLC clusters. (k) Cluster analysis of the molecules localized for the PALM images in h (k) andi (l). The threshold used for clusters required the local molecular density to be fivefold that of the average molecular density. TfR- clusters are plotted in red, PAGFP-CLC clusters in green and nonclustered molecules in black. PALM data in these images are limited to molecules that localized to o5 nm s. Scale bars, mm (a,b,d,e,g,h) and.5 mm (c,f,i). Red fluorescence Green fluorescence Merge Cluster analysis g(r) g(r) normalized 1 5 to fluoresce B1 frame (.1 s) longer than (Fig. 3c). Lastly, contrast comparisons made by observation and background quantification of frame 1, from 1,-frame experiments indicated no substantial difference between and tdeosfp in this property. We also compared and tdeosfp tagged to VSVG in PALM experiments. These data were similar to the TfR data and showed that VSVG- produced fewer photons but had a distribution of molecular localization uncertainties similar to that of VSVG-tdEosFP (Fig. 3d f). Evaluation of as a marker in two-color PALM TfR internalizes via clathrin-coated pits, which are reported 7 to have a similar size to the clusters of TfR we observed in images shown in Figure. To test the colocalization of TfR and clathrincoated pits, we made a PAGFP chimera with clathrin light chain (CLC), which localizes to clathrin-coated pits at the cytoplasmic face of the Golgi apparatus and the plasma membrane 8. We observed TfR- and PAGFP-CLC clusters in the TIRF microscopy image similar to those observed in the one-color experiments, with molecules located in clustered areas throughout TIRF PALM PALM a b c d e f g j k l 1..5 PAGFP-CLC TfR-.. r (µm)... r (µm).. h i NATURE METHODS VOL. NO. FEBRUARY 9 157
6 the plasma membrane (Figs. 3 and a f). In addition, TfR- was enriched in larger structures extending several micrometers (Fig. a,b), which may represent folds in the plasma membrane. We observed colocalization of TfR- with PAGFP- CLC in the TfR- TIRF microscopy image clusters as well as in PALM images (Fig. g i), but clusters of TfR and CLC also appeared either completely separate or in close proximity to each otherwithlittleoverlap.despitethelackofcompletecolocalization, pair correlation analysis confirmed similar molecular distributions and a characteristic cluster size of B nm, with more clustering of CLC, as evidenced by a higher y-axis intercept (Fig. j). Cluster analysis 5 identified many structures enriched in TfR- and PAGFP-CLC present in at least three different relative distributions (Fig. k,l). The shape and size of clusters observed here were often similar to those of clathrin-coated pits observed by electron microscopy 7 and the different TfR and CLC clusters may represent stages of previously observed and described clathrin-coated pit dynamics 8. PAGFP-CLC clusters lacking TfR- cargo may correspond to more mature clathrin-coated pits, whereas the TfR- clusters without PAGFP-CLC may represent mature uncoated vesicles close to the plasma membrane; clusters in which the two proteins colocalize may represent young pits that have captured the TfR- cargo. DISCUSSION Imaging multiple fluorophores in the same specimen is an important improvement of high-resolution molecular localization techniques. Two-color high-resolution localization experiments have been previously reported in which the green-to-red photoconvertible protein tdeosfp was paired with the photoswitchable fluorescent protein Dronpa,9, or in which the photoswitchable fluorescent protein rsfastlime was paired with Cy5 in PALMIRA 3. These studies produced images at B5 75 nm resolution on organelles, cytoskeletal elements and extracellular adhesion proteins. However, with exception of PSCFP paired with tdeosfp, photoswitching of one or both fluorophores between light and dark states is a necessary component for these two-color experiments, which may complicate the final results. If a photoactivated molecule switches off, it can be repeatedly switched on and can then be localized again, skewing the molecular distributions in the final high-resolution image. The use of rscherry proteins 1 can also result in skewed distributions. As it is not reliant on photoswitching behavior to maintain a low density of molecular signals in twocolor PALM studies, is a PAFP that can be used to avoid any potential complication associated with repeated localization of the same molecule. We developed in one of the first mutagenesis screens using single-molecule PALM characteristics in addition to ensemble photoactivation experiments as the selection criteria. lacks the capability for ratiometric imaging provided by the green-to-red photoconvertible fluorescent proteins. It performed substantially better than rscherry and rscherryrev in PALM, was similar to tdeosfp in most PALM comparisons, and did not hamper the trafficking and function of the TfR. The absence of a green emission state allows greater flexibility in one-color PALM experiments using a diffraction-limited green fluorescent molecule, in two-color PALM experiments using a dark-to-green PAFP and notably, will enable two-color live-cell and single-particle PALM. Compared to tdeosfp, the smaller monomeric is potentially less disruptive to tagged fusion partners. also has higher ph stability, displays better photostability, produces higher photoactivation contrast and exhibits faster chromophore formation at 37 1C. Thus, simultaneously addresses many disadvantages exhibited by other red PAFPs and, as shown here, meets the demands for use in both diffraction-limited microscopy and super-resolution PALM. METHODS Confocal and PALM imaging. Cell imaging was performed on either a Zeiss LSM51 or on a Leica SP laser scanning confocal microscope as described previously 3,13. Single-molecule imaging was performed as previously described,5 on an Olympus IX81 microscope using an 1.5 numerical aperture (NA) PlanApoN TIRF objective (Olympus) with the exception that a Dual- View imaging system (Photometrics) was used in the two-color PALM imaging. Additional methods. Descriptions of cloning, characterization of proteins in vitro, construction of plasmids, cell cultures and additional details on imaging are available in Supplementary Methods. Note: Supplementary information is available on the Nature Methods website. ACKNOWLEDGMENTS We thank R. Tsien (University of California at San Diego) for providing the prsetbmcherry plasmid, J. Wiedenmann (University of Ulm) for providing plasmids encoding EosFP variants, J. Zhang for assistance with flow cytometry, O. Subach for assistance with cell culture and imaging, and E. Betzig and H. Hess for assistance with PALM experiments, analysis and discussion. This work was supported by grants GM7358 and GM73913 from the US National Institutes of Health to V.V.V. AUTHOR CONTRIBUTIONS F.V.S. developed proteins and characterized them in vitro. G.H.P., S.M. and J.M.G. characterized proteins in mammalian cells. J.L.-S. and V.V.V. designed and planned the project. G.H.P. and V.V.V. wrote the manuscript. Published online at Reprints and permissions information is available online at 1. Lukyanov, K.A., Chudakov, D.M., Lukyanov, S. & Verkhusha, V.V. Innovation: Photoactivatable fluorescent proteins. Nat. Rev. Mol. Cell Biol., (5).. Hell, S.W. Far-field optical nanoscopy. Science 31, (7). 3. Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1 15 ().. Hess, S.T., Girirajan, T.P. & Mason, M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 58 7 (). 5. Rust, M.J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, ().. Egner, A. et al. Fluorescence nanoscopy in whole cells by asynchronous localization of photoswitching emitters. Biophys. J. 93, (7). 7. Flors, C. et al. A stroboscopic approach for fast photoactivation-localization microscopy with Dronpa mutants. J. Am. Chem. Soc. 19, (7). 8. Verkhusha, V.V. & Sorkin, A. Conversion of the monomeric red fluorescent protein into a photoactivatable probe. Chem. Biol. 1, (5). 9. Ando, R., Hama, H., Yamamoto-Hino, M., Mizuno, H. & Miyawaki, A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. USA 99, (). 1. Chudakov, D.M. et al. Kindling fluorescent proteins for precise in vivo photolabeling. Nat. Biotechnol. 1, (3). 11. Wiedenmann, J. et al. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci. USA 11, (). 158 VOL. NO. FEBRUARY 9 NATURE METHODS
