Lieberman-Aiden et al. (2009) Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 326:
|
|
|
- Curtis Hampton
- 6 years ago
- Views:
Transcription
1 Lieberman-Aiden et al. (2009) Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 326: : Understanding the 3D conformation of the genome can provide insight into chromatin structure, gene expression, and cellular processes. Imaging of painted chromosomes is laborious and only provides information about the spatial positions of a small number of markers at the time of imaging. Chromosome conformation capture (3C) can be used to identify pairs of loci that come into close contact, but is also locus-specific. This study introduces a new genome-wide version of 3C called Hi-C. Jay Taylor (ASU) APM Lecture 13 Fall / 19
2 Hi-C consists of six steps. Step 1: The DNA is crosslinked. Formaldehyde (CH 2 O) forms crosslinks between nucleic acids and proteins. Crosslinking mainly occurs between amino groups and nearby nitrogen atoms. Jay Taylor (ASU) APM Lecture 13 Fall / 19
3 Step 2: The crosslinked DNA is digested. Restriction endonucleases (REs) are bacterial enzymes that cut DNA at specific nucleotide sequences. This study used HindIII and NcoI which recognize the palindromic sequences 5 -A A G C T T-3 and 5 -C C A T G G-3. The human genome contains approximately 850,000 HindIII restriction sites. HindIII and NcoI both create DNA fragments with complementary 5 overhangs known as sticky ends. Jay Taylor (ASU) APM Lecture 13 Fall / 19
4 Step 3: The sticky ends are filled and marked with biotin. The reactive sticky ends are converted to less reactive blunt ends by the addition of nucleotides and DNA polymerase. The blunt ends are also marked with biotin (vitamin B7), which is incorporated as biotin-dctp. Jay Taylor (ASU) APM Lecture 13 Fall / 19
5 Step 4: The filled DNA ends are ligated. Ligases are enzymes that join dsdna strands together. T4 DNA ligase is specifically used to anneal blunt ends. By carrying out the reaction under very dilute conditions, ligation mainly occurs between the crosslinked DNA fragments. Ligation of HindIII fragments creates a new Nhel restriction site within the annealed region which is used for quality control. Jay Taylor (ASU) APM Lecture 13 Fall / 19
6 Step 5: The ligated DNA is purified and sheared into small fragments. Protein and RNA are separated from the DNA enzymatically and by centrifugation. Biotin is then removed from unligated ends using an exonuclease. The purified DNA is sheared into bp fragments using ultrasound. The biotinylated fragments are isolated by passage through a column containing streptavidin beads. Streptavidin is a bacterial enzyme with a very high affinity for biotin. Jay Taylor (ASU) APM Lecture 13 Fall / 19
7 Step 6: The purified fragments are sequenced with paired-ends. Paired-end sequencing works by sequencing each end of a short DNA fragment. The resulting reads are called paired ends and are bp long. This study used the Illumina platform with 76 bp reads. Each component of the ligated fragment was identified by aligning the corresponding end read to the reference human genome. Proximity and orientation relative to HindIII sites in the reference genome were used to confirm each alignment. Jay Taylor (ASU) APM Lecture 13 Fall / 19
8 Summary Statistics The authors created a Hi-C library using 25 million cells derived from an EBV-transformed human lymphoblastoid tissue culture. Sequencing of this library generated 8.4 million alignable read pairs. Of these, 6.7 million corresponded to long-range contacts between sites separated by > 20 kb (or on different chromosomes). Spatial proximity maps were constructed by dividing the genome into 1-Mb regions (loci) and setting m ij = number of ligation products between locus i and locus j. This overlap matrix averages over the spatial conformations of the genomes in the original sample of cells. Jay Taylor (ASU) APM Lecture 13 Fall / 19
9 Reproducibility The reproducibility of the Hi-C results was investigated by producing libraries for two separate experiments with HindIII and a third with NcoI. Since the contact matrices for all three libraries were highly-correlated (Pearson s r = and r = for C or D vs. B), the three data sets were combined. Jay Taylor (ASU) APM Lecture 13 Fall / 19
10 Chromosome Territories The contact matrix was used to estimate the contact probability I n (s) within each chromosome as a function of the genomic distance between loci. In each case, I n (s) decreases monotonically with s. However, intrachromosomal contact probabilities at all distances exceed the average contact probabilities between different chromosomes. The last observation is consistent with the existence of chromosome territories. Figure 2A Jay Taylor (ASU) APM Lecture 13 Fall / 19
11 Interchromosomal Contacts Fig. 2B is a heat map showing observed/expected numbers of interchromosomal contacts. Red indicates enrichment. Consistent with FISH studies, the map shows an excess of interactions between small, gene-rich chromosomes (16-17, 19-22). Chr. 18 is gene-poor and interacts less frequently with the other small chromosomes. FISH shows that chr. 18 localizes near the nuclear periphery. Figure 2B Jay Taylor (ASU) APM Lecture 13 Fall / 19
12 Spatial Compartmentalization of Chromosome 14 Fig. 3B plots the observed/expected contacts between loci in chr. 14 relative to the genome-wide averages (M ). Fig. 3C shows the Pearson correlation matrix C for the rows and columns of M. Red blocks are highly positively correlated. The plaid pattern suggests that chr. 14 is divided into two sets of loci such that contacts are enriched within sets but depleted between them. These sets were also identified using principal component analysis. Jay Taylor (ASU) APM Lecture 13 Fall / 19
13 Genome-Wide Spatial Compartmentalization Fig. 3D shows the correlation matrix for the intrachromosomal contact profiles of loci on chr. 14 versus chr. 20. The compartment labels A and B can be assigned to each chromosome so that sets of loci on different chromosomes carrying the same label have correlated contact profiles. This suggests that a common feature is responsible for compartmentalization of each chromosome. Jay Taylor (ASU) APM Lecture 13 Fall / 19
14 Loci belonging to the same compartment tend to be closer in space than those in different compartments. L1 and L3 are in A; L2 and L4 are in B. FISH shows that L3 is consistently closer to L1 than to L2. Likewise, L2 is closer to L4 than L3. Comparable results were found for 4 loci on chr. 22. Jay Taylor (ASU) APM Lecture 13 Fall / 19
15 The spatial compartments correspond to gene density and expression. Compartment A correlates with: gene density (ρ = 0.431); mrna expression (ρ = 0.476); DNAseI sensitivity (ρ = 0.651), which is a proxy for chromatin accessibility; H3K36 trimethylation (ρ = 0.601), which is an activating chromatin mark. In contrast, the Hi-C data shows that loci at a fixed genomic distance in compartment B interact more frequently than comparable loci in compartment A. Jay Taylor (ASU) APM Lecture 13 Fall / 19
16 Compartmentalization in Different Cell Types A separate Hi-C library was constructed for a sample of cells from a erythroleukemia cell line (K562) with an abnormal karyotype. The K562 compartments also are significantly correlated with open and closed chromatin states. Although the compartment patterns in K562 and GM06990 are similar (r = 0.732), many loci that are in the open compartment of one cell type are in the closed compartment of the other. Jay Taylor (ASU) APM Lecture 13 Fall / 19
17 The authors compare the results of the Hi-C experiment with the conformations predicted by two models of the structure of a chromosome. The equilibrium globule is a compact, highly-knotted configuration of a polymer at equilibrium in a poor solvent. The fractal globule is a non-equilibrium configuration formed by an unentangled polymer that passes through successive rounds of crumpling. Fig. 4C Jay Taylor (ASU) APM Lecture 13 Fall / 19
18 Chromatin packing is consistent with the fractal globule model. The intrachromosomal contact probability I (s) scales like s 1.08 for genomic distances between 500 kb and 7 Mb. 3D-FISH experiments indicate that physical distance scales like s 1/3 for genomic distances between 500 kb and 2 Mb. These results are consistent with simulated fractal globules but not with simulated equilibrium globules, e.g., I (s) s 1 for the former and like s 3/2 for the latter. Jay Taylor (ASU) APM Lecture 13 Fall / 19
19 Conclusions On the scale of several megabases, the Hi-C results are consistent with a fractal model of chromatin organization. More detailed analyses are needed to assess the extent to which this model describes human chromosomes (e.g., different models can give rise to the same power laws). The fractal globule model is attractive because it predicts that chromosomes are only weakly entangled or knotted, which would facilitate folding and unfolding during chromatin remodeling, gene activation, and the cell cycle. The Hi-C technique can be used to construct comprehensive interaction maps that should provide insight into gene regulation and chromosome crosstalk. Jay Taylor (ASU) APM Lecture 13 Fall / 19
WORKSHOP. Transcriptional circuitry and the regulatory conformation of the genome. Ofir Hakim Faculty of Life Sciences
 WORKSHOP Transcriptional circuitry and the regulatory conformation of the genome Ofir Hakim Faculty of Life Sciences Chromosome conformation capture (3C) Most GR Binding Sites Are Distant From Regulated
WORKSHOP Transcriptional circuitry and the regulatory conformation of the genome Ofir Hakim Faculty of Life Sciences Chromosome conformation capture (3C) Most GR Binding Sites Are Distant From Regulated
Synthetic Biology for
 Synthetic Biology for Plasmids and DNA Digestion Plasmids Plasmids are small DNA molecules that are separate from chromosomal DNA They are most commonly found as double stranded, circular DNA Typical plasmids
Synthetic Biology for Plasmids and DNA Digestion Plasmids Plasmids are small DNA molecules that are separate from chromosomal DNA They are most commonly found as double stranded, circular DNA Typical plasmids
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Computational Biology I LSM5191
 Computational Biology I LSM5191 Lecture 5 Notes: Genetic manipulation & Molecular Biology techniques Broad Overview of: Enzymatic tools in Molecular Biology Gel electrophoresis Restriction mapping DNA
Computational Biology I LSM5191 Lecture 5 Notes: Genetic manipulation & Molecular Biology techniques Broad Overview of: Enzymatic tools in Molecular Biology Gel electrophoresis Restriction mapping DNA
Recombinant DNA Technology
 Recombinant DNA Technology Common General Cloning Strategy Target DNA from donor organism extracted, cut with restriction endonuclease and ligated into a cloning vector cut with compatible restriction
Recombinant DNA Technology Common General Cloning Strategy Target DNA from donor organism extracted, cut with restriction endonuclease and ligated into a cloning vector cut with compatible restriction
T and B cell gene rearrangement October 17, Ram Savan
 T and B cell gene rearrangement October 17, 2016 Ram Savan savanram@uw.edu 441 Lecture #9 Slide 1 of 28 Three lectures on antigen receptors Part 1 (Last Friday): Structural features of the BCR and TCR
T and B cell gene rearrangement October 17, 2016 Ram Savan savanram@uw.edu 441 Lecture #9 Slide 1 of 28 Three lectures on antigen receptors Part 1 (Last Friday): Structural features of the BCR and TCR
Reading Lecture 8: Lecture 9: Lecture 8. DNA Libraries. Definition Types Construction
 Lecture 8 Reading Lecture 8: 96-110 Lecture 9: 111-120 DNA Libraries Definition Types Construction 142 DNA Libraries A DNA library is a collection of clones of genomic fragments or cdnas from a certain
Lecture 8 Reading Lecture 8: 96-110 Lecture 9: 111-120 DNA Libraries Definition Types Construction 142 DNA Libraries A DNA library is a collection of clones of genomic fragments or cdnas from a certain
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for :
 Transformation Insertion of DNA of interest Amplification Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for : DNA Sequence. Understand relatedness of genes and
Transformation Insertion of DNA of interest Amplification Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for : DNA Sequence. Understand relatedness of genes and
M Keramatipour 2. M Keramatipour 1. M Keramatipour 4. M Keramatipour 3. M Keramatipour 5. M Keramatipour
 Molecular Cloning Methods Mohammad Keramatipour MD, PhD keramatipour@tums.ac.ir Outline DNA recombinant technology DNA cloning co Cell based PCR PCR-based Some application of DNA cloning Genomic libraries
Molecular Cloning Methods Mohammad Keramatipour MD, PhD keramatipour@tums.ac.ir Outline DNA recombinant technology DNA cloning co Cell based PCR PCR-based Some application of DNA cloning Genomic libraries
Chapter 6 - Molecular Genetic Techniques
 Chapter 6 - Molecular Genetic Techniques Two objects of molecular & genetic technologies For analysis For generation Molecular genetic technologies! For analysis DNA gel electrophoresis Southern blotting
Chapter 6 - Molecular Genetic Techniques Two objects of molecular & genetic technologies For analysis For generation Molecular genetic technologies! For analysis DNA gel electrophoresis Southern blotting
HiChIP Protocol Mumbach et al. (CHANG), p. 1 Chang Lab, Stanford University. HiChIP Protocol
 HiChIP Protocol Mumbach et al. (CHANG), p. 1 HiChIP Protocol Citation: Mumbach et. al., HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nature Methods (2016). Cell Crosslinking
HiChIP Protocol Mumbach et al. (CHANG), p. 1 HiChIP Protocol Citation: Mumbach et. al., HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nature Methods (2016). Cell Crosslinking
Multiple choice questions (numbers in brackets indicate the number of correct answers)
 1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
Genetic Engineering & Recombinant DNA
 Genetic Engineering & Recombinant DNA Chapter 10 Copyright The McGraw-Hill Companies, Inc) Permission required for reproduction or display. Applications of Genetic Engineering Basic science vs. Applied
Genetic Engineering & Recombinant DNA Chapter 10 Copyright The McGraw-Hill Companies, Inc) Permission required for reproduction or display. Applications of Genetic Engineering Basic science vs. Applied
_ DNA absorbs light at 260 wave length and it s a UV range so we cant see DNA, we can see DNA only by staining it.