7 1. Gurskaya, N.G. et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat. Biotechnol., 1 5 (). 13. Patterson, G.H. & Lippincott-Schwartz, J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science 97, (). 1. Chudakov, D.M. et al. Photoswitchable cyan fluorescent protein for protein tracking. Nat. Biotechnol., (). 15. Ando, R., Mizuno, H. & Miyawaki, A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science 3, (). 1. Stiel, A.C. et al. Generation of monomeric reversibly switchable red fluorescent proteins for far-field fluorescence nanoscopy. Biophys. J. 95, (8). 17. Shaner, N.C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol., (). 18. Shu, X., Shaner, N.C., Yarbrough, C.A., Tsien, R.Y. & Remington, S.J. Novel chromophores and buried charges control color in mfruits. Biochemistry 5, (). 19. Verkhusha, V.V. & Lukyanov, K.A. The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat. Biotechnol., 89 9 ().. Huebers, H.A., Huebers, E., Josephson, B. & Csiba, E. A highly efficient chemical isolation procedure for the rat placental transferrin receptor. Biochim. Biophys. Acta 991, 3 35 (1989). 1. Gallione, C.J. & Rose, J.K. A single amino acid substitution in a hydrophobic domain causes temperature sensitive cell-surface transport of a mutant viral glycoprotein. J. Virol. 5, (1985).. McGraw, T.E. & Maxfield, F.R. Human transferrin receptor internalization is partially dependent upon an aromatic amino acid on the cytoplasmic domain. Cell Regul. 1, (199). 3. Hopkins, C.R. The appearance and internalization of transferrin receptors at the margins of spreading human tumor cells. Cell, (1985).. Shroff, H. et al. Dual-color super-resolution imaging of genetically expressed probes within individual adhesion complexes. Proc. Natl. Acad. Sci. USA 1, (7). 5. Manley, S. et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods 5, (8).. Pearse, B.M.F. & Robinson, M.S. Clathrin, adaptors, and sorting. Annu. Rev. Cell Biol., (199). 7. Heuser, J.E. & Anderson, R.G. Hypertonic media inhibit receptor-mediated endocytosis by blocking clathrin-coated pit formation. J. Cell Biol. 18, 389 (1989). 8. Kirchhausen, T. Clathrin. Annu. Rev. Biochem. 9, (). 9. Habuchi, S. et al. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc. Natl. Acad. Sci. USA 1, (5). 3. Bock, H. et al. Two-color far-field fluorescence nanoscopy based on photoswitchable emitters. Appl. Phys. B 88, (7). NATURE METHODS VOL. NO. FEBRUARY 9 159
Photoactivatable mcherry for high-resolution two-color fluorescence microscopy
 nature methods Photoactivatable mcherry for high-resolution two-color fluorescence microscopy Fedor V Subach, George H Patterson, Suliana Manley, Jennifer M Gillette, Jennifer Lippincott-Schwartz & Vladislav
nature methods Photoactivatable mcherry for high-resolution two-color fluorescence microscopy Fedor V Subach, George H Patterson, Suliana Manley, Jennifer M Gillette, Jennifer Lippincott-Schwartz & Vladislav
STORM/PALM. Super Resolution Microscopy 10/31/2011. Looking into microscopic world of life
 Super Resolution Microscopy STORM/PALM Bo Huang Department of Pharmaceutical Chemistry, UCSF CSHL Quantitative Microscopy, 1/31/211 Looking into microscopic world of life 1 µm 1 µm 1 nm 1 nm 1 nm 1 Å Naked
Super Resolution Microscopy STORM/PALM Bo Huang Department of Pharmaceutical Chemistry, UCSF CSHL Quantitative Microscopy, 1/31/211 Looking into microscopic world of life 1 µm 1 µm 1 nm 1 nm 1 nm 1 Å Naked
Feb No. 22. Dmitriy Chudakov, Timo Zimmermann
 Feb. 2007 No. 22 Using photoactivatable fluorescent proteins Dendra2 and PS-CFP2 to track protein movement with Leica TCS SP5 Dmitriy Chudakov, Timo Zimmermann LAS AF: FRAP Wizard Live Data Mode Photoconversion
Feb. 2007 No. 22 Using photoactivatable fluorescent proteins Dendra2 and PS-CFP2 to track protein movement with Leica TCS SP5 Dmitriy Chudakov, Timo Zimmermann LAS AF: FRAP Wizard Live Data Mode Photoconversion
PALM/STORM, BALM, STED
 PALM/STORM, BALM, STED Last class 2-photon Intro to PALM/STORM Cyanine dyes/dronpa This class Finish localization super-res BALM STED Localization microscopy Intensity Bins = pixels xx 2 = ss2 + aa 2 /12
PALM/STORM, BALM, STED Last class 2-photon Intro to PALM/STORM Cyanine dyes/dronpa This class Finish localization super-res BALM STED Localization microscopy Intensity Bins = pixels xx 2 = ss2 + aa 2 /12
Rational design of true monomeric and bright photoconvertible
 Rational design of true monomeric and bright photoconvertible fluorescent proteins Mingshu Zhang, Hao Chang, Yongdeng Zhang, Junwei Yu, Lijie Wu, Wei Ji, Juanjuan Chen, Bei Liu, Jingze Lu, Yingfang Liu,
Rational design of true monomeric and bright photoconvertible fluorescent proteins Mingshu Zhang, Hao Chang, Yongdeng Zhang, Junwei Yu, Lijie Wu, Wei Ji, Juanjuan Chen, Bei Liu, Jingze Lu, Yingfang Liu,
Bi177 - Lecture 13 Microscopy Outside the Box. Fluorescence Nanoscopy TIRF 4-pi STED STORM/PALM
 Bi177 - Lecture 13 Microscopy Outside the Box Fluorescence Nanoscopy TIRF 4-pi STED STORM/PALM The diffraction limit: Abbe s law The Problem Diffraction limit 100x larger than molecular scale! Green Fluorescent
Bi177 - Lecture 13 Microscopy Outside the Box Fluorescence Nanoscopy TIRF 4-pi STED STORM/PALM The diffraction limit: Abbe s law The Problem Diffraction limit 100x larger than molecular scale! Green Fluorescent
Fast, three-dimensional super-resolution imaging of live cells
 Nature Methods Fast, three-dimensional super-resolution imaging of live cells Sara A Jones, Sang-Hee Shim, Jiang He & Xiaowei Zhuang Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3
Nature Methods Fast, three-dimensional super-resolution imaging of live cells Sara A Jones, Sang-Hee Shim, Jiang He & Xiaowei Zhuang Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3
Tracking sub-microscopic protein organisation at the plasma membrane of live cells using triple-colour superresolution
 Tracking sub-microscopic protein organisation at the plasma membrane of live cells using triple-colour superresolution microscopy Jacob Piehler Division of Biophysics, University of Osnabrück, Germany
Tracking sub-microscopic protein organisation at the plasma membrane of live cells using triple-colour superresolution microscopy Jacob Piehler Division of Biophysics, University of Osnabrück, Germany
HHS Public Access Author manuscript Nat Methods. Author manuscript; available in PMC 2009 November 01.