 * GEL ELECTROPHORESIS : its a technique aim to separate DNA in agel based on size, in this technique we add a sample of DNA in a wells in the gel, then we turn on the electricity, the DNA will travel in
* GEL ELECTROPHORESIS : its a technique aim to separate DNA in agel based on size, in this technique we add a sample of DNA in a wells in the gel, then we turn on the electricity, the DNA will travel in
REGULATION OF PROTEIN SYNTHESIS. II. Eukaryotes
 REGULATION OF PROTEIN SYNTHESIS II. Eukaryotes Complexities of eukaryotic gene expression! Several steps needed for synthesis of mrna! Separation in space of transcription and translation! Compartmentation
REGULATION OF PROTEIN SYNTHESIS II. Eukaryotes Complexities of eukaryotic gene expression! Several steps needed for synthesis of mrna! Separation in space of transcription and translation! Compartmentation
Introduction to genome biology
 Introduction to genome biology Lisa Stubbs We ve found most genes; but what about the rest of the genome? Genome size* 12 Mb 95 Mb 170 Mb 1500 Mb 2700 Mb 3200 Mb #coding genes ~7000 ~20000 ~14000 ~26000
Introduction to genome biology Lisa Stubbs We ve found most genes; but what about the rest of the genome? Genome size* 12 Mb 95 Mb 170 Mb 1500 Mb 2700 Mb 3200 Mb #coding genes ~7000 ~20000 ~14000 ~26000
Structure meets function: Chromosome folding in mammals
 Transcription Regulation And Gene Expression in Eukaryotes FS 2016 Graduate Course G2 P Matthias and RG Clerc Pharmazentrum Hörsaal 2 16h15-18h00 Structure meets function: Chromosome folding in mammals
Transcription Regulation And Gene Expression in Eukaryotes FS 2016 Graduate Course G2 P Matthias and RG Clerc Pharmazentrum Hörsaal 2 16h15-18h00 Structure meets function: Chromosome folding in mammals
The Properties of Genome Conformation and Spatial Gene Interaction and Regulation Networks of Normal and Malignant Human Cell Types
 The Properties of Genome Conformation and Spatial Gene Interaction and Regulation Networks of Normal and Malignant Human Cell Types Zheng Wang 1, Renzhi Cao 1, Kristen Taylor 2, Aaron Briley 2 b, Charles
The Properties of Genome Conformation and Spatial Gene Interaction and Regulation Networks of Normal and Malignant Human Cell Types Zheng Wang 1, Renzhi Cao 1, Kristen Taylor 2, Aaron Briley 2 b, Charles
Friday, June 12, 15. Biotechnology Tools
 Biotechnology Tools Biotechnology: Tools and Techniques Science of biotechnology is based on recombining DNA of different organisms of another organism. Gene from one organism spliced into genome of another
Biotechnology Tools Biotechnology: Tools and Techniques Science of biotechnology is based on recombining DNA of different organisms of another organism. Gene from one organism spliced into genome of another
Chapter 10 (Part I) Gene Isolation and Manipulation
 Biology 234 J. G. Doheny Chapter 10 (Part I) Gene Isolation and Manipulation Practice Questions: Answer the following questions with one or two sentences. 1. From which types of organisms were most restriction
Biology 234 J. G. Doheny Chapter 10 (Part I) Gene Isolation and Manipulation Practice Questions: Answer the following questions with one or two sentences. 1. From which types of organisms were most restriction
Manipulation of Purified DNA
 Manipulation of Purified DNA To produce the recombinant DNA molecule, the vector, as well as the DNA to be cloned, must be cut at specific points and then joined together in a controlled manner by DNA
Manipulation of Purified DNA To produce the recombinant DNA molecule, the vector, as well as the DNA to be cloned, must be cut at specific points and then joined together in a controlled manner by DNA
DNA Technology. Asilomar Singer, Zinder, Brenner, Berg
 DNA Technology Asilomar 1973. Singer, Zinder, Brenner, Berg DNA Technology The following are some of the most important molecular methods we will be using in this course. They will be used, among other
DNA Technology Asilomar 1973. Singer, Zinder, Brenner, Berg DNA Technology The following are some of the most important molecular methods we will be using in this course. They will be used, among other
Basics of Recombinant DNA Technology Biochemistry 302. March 5, 2004 Bob Kelm
 Basics of Recombinant DNA Technology Biochemistry 302 March 5, 2004 Bob Kelm Applications of recombinant DNA technology Mapping and identifying genes (DNA cloning) Propagating genes (DNA subcloning) Modifying
Basics of Recombinant DNA Technology Biochemistry 302 March 5, 2004 Bob Kelm Applications of recombinant DNA technology Mapping and identifying genes (DNA cloning) Propagating genes (DNA subcloning) Modifying
2054, Chap. 14, page 1
 2054, Chap. 14, page 1 I. Recombinant DNA technology (Chapter 14) A. recombinant DNA technology = collection of methods used to perform genetic engineering 1. genetic engineering = deliberate modification
2054, Chap. 14, page 1 I. Recombinant DNA technology (Chapter 14) A. recombinant DNA technology = collection of methods used to perform genetic engineering 1. genetic engineering = deliberate modification
DNA Structure and Analysis. Chapter 4: Background
 DNA Structure and Analysis Chapter 4: Background Molecular Biology Three main disciplines of biotechnology Biochemistry Genetics Molecular Biology # Biotechnology: A Laboratory Skills Course explorer.bio-rad.com
DNA Structure and Analysis Chapter 4: Background Molecular Biology Three main disciplines of biotechnology Biochemistry Genetics Molecular Biology # Biotechnology: A Laboratory Skills Course explorer.bio-rad.com
Genome editing. Knock-ins
 Genome editing Knock-ins Experiment design? Should we even do it? In mouse or rat, the HR-mediated knock-in of homologous fragments derived from a donor vector functions well. However, HR-dependent knock-in
Genome editing Knock-ins Experiment design? Should we even do it? In mouse or rat, the HR-mediated knock-in of homologous fragments derived from a donor vector functions well. However, HR-dependent knock-in
Molecular Genetics Techniques. BIT 220 Chapter 20
 Molecular Genetics Techniques BIT 220 Chapter 20 What is Cloning? Recombinant DNA technologies 1. Producing Recombinant DNA molecule Incorporate gene of interest into plasmid (cloning vector) 2. Recombinant
Molecular Genetics Techniques BIT 220 Chapter 20 What is Cloning? Recombinant DNA technologies 1. Producing Recombinant DNA molecule Incorporate gene of interest into plasmid (cloning vector) 2. Recombinant
Genetics and Genomics in Medicine Chapter 3. Questions & Answers
 Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
Manipulating DNA. Nucleic acids are chemically different from other macromolecules such as proteins and carbohydrates.
 Lesson Overview 14.3 Studying the Human Genome Nucleic acids are chemically different from other macromolecules such as proteins and carbohydrates. Nucleic acids are chemically different from other macromolecules
Lesson Overview 14.3 Studying the Human Genome Nucleic acids are chemically different from other macromolecules such as proteins and carbohydrates. Nucleic acids are chemically different from other macromolecules
Overview of Human Genetics
 Overview of Human Genetics 1 Structure and function of nucleic acids. 2 Structure and composition of the human genome. 3 Mendelian genetics. Lander et al. (Nature, 2001) MAT 394 (ASU) Human Genetics Spring
Overview of Human Genetics 1 Structure and function of nucleic acids. 2 Structure and composition of the human genome. 3 Mendelian genetics. Lander et al. (Nature, 2001) MAT 394 (ASU) Human Genetics Spring
Nucleic acids. How DNA works. DNA RNA Protein. DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology
 Nucleic acid chemistry and basic molecular theory Nucleic acids DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology Cell cycle DNA RNA Protein Transcription Translation
Nucleic acid chemistry and basic molecular theory Nucleic acids DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology Cell cycle DNA RNA Protein Transcription Translation
Appendix A DNA and PCR in detail DNA: A Detailed Look
 Appendix A DNA and PCR in detail DNA: A Detailed Look A DNA molecule is a long polymer consisting of four different components called nucleotides. It is the various combinations of these four bases or
Appendix A DNA and PCR in detail DNA: A Detailed Look A DNA molecule is a long polymer consisting of four different components called nucleotides. It is the various combinations of these four bases or
Supplementary Figure 2
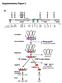 Supplementary Figure 2 a SBS-C1 SBS-C2 SBS-C3 SBS-C4 SBS-C5 SBS-C6 SBS-C7 SBS-C8 SBS-C9 LCR CNS-1 CNS-2 Il5 Rad50 Il13 Il4 Kif3a Sept8 0 50 100 150 200kb Sau3AI 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17
Supplementary Figure 2 a SBS-C1 SBS-C2 SBS-C3 SBS-C4 SBS-C5 SBS-C6 SBS-C7 SBS-C8 SBS-C9 LCR CNS-1 CNS-2 Il5 Rad50 Il13 Il4 Kif3a Sept8 0 50 100 150 200kb Sau3AI 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17
DNA Structure & the Genome. Bio160 General Biology
 DNA Structure & the Genome Bio160 General Biology Lecture Outline I. DNA A nucleic acid II. Chromosome Structure III. Chromosomes and Genes IV. DNA vs. RNA I. DNA A Nucleic Acid Structure of DNA: Remember:
DNA Structure & the Genome Bio160 General Biology Lecture Outline I. DNA A nucleic acid II. Chromosome Structure III. Chromosomes and Genes IV. DNA vs. RNA I. DNA A Nucleic Acid Structure of DNA: Remember:
3 Designing Primers for Site-Directed Mutagenesis
 3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
Impact of gdna Integrity on the Outcome of DNA Methylation Studies
 Impact of gdna Integrity on the Outcome of DNA Methylation Studies Application Note Nucleic Acid Analysis Authors Emily Putnam, Keith Booher, and Xueguang Sun Zymo Research Corporation, Irvine, CA, USA
Impact of gdna Integrity on the Outcome of DNA Methylation Studies Application Note Nucleic Acid Analysis Authors Emily Putnam, Keith Booher, and Xueguang Sun Zymo Research Corporation, Irvine, CA, USA
How Do You Clone a Gene?
 S-20 Edvo-Kit #S-20 How Do You Clone a Gene? Experiment Objective: The objective of this experiment is to gain an understanding of the structure of DNA, a genetically engineered clone, and how genes are
S-20 Edvo-Kit #S-20 How Do You Clone a Gene? Experiment Objective: The objective of this experiment is to gain an understanding of the structure of DNA, a genetically engineered clone, and how genes are
Lectures 9 and 10: Random Walks and the Structure of Macromolecules (contd.)