 Photoconversion in orange and red fluorescent proteins Gert-Jan Kremers 1, Kristin L. Hazelwood 2, Christopher S. Murphy 2, Michael W. Davidson 2, and David W. Piston 1 1 Department of Molecular Physiology
Photoconversion in orange and red fluorescent proteins Gert-Jan Kremers 1, Kristin L. Hazelwood 2, Christopher S. Murphy 2, Michael W. Davidson 2, and David W. Piston 1 1 Department of Molecular Physiology
Single-Particle Tracking Photoactivated Localization Microscopy for Mapping Single-Molecule Dynamics
 CHAPTER FIVE Single-Particle Tracking Photoactivated Localization Microscopy for Mapping Single-Molecule Dynamics Suliana Manley,* Jennifer M. Gillette, and Jennifer Lippincott-Schwartz Contents 1. Introduction
CHAPTER FIVE Single-Particle Tracking Photoactivated Localization Microscopy for Mapping Single-Molecule Dynamics Suliana Manley,* Jennifer M. Gillette, and Jennifer Lippincott-Schwartz Contents 1. Introduction
Localization Microscopy
 Localization Microscopy Theory, Sample Prep & Practical Considerations Patrina Pellett & Ann McEvoy Applications Scientist GE Healthcare, Cell Technologies May 27 th, 2015 Localization Microscopy Talk
Localization Microscopy Theory, Sample Prep & Practical Considerations Patrina Pellett & Ann McEvoy Applications Scientist GE Healthcare, Cell Technologies May 27 th, 2015 Localization Microscopy Talk
More on fluorescence
 More on fluorescence Last class Fluorescence Absorption emission Jablonski diagrams This class More on fluorescence Common fluorophores Jablonski diagrams to spectra Properties of fluorophores Excitation
More on fluorescence Last class Fluorescence Absorption emission Jablonski diagrams This class More on fluorescence Common fluorophores Jablonski diagrams to spectra Properties of fluorophores Excitation
A guide to choosing fluorescent proteins
 A guide to choosing fluorescent proteins Nathan C Shaner 1,2, Paul A Steinbach 1,3 & Roger Y Tsien 1,3,4 The recent explosion in the diversity of available fluorescent proteins (FPs) 1 16 promises a wide
A guide to choosing fluorescent proteins Nathan C Shaner 1,2, Paul A Steinbach 1,3 & Roger Y Tsien 1,3,4 The recent explosion in the diversity of available fluorescent proteins (FPs) 1 16 promises a wide
Fluorescence Nanoscopy
 Fluorescence Nanoscopy Keith A. Lidke University of New Mexico panda3.phys.unm.edu/~klidke/index.html Optical Microscopy http://en.wikipedia.org/wiki/k%c3%b6hler_illumination 30 µm Fluorescent Probes Michalet
Fluorescence Nanoscopy Keith A. Lidke University of New Mexico panda3.phys.unm.edu/~klidke/index.html Optical Microscopy http://en.wikipedia.org/wiki/k%c3%b6hler_illumination 30 µm Fluorescent Probes Michalet
Fluorescence microscopy
 Fluorescence microscopy 1 Fluorescence microscopies basic fluorescence, fluorophores Deconvolution Confocal Two-photon/multi-photon 4Pi Light sheet Total internal reflection STED FRAP/FLIP/FCS FRET PALM/STORM/iPALM
Fluorescence microscopy 1 Fluorescence microscopies basic fluorescence, fluorophores Deconvolution Confocal Two-photon/multi-photon 4Pi Light sheet Total internal reflection STED FRAP/FLIP/FCS FRET PALM/STORM/iPALM
F* techniques: FRAP, FLIP, FRET, FLIM,
 F* techniques: FRAP, FLIP, FRET, FLIM, FCS Antonia Göhler March 2015 Fluorescence explained in the Bohr model Absorption of light (blue) causes an electron to move to a higher energy orbit. After a particular
F* techniques: FRAP, FLIP, FRET, FLIM, FCS Antonia Göhler March 2015 Fluorescence explained in the Bohr model Absorption of light (blue) causes an electron to move to a higher energy orbit. After a particular
Localization-Based Super-Resolution Light Microscopy
 Kristin A. Gabor, 1,2,3 Mudalige S. Gunewardene, 1 David Santucci, 4 and Samuel T. Hess 1,3, * 1 Department of Physics and Astronomy, 120 Bennett Hall, University of Maine, Orono, ME 04469 2 Department
Kristin A. Gabor, 1,2,3 Mudalige S. Gunewardene, 1 David Santucci, 4 and Samuel T. Hess 1,3, * 1 Department of Physics and Astronomy, 120 Bennett Hall, University of Maine, Orono, ME 04469 2 Department
Confocal Microscopy Analyzes Cells
 Choosing Filters for Fluorescence A Laurin Publication Photonic Solutions for Biotechnology and Medicine November 2002 Confocal Microscopy Analyzes Cells Reprinted from the November 2002 issue of Biophotonics
Choosing Filters for Fluorescence A Laurin Publication Photonic Solutions for Biotechnology and Medicine November 2002 Confocal Microscopy Analyzes Cells Reprinted from the November 2002 issue of Biophotonics
Challenges to measuring intracellular Ca 2+ Calmodulin: nature s Ca 2+ sensor
 Calcium Signals in Biological Systems Lecture 3 (2/9/0) Measuring intracellular Ca 2+ signals II: Genetically encoded Ca 2+ sensors Henry M. Colecraft, Ph.D. Challenges to measuring intracellular Ca 2+
Calcium Signals in Biological Systems Lecture 3 (2/9/0) Measuring intracellular Ca 2+ signals II: Genetically encoded Ca 2+ sensors Henry M. Colecraft, Ph.D. Challenges to measuring intracellular Ca 2+
Q&A: Single-molecule localization microscopy for biological imaging
 Q U E S T I O N & ANSWER Q&A: Single-molecule localization microscopy for biological imaging Ann L McEvoy 1, Derek Greenfield 1,2,5, Mark Bates 3 and Jan Liphardt 1,2,4 * Open Access Why is it important
Q U E S T I O N & ANSWER Q&A: Single-molecule localization microscopy for biological imaging Ann L McEvoy 1, Derek Greenfield 1,2,5, Mark Bates 3 and Jan Liphardt 1,2,4 * Open Access Why is it important
SIM SSIM. nanoscopy RESOLFT. Super-resolution STORM GSDIM. dstorm PALMIRA FPALM PALM PAINT SPRAIPAINT SOFI BALM CALM. Bo Huang
 STEDGSD STORM SOFI nanoscopy GSDIM PALMIRA SMACM BBB PAINT SPRAIPAINT CALM RESOLFT BALM SIM SSIM Super-resolution Bo Huang 2013.08.01 dstorm FPALM PALM 50 years to extend the resolution Confocal microscopy
STEDGSD STORM SOFI nanoscopy GSDIM PALMIRA SMACM BBB PAINT SPRAIPAINT CALM RESOLFT BALM SIM SSIM Super-resolution Bo Huang 2013.08.01 dstorm FPALM PALM 50 years to extend the resolution Confocal microscopy
Super-resolution Microscopy
 Semr oc kwhi t epaperser i es : 1. Introduction Super-resolution Microscopy Fluorescence microscopy has revolutionized the study of biological samples. Ever since the invention of fluorescence microscopy
Semr oc kwhi t epaperser i es : 1. Introduction Super-resolution Microscopy Fluorescence microscopy has revolutionized the study of biological samples. Ever since the invention of fluorescence microscopy
Super-resolution imaging: early days w/ Video-enhanced DIC, TIRF, PALM, STORM, etc.
 15/05/2012 Super-resolution imaging: early days w/ Video-enhanced DIC, TIRF, PALM, STORM, etc. Prof. Dr. Rainer Duden duden@bio.uni-luebeck.de 1 Using conventional light microscopy resolution is limited
15/05/2012 Super-resolution imaging: early days w/ Video-enhanced DIC, TIRF, PALM, STORM, etc. Prof. Dr. Rainer Duden duden@bio.uni-luebeck.de 1 Using conventional light microscopy resolution is limited
Improving the photostability of bright monomeric orange and red fluorescent proteins
 ARTICLES Improving the photostability of bright monomeric orange and red fluorescent proteins 2008 Nature Publishing Group h ttp://www.nature.com/natur e method s Nathan C Shaner 1,5, Michael Z Lin 1,2,
ARTICLES Improving the photostability of bright monomeric orange and red fluorescent proteins 2008 Nature Publishing Group h ttp://www.nature.com/natur e method s Nathan C Shaner 1,5, Michael Z Lin 1,2,
Fluorescent protein. Origin of fluorescent proteins. Structural and biophysical analysis of FPs. Application in molecular biology and biotechnology
 Origin of fluorescent proteins types of FPs mechanism of fluorescence Fluorescent protein Structural and biophysical analysis of FPs Application in molecular biology and biotechnology Variety of organisms
Origin of fluorescent proteins types of FPs mechanism of fluorescence Fluorescent protein Structural and biophysical analysis of FPs Application in molecular biology and biotechnology Variety of organisms
Fluorescent probes for superresolution imaging in living cells
 Fluorescent probes for superresolution imaging in living cells Marta Fernández-Suárez* and Alice Y. Ting* Abstract In 1873, Ernst Abbe discovered that features closer than ~200 nm cannot be resolved by
Fluorescent probes for superresolution imaging in living cells Marta Fernández-Suárez* and Alice Y. Ting* Abstract In 1873, Ernst Abbe discovered that features closer than ~200 nm cannot be resolved by
Confocal Microscopy & Imaging Technology. Yan Wu
 Confocal Microscopy & Imaging Technology Yan Wu Dec. 05, 2014 Cells under the microscope What we use to see the details of the cell? Light and Electron Microscopy - Bright light / fluorescence microscopy
Confocal Microscopy & Imaging Technology Yan Wu Dec. 05, 2014 Cells under the microscope What we use to see the details of the cell? Light and Electron Microscopy - Bright light / fluorescence microscopy
Reviewer #1 (Remarks to the Author):
 Reviewer #1 (Remarks to the Author): This manuscript describes a novel application of dye-labeled DNA-conjugated AuNPs in visualizing intracellular transport within living cells. The most important result
Reviewer #1 (Remarks to the Author): This manuscript describes a novel application of dye-labeled DNA-conjugated AuNPs in visualizing intracellular transport within living cells. The most important result
Fluorescence Light Microscopy for Cell Biology
 Fluorescence Light Microscopy for Cell Biology Why use light microscopy? Traditional questions that light microscopy has addressed: Structure within a cell Locations of specific molecules within a cell
Fluorescence Light Microscopy for Cell Biology Why use light microscopy? Traditional questions that light microscopy has addressed: Structure within a cell Locations of specific molecules within a cell
Stochastic Optical Reconstruction Microscopy (STORM): A Method for Superresolution Fluorescence Imaging
 Topic Introduction Stochastic Optical Reconstruction Microscopy (STORM): A Method for Superresolution Fluorescence Imaging Mark Bates, Sara A. Jones, and Xiaowei Zhuang The relatively low spatial resolution
Topic Introduction Stochastic Optical Reconstruction Microscopy (STORM): A Method for Superresolution Fluorescence Imaging Mark Bates, Sara A. Jones, and Xiaowei Zhuang The relatively low spatial resolution
Nanoscale measurement examples in biology: Sub-diffractive imaging & the fly brain challenge
 Nanoscale measurement examples in biology: Sub-diffractive imaging & the fly brain challenge Harald Hess, HHMI Janelia Farm Introduction to HHMI Janelia Farms Sample: Nerves, Worm Brains, Fly Brains, Rodent
Nanoscale measurement examples in biology: Sub-diffractive imaging & the fly brain challenge Harald Hess, HHMI Janelia Farm Introduction to HHMI Janelia Farms Sample: Nerves, Worm Brains, Fly Brains, Rodent
Measure of surface protein mobility with u-paint technique
 Measure of surface protein mobility with u-paint technique How dynamic image can solve the situation? Random distribution or cluster? Why live super-resolution microscopy can solve the situation With mobility
Measure of surface protein mobility with u-paint technique How dynamic image can solve the situation? Random distribution or cluster? Why live super-resolution microscopy can solve the situation With mobility
Paper presentation PNAS October 20, 2009 vol. 106 no
 Paper presentation Continuous imaging of plasmon rulers in live cells reveals early-stage caspase-3 activation at the single-molecule level Young-wook Jun a, Sassan Sheikholeslami a,1, Daniel R. Hostetter
Paper presentation Continuous imaging of plasmon rulers in live cells reveals early-stage caspase-3 activation at the single-molecule level Young-wook Jun a, Sassan Sheikholeslami a,1, Daniel R. Hostetter
PALM Microscope User Manual
 PALM Microscope User Manual Stephan Uphoff, Mathew Stracy, Federico Garza de Leon Microscope components: The laser excitation is delivered from a Toptica Multi Laser Engine with 405 nm, 488 nm, 561 nm,
PALM Microscope User Manual Stephan Uphoff, Mathew Stracy, Federico Garza de Leon Microscope components: The laser excitation is delivered from a Toptica Multi Laser Engine with 405 nm, 488 nm, 561 nm,
Supplementary Information. A superfolding Spinach2 reveals the dynamic nature of. trinucleotide repeat RNA
 Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
Post-expansion antibody delivery, after epitope-preserving homogenization.