 Lectures 9 and 10: Random Walks and the Structure of Macromolecules (contd.) Lecturer: Brigita Urbanc Office: 12 909 (E mail: brigita@drexel.edu) Course website: www.physics.drexel.edu/~brigita/courses/biophys_2011
Lectures 9 and 10: Random Walks and the Structure of Macromolecules (contd.) Lecturer: Brigita Urbanc Office: 12 909 (E mail: brigita@drexel.edu) Course website: www.physics.drexel.edu/~brigita/courses/biophys_2011
Ren Lab ENCODE in situ HiC Protocol for Tissue
 Ren Lab ENCODE in situ HiC Protocol for Tissue Pulverization, Crosslinking of Tissue Note: Ensure the samples are kept frozen on dry ice throughout pulverization. 1. Pour liquid nitrogen into a mortar
Ren Lab ENCODE in situ HiC Protocol for Tissue Pulverization, Crosslinking of Tissue Note: Ensure the samples are kept frozen on dry ice throughout pulverization. 1. Pour liquid nitrogen into a mortar
Transcription. Unit: DNA. Central Dogma. 2. Transcription converts DNA into RNA. What is a gene? What is transcription? 1/7/2016
 Warm Up Questions 1. Where is DNA located? 2. Name the 3 parts of a nucleotide. 3. Enzymes can catalyze many different reactions (T or F) 4. How many variables should you have in an experiment? 5. A red
Warm Up Questions 1. Where is DNA located? 2. Name the 3 parts of a nucleotide. 3. Enzymes can catalyze many different reactions (T or F) 4. How many variables should you have in an experiment? 5. A red
Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C
 Ay et al. BMC Genomics (2015) 16:121 DOI 10.1186/s12864-015-1236-7 RESEARCH ARTICLE Open Access Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C Ferhat Ay 1, Thanh H
Ay et al. BMC Genomics (2015) 16:121 DOI 10.1186/s12864-015-1236-7 RESEARCH ARTICLE Open Access Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C Ferhat Ay 1, Thanh H
Thebiotutor.com A2 Biology OCR Unit F215: Control, genomes and environment Module 2.3 Genomes and gene technologies Answers
 Thebiotutor.com A2 Biology OCR Unit F215: Control, genomes and environment Module 2.3 Genomes and gene technologies Answers Andy Todd 1 1. (i) plasmid cut by restriction enzyme; at specific sequence; same
Thebiotutor.com A2 Biology OCR Unit F215: Control, genomes and environment Module 2.3 Genomes and gene technologies Answers Andy Todd 1 1. (i) plasmid cut by restriction enzyme; at specific sequence; same
Recombinant DNA Technology
 History of recombinant DNA technology Recombinant DNA Technology (DNA cloning) Majid Mojarrad Recombinant DNA technology is one of the recent advances in biotechnology, which was developed by two scientists
History of recombinant DNA technology Recombinant DNA Technology (DNA cloning) Majid Mojarrad Recombinant DNA technology is one of the recent advances in biotechnology, which was developed by two scientists
Molecular Genetics II - Genetic Engineering Course (Supplementary notes)
 1 von 12 21.02.2015 15:13 Molecular Genetics II - Genetic Engineering Course (Supplementary notes) Figures showing examples of cdna synthesis (currently 11 figures) cdna is a DNA copy synthesized from
1 von 12 21.02.2015 15:13 Molecular Genetics II - Genetic Engineering Course (Supplementary notes) Figures showing examples of cdna synthesis (currently 11 figures) cdna is a DNA copy synthesized from
NUCLEIC ACIDS Genetic material of all known organisms DNA: deoxyribonucleic acid RNA: ribonucleic acid (e.g., some viruses)
 NUCLEIC ACIDS Genetic material of all known organisms DNA: deoxyribonucleic acid RNA: ribonucleic acid (e.g., some viruses) Consist of chemically linked sequences of nucleotides Nitrogenous base Pentose-
NUCLEIC ACIDS Genetic material of all known organisms DNA: deoxyribonucleic acid RNA: ribonucleic acid (e.g., some viruses) Consist of chemically linked sequences of nucleotides Nitrogenous base Pentose-
Recombinant DNA Technology. The Role of Recombinant DNA Technology in Biotechnology. yeast. Biotechnology. Recombinant DNA technology.
 PowerPoint Lecture Presentations prepared by Mindy Miller-Kittrell, North Carolina State University C H A P T E R 8 Recombinant DNA Technology The Role of Recombinant DNA Technology in Biotechnology Biotechnology?
PowerPoint Lecture Presentations prepared by Mindy Miller-Kittrell, North Carolina State University C H A P T E R 8 Recombinant DNA Technology The Role of Recombinant DNA Technology in Biotechnology Biotechnology?
Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter
 Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter 9/16/2008 1 Learning Objectives 1. List and explain how DNA
Plant Molecular and Cellular Biology Lecture 9: Nuclear Genome Organization: Chromosome Structure, Chromatin, DNA Packaging, Mitosis Gary Peter 9/16/2008 1 Learning Objectives 1. List and explain how DNA
CHAPTER 9 DNA Technologies
 CHAPTER 9 DNA Technologies Recombinant DNA Artificially created DNA that combines sequences that do not occur together in the nature Basis of much of the modern molecular biology Molecular cloning of genes
CHAPTER 9 DNA Technologies Recombinant DNA Artificially created DNA that combines sequences that do not occur together in the nature Basis of much of the modern molecular biology Molecular cloning of genes
Supplementary Figure 1. HiChIP provides confident 1D factor binding information.
 Supplementary Figure 1 HiChIP provides confident 1D factor binding information. a, Reads supporting contacts called using the Mango pipeline 19 for GM12878 Smc1a HiChIP and GM12878 CTCF Advanced ChIA-PET
Supplementary Figure 1 HiChIP provides confident 1D factor binding information. a, Reads supporting contacts called using the Mango pipeline 19 for GM12878 Smc1a HiChIP and GM12878 CTCF Advanced ChIA-PET
Control of Eukaryotic Genes. AP Biology
 Control of Eukaryotic Genes The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different, specialized functions? Evolution
Control of Eukaryotic Genes The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different, specialized functions? Evolution
Three-dimensional organization of genomes: interpreting chromatin interaction data
 3C to Three-dimensional organization of genomes: interpreting chromatin interaction data 3C to Institut für Medizinische tik und Humangenetik Charité Universitätsmedizin Berlin Genomics: Lecture #15 Outline
3C to Three-dimensional organization of genomes: interpreting chromatin interaction data 3C to Institut für Medizinische tik und Humangenetik Charité Universitätsmedizin Berlin Genomics: Lecture #15 Outline