 Supplementary Figure 1 Post-expansion antibody delivery, after epitope-preserving homogenization. (a, b) Wide-field fluorescence images of Thy1-YFP-expressing mouse brain hemisphere slice before expansion
Supplementary Figure 1 Post-expansion antibody delivery, after epitope-preserving homogenization. (a, b) Wide-field fluorescence images of Thy1-YFP-expressing mouse brain hemisphere slice before expansion
FLUORESCENCE. Matyas Molnar and Dirk Pacholsky
 FLUORESCENCE Matyas Molnar and Dirk Pacholsky 1 Information This lecture contains images and information from the following internet homepages http://micro.magnet.fsu.edu/primer/index.html http://www.microscopyu.com/
FLUORESCENCE Matyas Molnar and Dirk Pacholsky 1 Information This lecture contains images and information from the following internet homepages http://micro.magnet.fsu.edu/primer/index.html http://www.microscopyu.com/
Module 2 overview SPRING BREAK
 1 Module 2 overview lecture lab 1. Introduction to the module 1. Start-up protein eng. 2. Rational protein design 2. Site-directed mutagenesis 3. Fluorescence and sensors 3. DNA amplification 4. Protein
1 Module 2 overview lecture lab 1. Introduction to the module 1. Start-up protein eng. 2. Rational protein design 2. Site-directed mutagenesis 3. Fluorescence and sensors 3. DNA amplification 4. Protein
Microscopy from Carl Zeiss. DirectFRAP. News from the Cell. The New Class of Laser Manipulation for the Analysis of Cell Dynamics
 Microscopy from Carl Zeiss DirectFRAP News from the Cell The New Class of Laser Manipulation for the Analysis of Cell Dynamics DirectFRAP. New Insights into Cell Dynamics. Fluorescence breaks new ground:
Microscopy from Carl Zeiss DirectFRAP News from the Cell The New Class of Laser Manipulation for the Analysis of Cell Dynamics DirectFRAP. New Insights into Cell Dynamics. Fluorescence breaks new ground:
A Brief History of Light Microscopy And How It Transformed Biomedical Research
 A Brief History of Light Microscopy And How It Transformed Biomedical Research Suewei Lin Office: Interdisciplinary Research Building 8A08 Email: sueweilin@gate.sinica.edu.tw TEL: 2789-9315 Microscope
A Brief History of Light Microscopy And How It Transformed Biomedical Research Suewei Lin Office: Interdisciplinary Research Building 8A08 Email: sueweilin@gate.sinica.edu.tw TEL: 2789-9315 Microscope
D e c N o. 2 8
 D e c. 2 0 0 7 N o. 2 8 CONFOCAL APPLICATION LETTER resolution FRET Acceptor Photobleaching LAS AF Application Wizard FRET with Leica TCS SP5 LAS AF Version 1.7.0 Introduction Fluorescence Resonance Energy
D e c. 2 0 0 7 N o. 2 8 CONFOCAL APPLICATION LETTER resolution FRET Acceptor Photobleaching LAS AF Application Wizard FRET with Leica TCS SP5 LAS AF Version 1.7.0 Introduction Fluorescence Resonance Energy
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
The Green Fluorescent Protein. w.chem.uwec.edu/chem412_s99/ppt/green.ppt
 The Green Fluorescent Protein w.chem.uwec.edu/chem412_s99/ppt/green.ppt www.chem.uwec.edu/chem412_s99/ppt/green.ppt Protein (gene) is from a jellyfish: Aequorea victoria www.chem.uwec.edu/chem412_s99/ppt/green.ppt
The Green Fluorescent Protein w.chem.uwec.edu/chem412_s99/ppt/green.ppt www.chem.uwec.edu/chem412_s99/ppt/green.ppt Protein (gene) is from a jellyfish: Aequorea victoria www.chem.uwec.edu/chem412_s99/ppt/green.ppt
Improving the photostability of bright monomeric orange and red fluorescent proteins
 Improving the photostability of bright monomeric orange and red fluorescent proteins Nathan C Shaner 1,5, Michael Z Lin 1,2, Michael R McKeown 1,2, Paul A Steinbach 1,2, Kristin L Hazelwood 4, Michael
Improving the photostability of bright monomeric orange and red fluorescent proteins Nathan C Shaner 1,5, Michael Z Lin 1,2, Michael R McKeown 1,2, Paul A Steinbach 1,2, Kristin L Hazelwood 4, Michael
Introduction to N-STORM
 Introduction to N-STORM Dan Metcalf Advanced Imaging Manager Outline Introduction Principles of STORM Applications N-STORM overview Biological Scale Mitochondrion Microtubule Amino Acid 1Å Kinesin 1nm
Introduction to N-STORM Dan Metcalf Advanced Imaging Manager Outline Introduction Principles of STORM Applications N-STORM overview Biological Scale Mitochondrion Microtubule Amino Acid 1Å Kinesin 1nm
SUPPLEMENTARY INFORMATION
 a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
Lab 5: Optical trapping and single molecule fluorescence
 Lab 5: Optical trapping and single molecule fluorescence PI: Matt Lang Lab Instructor: Jorge Ferrer Summary Optical tweezers are an excellent experimental tool to study the biophysics of single molecule
Lab 5: Optical trapping and single molecule fluorescence PI: Matt Lang Lab Instructor: Jorge Ferrer Summary Optical tweezers are an excellent experimental tool to study the biophysics of single molecule
Dino-Lite knowledge & education. Fluorescence Microscopes
 Dino-Lite knowledge & education Fluorescence Microscopes Dino-Lite Fluorescence models Smallest fluorescence microscope in the world Revolution to biomedical and educational applications Flexible Easy
Dino-Lite knowledge & education Fluorescence Microscopes Dino-Lite Fluorescence models Smallest fluorescence microscope in the world Revolution to biomedical and educational applications Flexible Easy
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and
 Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Photoswitchable cyan fluorescent protein for protein tracking
 24 Nature Publishing Group http://www.nature.com/naturebiotechnology Photoswitchable cyan fluorescent protein for protein tracking Dmitriy M Chudakov 1,4, Vladislav V Verkhusha 2,4, Dmitry B Staroverov
24 Nature Publishing Group http://www.nature.com/naturebiotechnology Photoswitchable cyan fluorescent protein for protein tracking Dmitriy M Chudakov 1,4, Vladislav V Verkhusha 2,4, Dmitry B Staroverov
Live cell microscopy
 Live cell microscopy 1. Why do live cell microscopy? 2. Maintaining living cells on a microscope stage. 3. Considerations for imaging living cells. 4. Fluorescence labeling of living cells. 5. Imaging
Live cell microscopy 1. Why do live cell microscopy? 2. Maintaining living cells on a microscope stage. 3. Considerations for imaging living cells. 4. Fluorescence labeling of living cells. 5. Imaging
Tracking Cellular Protein Localization and Movement in Cells with a Flexible Fluorescent Labeling Technology. Chad Zimprich January 2015
 Tracking Cellular Protein Localization and Movement in Cells with a Flexible Fluorescent Labeling Technology Chad Zimprich January 2015 Presentation verview HaloTag Fusion Technology Design Functionality
Tracking Cellular Protein Localization and Movement in Cells with a Flexible Fluorescent Labeling Technology Chad Zimprich January 2015 Presentation verview HaloTag Fusion Technology Design Functionality
Supporting Information for. Electrical control of Förster energy transfer.
 1 Supporting Information for Electrical control of Förster energy transfer. Klaus Becker 1, John M. Lupton 1*, Josef Müller 1, Andrey. L. Rogach 1, Dmitri V. Talapin, Horst Weller & Jochen Feldmann 1 1
1 Supporting Information for Electrical control of Förster energy transfer. Klaus Becker 1, John M. Lupton 1*, Josef Müller 1, Andrey. L. Rogach 1, Dmitri V. Talapin, Horst Weller & Jochen Feldmann 1 1
Supplementary Figure 1. Design of linker truncation library between Nluc and either Venus or mneongreen
 1 2 3 Supplementary Figure 1. Design of linker truncation library between Nluc and either Venus or mneongreen 4 5 6 7 8 9 10 The cdna of C-terminally deleted FPs (mneongreen or Venus) mutants and N-terminally
1 2 3 Supplementary Figure 1. Design of linker truncation library between Nluc and either Venus or mneongreen 4 5 6 7 8 9 10 The cdna of C-terminally deleted FPs (mneongreen or Venus) mutants and N-terminally
Photoactivatable. Fluorescent Proteins. Effective tool to monitor cellular events in real time
 Section B Photoactivatable Fluorescent Proteins Effective tool to monitor cellular events in real time t r a c k i n g c e l l m o v e m e n t m o n i t o r i n g c o m m u n i c a t i o n p a t h w a
Section B Photoactivatable Fluorescent Proteins Effective tool to monitor cellular events in real time t r a c k i n g c e l l m o v e m e n t m o n i t o r i n g c o m m u n i c a t i o n p a t h w a
Biophotonics?? Biophotonics. technology in biomedical engineering. Advantages of the lightwave
 Biophotonics - Imaging: X-ray, OCT, polarimetry, DOT, TIRF, photon migration, endoscopy, confocal microscopy, multiphoton microscopy, multispectral imaging - Biosensing: IR spectroscopy, fluorescence,
Biophotonics - Imaging: X-ray, OCT, polarimetry, DOT, TIRF, photon migration, endoscopy, confocal microscopy, multiphoton microscopy, multispectral imaging - Biosensing: IR spectroscopy, fluorescence,
Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging
 Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging Graham T Dempsey 1,6, Joshua C Vaughan 2,3,6, Kok Hao Chen 3,6, Mark Bates 4 & Xiaowei Zhuang 2,3,5 211
Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging Graham T Dempsey 1,6, Joshua C Vaughan 2,3,6, Kok Hao Chen 3,6, Mark Bates 4 & Xiaowei Zhuang 2,3,5 211
Product Manual. RFP ELISA Kit. Catalog Number. FOR RESEARCH USE ONLY Not for use in diagnostic procedures
 Product Manual RFP ELISA Kit Catalog Number AKR-122 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Red fluorescent protein (DsRed) is a spontaneously fluorescent protein
Product Manual RFP ELISA Kit Catalog Number AKR-122 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Red fluorescent protein (DsRed) is a spontaneously fluorescent protein
The most extensively used technique for tissue analysis is light microscopy.
 Fluorescence Theory Quantum yield Wavelength shift Ligand interactions Membrane interactions Using quenchning effects Fluorescence in-vivo Localization Distance measurements FRET The most extensively used
Fluorescence Theory Quantum yield Wavelength shift Ligand interactions Membrane interactions Using quenchning effects Fluorescence in-vivo Localization Distance measurements FRET The most extensively used
Contact Details. Dr Alexander Galkin. Office: MBC Room 186. Tel: (028) Frequency and wavelength.
 Contact Details The electromagnetic spectrum Biological Spectroscopy Dr Alexander Galkin Email: a.galkin@qub.ac.uk Dr Alexander Galkin MSc Biomolecular Function - BBC8045 Office: MBC Room 186 Tel: (028)
Contact Details The electromagnetic spectrum Biological Spectroscopy Dr Alexander Galkin Email: a.galkin@qub.ac.uk Dr Alexander Galkin MSc Biomolecular Function - BBC8045 Office: MBC Room 186 Tel: (028)
What to look for in a fluorophore. What to do with a fluorophore. Types of fluorochromes
 What to do with a fluorophore Intracellular localization (ER, Golgi, PM, nuclear, lysosome, MT, actin,...) Dynamic processes (protein synthesis, trafficking, turnover, DNA replication, cytoskeletal remodeling,
What to do with a fluorophore Intracellular localization (ER, Golgi, PM, nuclear, lysosome, MT, actin,...) Dynamic processes (protein synthesis, trafficking, turnover, DNA replication, cytoskeletal remodeling,
New generation of sensors to map ph dynamics and Chloride transport in organelles
 New generation of sensors to map ph dynamics and Chloride transport in organelles TJC 11.08.2015 Rochat Mary-aude Subcellular ph >Cellular Compartimentalization: Segregation of specific function in organelles
New generation of sensors to map ph dynamics and Chloride transport in organelles TJC 11.08.2015 Rochat Mary-aude Subcellular ph >Cellular Compartimentalization: Segregation of specific function in organelles
Imaging of endocrine organs
 Imaging of endocrine organs Helen Christian Department of Physiology, Anatomy & Genetics St Anne s College, University of Oxford Diabetesforum, Stockholm 2017 Islets of Langerhan Pituitary gland Renin
Imaging of endocrine organs Helen Christian Department of Physiology, Anatomy & Genetics St Anne s College, University of Oxford Diabetesforum, Stockholm 2017 Islets of Langerhan Pituitary gland Renin
Special Techniques 1. Mark Scott FILM Facility
 Special Techniques 1 Mark Scott FILM Facility SPECIAL TECHNIQUES Multi-photon microscopy Second Harmonic Generation FRAP FRET FLIM In-vivo imaging TWO-PHOTON MICROSCOPY Alternative to confocal and deconvolution
Special Techniques 1 Mark Scott FILM Facility SPECIAL TECHNIQUES Multi-photon microscopy Second Harmonic Generation FRAP FRET FLIM In-vivo imaging TWO-PHOTON MICROSCOPY Alternative to confocal and deconvolution
Supplementary Material
 Supplementary Material Supplementary Table 1. Definitions of variants described in this work. Protein variant Substitutions and modifications relative to parent mtfp1 a = cfp484 b + delete all residues
Supplementary Material Supplementary Table 1. Definitions of variants described in this work. Protein variant Substitutions and modifications relative to parent mtfp1 a = cfp484 b + delete all residues
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10016 Supplementary discussion on binding site density for protein complexes on the surface: The density of biotin sites on the chip is ~10 3 biotin-peg per µm 2. The biotin sites are
doi:10.1038/nature10016 Supplementary discussion on binding site density for protein complexes on the surface: The density of biotin sites on the chip is ~10 3 biotin-peg per µm 2. The biotin sites are
Supplementary Table 1. Components of an FCS setup (1PE and 2PE)
 Supplementary Table 1. Components of an FCS setup (1PE and 2PE) Component and function Laser source Excitation of fluorophores Microscope with xy-translation stage mounted on vibration isolated optical
Supplementary Table 1. Components of an FCS setup (1PE and 2PE) Component and function Laser source Excitation of fluorophores Microscope with xy-translation stage mounted on vibration isolated optical