Figure S1. Unrearranged locus. Rearranged locus. Concordant read pairs. Region1. Region2. Cluster of discordant read pairs, bundle
 Figure S1 a Unrearranged locus Rearranged locus Concordant read pairs Region1 Concordant read pairs Cluster of discordant read pairs, bundle Region2 Concordant read pairs b Physical coverage 5 4 3 2 1
Figure S1 a Unrearranged locus Rearranged locus Concordant read pairs Region1 Concordant read pairs Cluster of discordant read pairs, bundle Region2 Concordant read pairs b Physical coverage 5 4 3 2 1
Mate-pair library data improves genome assembly
 De Novo Sequencing on the Ion Torrent PGM APPLICATION NOTE Mate-pair library data improves genome assembly Highly accurate PGM data allows for de Novo Sequencing and Assembly For a draft assembly, generate
De Novo Sequencing on the Ion Torrent PGM APPLICATION NOTE Mate-pair library data improves genome assembly Highly accurate PGM data allows for de Novo Sequencing and Assembly For a draft assembly, generate
Lecture 25 (11/15/17)
 Lecture 25 (11/15/17) Reading: Ch9; 328-332 Ch25; 990-995, 1005-1012 Problems: Ch9 (study-guide: applying); 1,2 Ch9 (study-guide: facts); 7,8 Ch25 (text); 1-3,5-7,9,10,13-15 Ch25 (study-guide: applying);
Lecture 25 (11/15/17) Reading: Ch9; 328-332 Ch25; 990-995, 1005-1012 Problems: Ch9 (study-guide: applying); 1,2 Ch9 (study-guide: facts); 7,8 Ch25 (text); 1-3,5-7,9,10,13-15 Ch25 (study-guide: applying);
Polymerase chain reaction
 Core course BMS361N Genetic Engineering Polymerase chain reaction Prof. Narkunaraja Shanmugam Dept. Of Biomedical Science School of Basic Medical Sciences Bharathidasan University The polymerase chain
Core course BMS361N Genetic Engineering Polymerase chain reaction Prof. Narkunaraja Shanmugam Dept. Of Biomedical Science School of Basic Medical Sciences Bharathidasan University The polymerase chain
Bio 101 Sample questions: Chapter 10
 Bio 101 Sample questions: Chapter 10 1. Which of the following is NOT needed for DNA replication? A. nucleotides B. ribosomes C. Enzymes (like polymerases) D. DNA E. all of the above are needed 2 The information
Bio 101 Sample questions: Chapter 10 1. Which of the following is NOT needed for DNA replication? A. nucleotides B. ribosomes C. Enzymes (like polymerases) D. DNA E. all of the above are needed 2 The information
Regulation of enzyme synthesis
 Regulation of enzyme synthesis The lac operon is an example of an inducible operon - it is normally off, but when a molecule called an inducer is present, the operon turns on. The trp operon is an example
Regulation of enzyme synthesis The lac operon is an example of an inducible operon - it is normally off, but when a molecule called an inducer is present, the operon turns on. The trp operon is an example
Chapter 5 DNA and Chromosomes
 Chapter 5 DNA and Chromosomes DNA as the genetic material Heat-killed bacteria can transform living cells S Smooth R Rough Fred Griffith, 1920 DNA is the genetic material Oswald Avery Colin MacLeod Maclyn
Chapter 5 DNA and Chromosomes DNA as the genetic material Heat-killed bacteria can transform living cells S Smooth R Rough Fred Griffith, 1920 DNA is the genetic material Oswald Avery Colin MacLeod Maclyn
Genome Sequence Assembly
 Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
Group Members: Lab Station: BIOTECHNOLOGY: Gel Electrophoresis
 BIOTECHNOLOGY: Gel Electrophoresis Group Members: Lab Station: Restriction Enzyme Analysis Standard: AP Big Idea #3, SB2 How can we use genetic information to identify and profile individuals? Lab Specific
BIOTECHNOLOGY: Gel Electrophoresis Group Members: Lab Station: Restriction Enzyme Analysis Standard: AP Big Idea #3, SB2 How can we use genetic information to identify and profile individuals? Lab Specific
Data and Metadata Models Recommendations Version 1.2 Developed by the IHEC Metadata Standards Workgroup
 Data and Metadata Models Recommendations Version 1.2 Developed by the IHEC Metadata Standards Workgroup 1. Introduction The data produced by IHEC is illustrated in Figure 1. Figure 1. The space of epigenomic
Data and Metadata Models Recommendations Version 1.2 Developed by the IHEC Metadata Standards Workgroup 1. Introduction The data produced by IHEC is illustrated in Figure 1. Figure 1. The space of epigenomic
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Plasmids. BIL 333 Lecture I. Plasmids. Useful Plasmids. Useful Plasmids. Useful Plasmids. ( Transfection ) v Small, circular, double-stranded DNA
 BIL 333 Lecture I Plasmids v Small, circular, double-stranded DNA v Exogenous to genome! v Origin of Replication v Marker Gene v ( Reporter Gene ) Plasmids v Marker Gene Changes Phenotype Of Host v (Antibiotic
BIL 333 Lecture I Plasmids v Small, circular, double-stranded DNA v Exogenous to genome! v Origin of Replication v Marker Gene v ( Reporter Gene ) Plasmids v Marker Gene Changes Phenotype Of Host v (Antibiotic
Applications of Cas9 nickases for genome engineering
 application note genome editing Applications of Cas9 nickases for genome engineering Shuqi Yan, Mollie Schubert, Maureen Young, Brian Wang Integrated DNA Technologies, 17 Commercial Park, Coralville, IA,
application note genome editing Applications of Cas9 nickases for genome engineering Shuqi Yan, Mollie Schubert, Maureen Young, Brian Wang Integrated DNA Technologies, 17 Commercial Park, Coralville, IA,
Learning Objectives :
 Learning Objectives : Understand the basic differences between genomic and cdna libraries Understand how genomic libraries are constructed Understand the purpose for having overlapping DNA fragments in
Learning Objectives : Understand the basic differences between genomic and cdna libraries Understand how genomic libraries are constructed Understand the purpose for having overlapping DNA fragments in
Problem Set 8. Answer Key
 MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
Chapter 13: Biotechnology
 Chapter Review 1. Explain why the brewing of beer is considered to be biotechnology. The United Nations defines biotechnology as any technological application that uses biological system, living organism,
Chapter Review 1. Explain why the brewing of beer is considered to be biotechnology. The United Nations defines biotechnology as any technological application that uses biological system, living organism,
Functional Genomics II
 Functional Genomics II Jay Hesselberth PhD University of Colorado SOM Biochemistry & Molecular Genetics jay.hesselberth@gmail.com Outline Tues, May. Definition. Yeast genome 3. Gene-centric resources and