Principles of flow cytometry: overview of flow cytometry and its uses for cell analysis and sorting. Shoreline Community College BIOL 288
 Principles of flow cytometry: overview of flow cytometry and its uses for cell analysis and sorting Shoreline Community College BIOL 288 Flow Cytometry What is Flow Cytometry? Measurement of cells or particles
Principles of flow cytometry: overview of flow cytometry and its uses for cell analysis and sorting Shoreline Community College BIOL 288 Flow Cytometry What is Flow Cytometry? Measurement of cells or particles
Chapter 4. the biological community to assay for protein-protein interactions. FRET describes the
 31 Chapter 4 Determination of nachr stoichiometry using Normalized Försters Resonance Energy Transfer (NFRET) Försters resonance energy transfer (FRET) has become a technique widely used in the biological
31 Chapter 4 Determination of nachr stoichiometry using Normalized Försters Resonance Energy Transfer (NFRET) Försters resonance energy transfer (FRET) has become a technique widely used in the biological
 Introduction to Fluorescent Proteins The discovery of green fluorescent protein in the early 1960s ultimately heralded a new era in cell biology by enabling investigators to apply molecular cloning methods,
Introduction to Fluorescent Proteins The discovery of green fluorescent protein in the early 1960s ultimately heralded a new era in cell biology by enabling investigators to apply molecular cloning methods,
Self-labelling enzymes as universal tags for fluorescence microscopy, superresolution microscopy and electron microscopy
 Supplementary Materials Self-labelling enzymes as universal tags for fluorescence microscopy, superresolution microscopy and electron microscopy Viktoria Liss, Britta Barlag, Monika Nietschke and Michael
Supplementary Materials Self-labelling enzymes as universal tags for fluorescence microscopy, superresolution microscopy and electron microscopy Viktoria Liss, Britta Barlag, Monika Nietschke and Michael
Spectral Separation of Multifluorescence Labels with the LSM 510 META
 Microscopy from Carl Zeiss Spectral Separation of Multifluorescence Labels with the LSM 510 META Indians living in the South American rain forest can distinguish between almost 200 hues of green in their
Microscopy from Carl Zeiss Spectral Separation of Multifluorescence Labels with the LSM 510 META Indians living in the South American rain forest can distinguish between almost 200 hues of green in their
Absorption of an electromagnetic wave
 In vivo optical imaging?? Absorption of an electromagnetic wave Tissue absorption spectrum Extinction = Absorption + Scattering Absorption of an electromagnetic wave Scattering of an electromagnetic wave
In vivo optical imaging?? Absorption of an electromagnetic wave Tissue absorption spectrum Extinction = Absorption + Scattering Absorption of an electromagnetic wave Scattering of an electromagnetic wave
Super-resolution microscopy at a glance
 ARTICLE SERIES: Imaging Cell Science at a Glance 1607 Super-resolution microscopy at a glance Catherine G. Galbraith 1, * and James A. Galbraith 2, * 1 National Institute of Health, 1 NICHD and 2 NINDS,
ARTICLE SERIES: Imaging Cell Science at a Glance 1607 Super-resolution microscopy at a glance Catherine G. Galbraith 1, * and James A. Galbraith 2, * 1 National Institute of Health, 1 NICHD and 2 NINDS,
Widefield Microscopy Bleed-Through
 In widefield microscopy the excitation wavelengths which illuminate the sample, and the emission wavelengths which reach the CCD camera are selected throughout a filter cube. A filter cube consists of
In widefield microscopy the excitation wavelengths which illuminate the sample, and the emission wavelengths which reach the CCD camera are selected throughout a filter cube. A filter cube consists of
Fluorescence Microscopy
 Fluorescence Microscopy Dr. Arne Seitz Swiss Institute of Technology (EPFL) Faculty of Life Sciences Head of BIOIMAGING AND OPTICS BIOP arne.seitz@epfl.ch Fluorescence Microscopy Why do we need fluorescence
Fluorescence Microscopy Dr. Arne Seitz Swiss Institute of Technology (EPFL) Faculty of Life Sciences Head of BIOIMAGING AND OPTICS BIOP arne.seitz@epfl.ch Fluorescence Microscopy Why do we need fluorescence
λ N -GFP: an RNA reporter system for live-cell imaging
 λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
Supporting Information
 Supporting Information Azido Push Pull Fluorogens Photoactivate to Produce Bright Fluorescent Labels Samuel J. Lord, Hsiao-lu D. Lee, Reichel Samuel, Ryan Weber, Na Liu, Nicholas R. Conley, Michael A.
Supporting Information Azido Push Pull Fluorogens Photoactivate to Produce Bright Fluorescent Labels Samuel J. Lord, Hsiao-lu D. Lee, Reichel Samuel, Ryan Weber, Na Liu, Nicholas R. Conley, Michael A.
Workshop advanced light microscopy
 Workshop advanced light microscopy Multi-mode confocal laser scanning microscope Jan Willem Borst Laboratory of Biochemistry Biomolecular Networks www.bic.wur.nl MicroSpectroscopy Centre Wageningen Microspectroscopy
Workshop advanced light microscopy Multi-mode confocal laser scanning microscope Jan Willem Borst Laboratory of Biochemistry Biomolecular Networks www.bic.wur.nl MicroSpectroscopy Centre Wageningen Microspectroscopy
Introduction CHAPTER 1
 CHAPTER 1 Introduction 1.1 Light Microscopy The light microscope is one of the significant inventions in the history of humankind that, along with the telescope, played a central role in the Scientific
CHAPTER 1 Introduction 1.1 Light Microscopy The light microscope is one of the significant inventions in the history of humankind that, along with the telescope, played a central role in the Scientific
In vitro EGFP-CALI comprehensive analysis. Liang CAI. David Humphrey. Jessica Wilson. Ken Jacobson. Dept. of Cell and Developmental Biology
 In vitro EGFP-CALI comprehensive analysis Liang CAI David Humphrey Jessica Wilson Ken Jacobson Dept. of Cell and Developmental Biology 1/15 Liang s Rotation in Ken s Lab, Sep-Nov, 2003 1. CALI background
In vitro EGFP-CALI comprehensive analysis Liang CAI David Humphrey Jessica Wilson Ken Jacobson Dept. of Cell and Developmental Biology 1/15 Liang s Rotation in Ken s Lab, Sep-Nov, 2003 1. CALI background
Rice/TCU REU on Computational Neuroscience. Fundamentals of Molecular Imaging
 Rice/TCU REU on Computational Neuroscience Fundamentals of Molecular Imaging June 2, 2009 Neal Waxham 713-500-5621 m.n.waxham@uth.tmc.edu Objectives Introduction to resolution in light microscopy Brief
Rice/TCU REU on Computational Neuroscience Fundamentals of Molecular Imaging June 2, 2009 Neal Waxham 713-500-5621 m.n.waxham@uth.tmc.edu Objectives Introduction to resolution in light microscopy Brief
Monomeric fluorescent timers that change color from blue to red report on cellular trafficking
 Monomeric fluorescent timers that change color from blue to red report on cellular trafficking Fedor V. Subach, Oksana M. Subach, Illia S. Gundorov, Kateryna S. Morozova, Kiryl D. Piatkevich, Ana Maria