Functional Genomics II Jay Hesselberth PhD University of Colorado SOM Biochemistry & Molecular Genetics jay.hesselberth@gmail.com Outline Tues, May. Definition. Yeast genome 3. Gene-centric resources and
DNA AND CHROMOSOMES. Genetica per Scienze Naturali a.a prof S. Presciuttini
 DNA AND CHROMOSOMES This document is licensed under the Attribution-NonCommercial-ShareAlike 2.5 Italy license, available at http://creativecommons.org/licenses/by-nc-sa/2.5/it/ 1. The Building Blocks
DNA AND CHROMOSOMES This document is licensed under the Attribution-NonCommercial-ShareAlike 2.5 Italy license, available at http://creativecommons.org/licenses/by-nc-sa/2.5/it/ 1. The Building Blocks
DNA REPLICATION & REPAIR
 DNA REPLICATION & REPAIR Table of contents 1. DNA Replication Model 2. DNA Replication Mechanism 3. DNA Repair: Proofreading 1. DNA Replication Model Replication in the cell cycle 3 models of DNA replication
DNA REPLICATION & REPAIR Table of contents 1. DNA Replication Model 2. DNA Replication Mechanism 3. DNA Repair: Proofreading 1. DNA Replication Model Replication in the cell cycle 3 models of DNA replication
Biotechnology. Chapter 20. Biology Eighth Edition Neil Campbell and Jane Reece. PowerPoint Lecture Presentations for
 Chapter 20 Biotechnology PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Copyright
Chapter 20 Biotechnology PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Copyright
GENERATION OF HIC LIBRARY
 GENERATION OF HIC LIBRARY Gel checking points during one HiC experiment ------ % gel Step Checking list 1 0.8 After cells are lysed Pellet degradation check 2 0.8 After template generation 1 template quality
GENERATION OF HIC LIBRARY Gel checking points during one HiC experiment ------ % gel Step Checking list 1 0.8 After cells are lysed Pellet degradation check 2 0.8 After template generation 1 template quality
THE CELLULAR AND MOLECULAR BASIS OF INHERITANCE
 Umm AL Qura University THE CELLULAR AND MOLECULAR BASIS OF INHERITANCE Dr. Neda Bogari www.bogari.net EMERY'S ELEMENTS OF MEDICAL GENETICS Peter Turnpenny and Sian Ellard 13 th edition 2008 COURSE SYLLABUS
Umm AL Qura University THE CELLULAR AND MOLECULAR BASIS OF INHERITANCE Dr. Neda Bogari www.bogari.net EMERY'S ELEMENTS OF MEDICAL GENETICS Peter Turnpenny and Sian Ellard 13 th edition 2008 COURSE SYLLABUS
Subtractive hybridization is a powerful technique particularly well-suited for the identification of target cdnas that correspond to rare
 Subtractive hybridization is a powerful technique particularly well-suited for the identification of target cdnas that correspond to rare transcripts, that are expressed in one population but not in the
Subtractive hybridization is a powerful technique particularly well-suited for the identification of target cdnas that correspond to rare transcripts, that are expressed in one population but not in the
DNA vs. RNA DNA: deoxyribonucleic acid (double stranded) RNA: ribonucleic acid (single stranded) Both found in most bacterial and eukaryotic cells RNA
 DNA Replication DNA vs. RNA DNA: deoxyribonucleic acid (double stranded) RNA: ribonucleic acid (single stranded) Both found in most bacterial and eukaryotic cells RNA molecule can assume different structures
DNA Replication DNA vs. RNA DNA: deoxyribonucleic acid (double stranded) RNA: ribonucleic acid (single stranded) Both found in most bacterial and eukaryotic cells RNA molecule can assume different structures
Methods of Biomaterials Testing Lesson 3-5. Biochemical Methods - Molecular Biology -
 Methods of Biomaterials Testing Lesson 3-5 Biochemical Methods - Molecular Biology - Chromosomes in the Cell Nucleus DNA in the Chromosome Deoxyribonucleic Acid (DNA) DNA has double-helix structure The
Methods of Biomaterials Testing Lesson 3-5 Biochemical Methods - Molecular Biology - Chromosomes in the Cell Nucleus DNA in the Chromosome Deoxyribonucleic Acid (DNA) DNA has double-helix structure The
Chapter 9: DNA: The Molecule of Heredity
 Chapter 9: DNA: The Molecule of Heredity What is DNA? Answer: Molecule that carries the blueprint of life General Features: DNA is packages in chromosomes (DNA + Proteins) Gene = Functional segment of
Chapter 9: DNA: The Molecule of Heredity What is DNA? Answer: Molecule that carries the blueprint of life General Features: DNA is packages in chromosomes (DNA + Proteins) Gene = Functional segment of
Human Fibroblast IMR90 Hi-C Data (Dixon et al.)
 Human Fibroblast IMR90 Hi-C Data (Dixon et al.) Nicolas Servant October 31, 2017 1 Introduction The Hi-C technic was first introduced by Lieberman-Aiden et al. [2009]. In the continuity with 3C, 4C and
Human Fibroblast IMR90 Hi-C Data (Dixon et al.) Nicolas Servant October 31, 2017 1 Introduction The Hi-C technic was first introduced by Lieberman-Aiden et al. [2009]. In the continuity with 3C, 4C and
Nucleic acids deoxyribonucleic acid (DNA) ribonucleic acid (RNA) nucleotide
 Nucleic Acids Nucleic acids are molecules that store information for cellular growth and reproduction There are two types of nucleic acids: - deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) These
Nucleic Acids Nucleic acids are molecules that store information for cellular growth and reproduction There are two types of nucleic acids: - deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) These
Chromatin. Structure and modification of chromatin. Chromatin domains
 Chromatin Structure and modification of chromatin Chromatin domains 2 DNA consensus 5 3 3 DNA DNA 4 RNA 5 ss RNA forms secondary structures with ds hairpins ds forms 6 of nucleic acids Form coiling bp/turn
Chromatin Structure and modification of chromatin Chromatin domains 2 DNA consensus 5 3 3 DNA DNA 4 RNA 5 ss RNA forms secondary structures with ds hairpins ds forms 6 of nucleic acids Form coiling bp/turn
Lecture 3 (FW) January 28, 2009 Cloning of DNA; PCR amplification Reading assignment: Cloning, ; ; 330 PCR, ; 329.
 Lecture 3 (FW) January 28, 2009 Cloning of DNA; PCR amplification Reading assignment: Cloning, 240-245; 286-87; 330 PCR, 270-274; 329. Take Home Lesson(s) from Lecture 2: 1. DNA is a double helix of complementary
Lecture 3 (FW) January 28, 2009 Cloning of DNA; PCR amplification Reading assignment: Cloning, 240-245; 286-87; 330 PCR, 270-274; 329. Take Home Lesson(s) from Lecture 2: 1. DNA is a double helix of complementary
Biology 201 (Genetics) Exam #3 120 points 20 November Read the question carefully before answering. Think before you write.
 Name KEY Section Biology 201 (Genetics) Exam #3 120 points 20 November 2006 Read the question carefully before answering. Think before you write. You will have up to 50 minutes to take this exam. After
Name KEY Section Biology 201 (Genetics) Exam #3 120 points 20 November 2006 Read the question carefully before answering. Think before you write. You will have up to 50 minutes to take this exam. After
Cloning and Genetic Engineering
 Cloning and Genetic Engineering Bởi: OpenStaxCollege Biotechnology is the use of artificial methods to modify the genetic material of living organisms or cells to produce novel compounds or to perform
Cloning and Genetic Engineering Bởi: OpenStaxCollege Biotechnology is the use of artificial methods to modify the genetic material of living organisms or cells to produce novel compounds or to perform
DNA Replication. The Organization of DNA. Recall:
 Recall: The Organization of DNA DNA Replication Chromosomal form appears only during mitosis, and is used in karyotypes. folded back upon itself (chromosomes) coiled around itself (chromatin) wrapped around
Recall: The Organization of DNA DNA Replication Chromosomal form appears only during mitosis, and is used in karyotypes. folded back upon itself (chromosomes) coiled around itself (chromatin) wrapped around
The Genetic Code and Transcription. Chapter 12 Honors Genetics Ms. Susan Chabot
 The Genetic Code and Transcription Chapter 12 Honors Genetics Ms. Susan Chabot TRANSCRIPTION Copy SAME language DNA to RNA Nucleic Acid to Nucleic Acid TRANSLATION Copy DIFFERENT language RNA to Amino
The Genetic Code and Transcription Chapter 12 Honors Genetics Ms. Susan Chabot TRANSCRIPTION Copy SAME language DNA to RNA Nucleic Acid to Nucleic Acid TRANSLATION Copy DIFFERENT language RNA to Amino
12 2 Chromosomes and DNA Replication
 DNA Replication 12-2 Chromosomes and 1 of 21 DNA and Chromosomes DNA and Chromosomes In prokaryotic cells, DNA is located in the cytoplasm. Most prokaryotes have a single DNA molecule containing nearly
DNA Replication 12-2 Chromosomes and 1 of 21 DNA and Chromosomes DNA and Chromosomes In prokaryotic cells, DNA is located in the cytoplasm. Most prokaryotes have a single DNA molecule containing nearly
Bioinformatics of Transcriptional Regulation
 Bioinformatics of Transcriptional Regulation Carl Herrmann IPMB & DKFZ c.herrmann@dkfz.de Wechselwirkung von Maßnahmen und Auswirkungen Einflussmöglichkeiten in einem Dialog From genes to active compounds
Bioinformatics of Transcriptional Regulation Carl Herrmann IPMB & DKFZ c.herrmann@dkfz.de Wechselwirkung von Maßnahmen und Auswirkungen Einflussmöglichkeiten in einem Dialog From genes to active compounds
Control of Eukaryotic Genes
 Control of Eukaryotic Genes 2007-2008 The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different, specialized functions?
Control of Eukaryotic Genes 2007-2008 The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different, specialized functions?
AP Biology Day 34. Monday, November 14, 2016
 AP Biology Day 34 Monday, November 14, 2016 Essen%al knowledge standards 3.A.1: DNA, and in some cases RNA, is the primary source of heritable informa%on 3.A.1.e: Gene%c engineering techniques can manipulate
AP Biology Day 34 Monday, November 14, 2016 Essen%al knowledge standards 3.A.1: DNA, and in some cases RNA, is the primary source of heritable informa%on 3.A.1.e: Gene%c engineering techniques can manipulate
Big Idea 3C Basic Review
 Big Idea 3C Basic Review 1. A gene is a. A sequence of DNA that codes for a protein. b. A sequence of amino acids that codes for a protein. c. A sequence of codons that code for nucleic acids. d. The end
Big Idea 3C Basic Review 1. A gene is a. A sequence of DNA that codes for a protein. b. A sequence of amino acids that codes for a protein. c. A sequence of codons that code for nucleic acids. d. The end
Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Supplementary Material
 Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions Joshua N. Burton 1, Andrew Adey 1, Rupali P. Patwardhan 1, Ruolan Qiu 1, Jacob O. Kitzman 1, Jay Shendure 1 1 Department
Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions Joshua N. Burton 1, Andrew Adey 1, Rupali P. Patwardhan 1, Ruolan Qiu 1, Jacob O. Kitzman 1, Jay Shendure 1 1 Department
Molecular Biology (1)
 Molecular Biology (1) DNA structure and basic applications Mamoun Ahram, PhD Second semester, 2017-2018 Resources This lecture Cooper, pp. 49-52, 118-119, 130 What is molecular biology? Central dogma
Molecular Biology (1) DNA structure and basic applications Mamoun Ahram, PhD Second semester, 2017-2018 Resources This lecture Cooper, pp. 49-52, 118-119, 130 What is molecular biology? Central dogma
NOTES Gene Expression ACP Biology, NNHS
 Name Date Block NOTES Gene Expression ACP Biology, NNHS Model 1: Transcription the process of genes in DNA being copied into a messenger RNA 1. Where in the cell is DNA found? 2. Where in the cell does
Name Date Block NOTES Gene Expression ACP Biology, NNHS Model 1: Transcription the process of genes in DNA being copied into a messenger RNA 1. Where in the cell is DNA found? 2. Where in the cell does
Chapter 12 Packet DNA 1. What did Griffith conclude from his experiment? 2. Describe the process of transformation.
 Chapter 12 Packet DNA and RNA Name Period California State Standards covered by this chapter: Cell Biology 1. The fundamental life processes of plants and animals depend on a variety of chemical reactions
Chapter 12 Packet DNA and RNA Name Period California State Standards covered by this chapter: Cell Biology 1. The fundamental life processes of plants and animals depend on a variety of chemical reactions
P HENIX. PHENIX PCR Enzyme Guide Tools For Life Science Discovery RESEARCH PRODUCTS
 PHENIX PCR Enzyme Guide PHENIX offers a broad line of premium quality PCR Enzymes. This PCR Enzyme Guide will help simplify your polymerase selection process. Each DNA Polymerase has different characteristics
PHENIX PCR Enzyme Guide PHENIX offers a broad line of premium quality PCR Enzymes. This PCR Enzyme Guide will help simplify your polymerase selection process. Each DNA Polymerase has different characteristics
Chapter 12. DNA TRANSCRIPTION and TRANSLATION
 Chapter 12 DNA TRANSCRIPTION and TRANSLATION 12-3 RNA and Protein Synthesis WARM UP What are proteins? Where do they come from? From DNA to RNA to Protein DNA in our cells carry the instructions for making
Chapter 12 DNA TRANSCRIPTION and TRANSLATION 12-3 RNA and Protein Synthesis WARM UP What are proteins? Where do they come from? From DNA to RNA to Protein DNA in our cells carry the instructions for making
In other words there are 4.0 x10 5 phosphodiester groups in the basic form to one in the acidic form at ph 7.0.
 In other words there are 4.0 x10 5 phosphodiester groups in the basic form to one in the acidic form at ph 7.0. There are a number of shorthand abbreviations a linear polymer of deoxyribonucleotides. One
In other words there are 4.0 x10 5 phosphodiester groups in the basic form to one in the acidic form at ph 7.0. There are a number of shorthand abbreviations a linear polymer of deoxyribonucleotides. One
4/26/2015. Cut DNA either: Cut DNA either:
 Ch.20 Enzymes that cut DNA at specific sequences (restriction sites) resulting in segments of DNA (restriction fragments) Typically 4-8 bp in length & often palindromic Isolated from bacteria (Hundreds
Ch.20 Enzymes that cut DNA at specific sequences (restriction sites) resulting in segments of DNA (restriction fragments) Typically 4-8 bp in length & often palindromic Isolated from bacteria (Hundreds