Monomeric fluorescent timers that change color from blue to red report on cellular trafficking Fedor V. Subach, Oksana M. Subach, Illia S. Gundorov, Kateryna S. Morozova, Kiryl D. Piatkevich, Ana Maria
Chapter One. Construction of a Fluorescent α5 Subunit. Elucidation of the unique contribution of the α5 subunit is complicated by several factors
 4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
Fluorescence Nanoscopy 高甫仁 ) Institute of Biophotonics, National Yang Ming University. Outline
 Fluorescence Nanoscopy 高甫仁 ) Fu-Jen Kao ( 高甫仁 Institute of Biophotonics, National Yang Ming University Outline The Abbe s (diffraction) limit and nanoscopy Fundamentals and opportunities of FLIM/FRET Visualizing
Fluorescence Nanoscopy 高甫仁 ) Fu-Jen Kao ( 高甫仁 Institute of Biophotonics, National Yang Ming University Outline The Abbe s (diffraction) limit and nanoscopy Fundamentals and opportunities of FLIM/FRET Visualizing
Aoki et al.,
 JCB: SUPPLEMENTAL MATERIAL Aoki et al., http://www.jcb.org/cgi/content/full/jcb.200609017/dc1 Supplemental materials and methods Calculation of the raw number of translocated proteins First, the average
JCB: SUPPLEMENTAL MATERIAL Aoki et al., http://www.jcb.org/cgi/content/full/jcb.200609017/dc1 Supplemental materials and methods Calculation of the raw number of translocated proteins First, the average
Super Resolution Imaging Solution Provider. Imaging Future
 Super Resolution Imaging Solution Provider Imaging Future Imaging Solution More Than Equipment NanoBioImaging(NBI) is the Industrial Partner of HKUST Super Resolution Imaging Center (SRIC). NBI aims to
Super Resolution Imaging Solution Provider Imaging Future Imaging Solution More Than Equipment NanoBioImaging(NBI) is the Industrial Partner of HKUST Super Resolution Imaging Center (SRIC). NBI aims to
Biophysics of Macromolecules
 Biophysics of Macromolecules Lecture 18: In vivo Methods Braun/Lipfert SS 2015 How to create methods to probe macromolecules in vivo? 6. July 2015 Crowding alters Biochemical Equilibria Excluded volume
Biophysics of Macromolecules Lecture 18: In vivo Methods Braun/Lipfert SS 2015 How to create methods to probe macromolecules in vivo? 6. July 2015 Crowding alters Biochemical Equilibria Excluded volume
Supplementary Figures
 Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
Single molecule evaluation of fluorescent protein photoactivation efficiency using an in vivo nanotemplate
 Single molecule evaluation of fluorescent protein photoactivation efficiency using an in vivo nanotemplate Nela Durisic 1, Lara Laparra-Cuervo 1, Ángel Sandoval-Álvarez 1, Joseph Steven Borbely 1, Melike
Single molecule evaluation of fluorescent protein photoactivation efficiency using an in vivo nanotemplate Nela Durisic 1, Lara Laparra-Cuervo 1, Ángel Sandoval-Álvarez 1, Joseph Steven Borbely 1, Melike
Optically Controlled Reversible Protein Hydrogels Based on Photoswitchable Fluorescent Protein Dronpa. Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Optically Controlled Reversible Protein Hydrogels Based on Photoswitchable Fluorescent Protein
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Optically Controlled Reversible Protein Hydrogels Based on Photoswitchable Fluorescent Protein
SUPPLEMENTARY INFORMATION. Tolerance of a knotted near infrared fluorescent protein to random circular permutation
 SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
Imaging of BacMam Transfected U-2 OS Cells
 A p p l i c a t i o n N o t e Imaging of BacMam Transfected U-2 OS Cells Optimization of Transfection Conditions Using the Cytation 3 Multi- Mode Reader and Gen5 Data Analysis Software Paul Held Ph. D.
A p p l i c a t i o n N o t e Imaging of BacMam Transfected U-2 OS Cells Optimization of Transfection Conditions Using the Cytation 3 Multi- Mode Reader and Gen5 Data Analysis Software Paul Held Ph. D.
APPLICATION NOTE Rev. 7/2017, v4.0 Fluorescent Nanodiamonds: Bio-applications. Physical and Fluorescence Properties
 APPLICATION NOTE Rev. 7/2017, v4.0 Fluorescent Nanodiamonds: Bio-applications Fluorescent nanodiamonds (FNDs) offer a unique alternative to currently existing fluorescent biomarkers. With exceptional photo
APPLICATION NOTE Rev. 7/2017, v4.0 Fluorescent Nanodiamonds: Bio-applications Fluorescent nanodiamonds (FNDs) offer a unique alternative to currently existing fluorescent biomarkers. With exceptional photo
SURFACE ENHANCED RAMAN SCATTERING NANOPARTICLES AS AN ALTERNATIVE TO FLUORESCENT PROBES AN EVALUATION
 APPLICATION NOTE SURFACE ENHANCED RAMAN SCATTERING NANOPARTICLES AS AN ALTERNATIVE TO FLUORESCENT PROBES AN EVALUATION Summary: Interest in using nanoparticles specifically, Surface Enhanced Raman Scattering
APPLICATION NOTE SURFACE ENHANCED RAMAN SCATTERING NANOPARTICLES AS AN ALTERNATIVE TO FLUORESCENT PROBES AN EVALUATION Summary: Interest in using nanoparticles specifically, Surface Enhanced Raman Scattering
CENTRAL FACILITY BIOOPTICS MFPL
 CENTRAL FACILITY BIOOPTICS MFPL LIVE IMAGING CONSIDERATIONS Why Live Imaging? You can (somehow) believe what you see no fixation artefacts Follow the order of events in real-time Monitor dynamics: kinetics,
CENTRAL FACILITY BIOOPTICS MFPL LIVE IMAGING CONSIDERATIONS Why Live Imaging? You can (somehow) believe what you see no fixation artefacts Follow the order of events in real-time Monitor dynamics: kinetics,
HHS Public Access Author manuscript Phys Rev Lett. Author manuscript; available in PMC 2014 March 18.
 Photo-imprint Photoacoustic Microscopy for Three-dimensional Label-free Sub-diffraction Imaging Junjie Yao, Lidai Wang, Chiye Li, Chi Zhang, and Lihong V. Wang * Optical Imaging Laboratory, Department
Photo-imprint Photoacoustic Microscopy for Three-dimensional Label-free Sub-diffraction Imaging Junjie Yao, Lidai Wang, Chiye Li, Chi Zhang, and Lihong V. Wang * Optical Imaging Laboratory, Department
Lab module 6b Receptor-mediated endocytosis
 Goal for the module Lab module 6b Receptor-mediated endocytosis To follow the movement of a degraded ligand, LDL, and a recycled ligand, transferrin, as they undergo endocytic processing. Pre-lab homework
Goal for the module Lab module 6b Receptor-mediated endocytosis To follow the movement of a degraded ligand, LDL, and a recycled ligand, transferrin, as they undergo endocytic processing. Pre-lab homework
Supplementary Information
 Supplementary Information STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA Iddo Heller, Gerrit Sitters, Onno D. Broekmans, Géraldine Farge, Carolin Menges,
Supplementary Information STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA Iddo Heller, Gerrit Sitters, Onno D. Broekmans, Géraldine Farge, Carolin Menges,
Introduction to Fluorescence Jablonski Diagram
 ntroduction to Fluorescence Jablonski Diagram Excited Singlet Manifold S1 internal conversion S2 k -isc k isc Excited riplet Manifold 1 S0 k nr k k' f nr fluorescence k p phosphorescence Singlet round
ntroduction to Fluorescence Jablonski Diagram Excited Singlet Manifold S1 internal conversion S2 k -isc k isc Excited riplet Manifold 1 S0 k nr k k' f nr fluorescence k p phosphorescence Singlet round
