Supplementary Methods
|
|
|
- Verity Hodges
- 5 years ago
- Views:
Transcription
1 Supplementary Methods Microarray Data Analysis Gene expression data were obtained by hybridising a total of 24 samples from 6 experimental groups (n=4 per group) to Illumina HumanHT-12 Expression BeadChips. Raw data were exported from the Illumina GenomeStudio software (v1..6) for further processing and analysis using R statistical software 1 (v2.1) and BioConductor packages. Raw signal intensities were background corrected using array-specific measures of background intensity based on negative control probes, prior to being transformed and normalised using the vsn package 2. Quality control analyses did not reveal any outlier samples. The dataset was then filtered to remove probes not detected (detection score <.95) in any of the samples, resulting in a final dataset of 25,62 probes. Statistical analysis was performed using the Linear Models for Microarray Analysis (limma) package 3. Differential expression between the experimental groups was assessed by generating relevant contrasts corresponding to the relevant comparisons. Raw p-values were corrected for multiple testing using the false discovery rate controlling procedure of Benjamini and Hochberg 4, adjusted p-values below.1 were considered significant. Significant probe lists were then annotated using the relevant annotation file (HumanHT-12_V3 R2_ _A) that was downloaded from the Illumina website ( for further biological investigation. Bioinformatics and statistical analyses The nucleotide sequence were inspected with transcription factor binding site searching software JASPAR ( 5 and Genomatrix ( for the presence of putative ISRE sites (Supplementary Table S1). Statistical analysis was performed using One-way ANOVA with Dunnett s multiple comparison post test or Student s T-test where appropriate (p<.5, p<.1, p<.1). References 1. Team, R.D.C. in R Foundation for Statistical Computing, Vol. Vienna Austria, 2121). 2. Huber, W., von Heydebreck, A., Sultmann, H., Poustka, A. & Vingron, M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 Suppl 1, S96-14 (22). 3. Smyth, G.K. et al. Limma: linear models for microarray data, in Bioinformatics and Computational Biology Solutions using R and Bioconductor (Springer, New York, 25). 4. Benjamini, Y. & Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, (1995). 5. Vlieghe, D. et al. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res 34, D95-97 (26). Nature Immunology: doi:1.138/ni.199
2 Supplementary Figure legends Figure S1: expression is induced by M1 macrophage maturation protocols (a) M1 and M2 macrophages from the same donor were stimulated with LPS (1ng/ml) for 24h and the secretion of IL-12p7, IL-23 and IL-1 was determined by ELISA. Data shown are the mean ± SEM from 4 independent experiments each using macrophages derived from a different donor: p<.5, p<.1 (One-way ANOVA). (b) protein expression was analysed in total cell lysates of monocytes, M1 and M2 macrophages by Western blotting. Densitometric analysis was performed using Quantity One software and data were normalised to actin. Shown are the mean ± SEM from 3 independent experiments presented as % of increase in protein levels relative to monocytes. p<.5 (One-way ANOVA with Dunnett's Multiple Comparison Post Test). (c) M2 macrophages were left untreated or treated with (5ng/ml), IFN- (5ng/ml), or LPS (1ng/ml) plus IFN- for 24h and total protein extracts were subjected to Western blot analysis followed by densitometry. Data shown are the mean ± SEM from 6 independent experiments presented as % of increase in (protein levels relative to untreated cells. p<.1 (One-way ANOVA with Dunnett's Multiple Comparison Post Test). (d) p5 protein expression was analysed in total cell lysates of monocytes, M1 and M2 macrophages by Western blotting. Actin was used as a loading control. Representative blots of at least 4 independent experiments, each using cells derived from a different donor are shown. (e) M2 macrophages were left untreated or treated with (5ng/ml), IFN- (5ng/ml), or LPS (1ng/ml) plus IFN- for 24h and total protein extracts were subjected to Western blot analysis followed by densitometry. Data shown are the mean ± SEM from 6 independent experiments presented as % of increase in p5 protein levels relative to untreated cells. Figure S2: Plasticity of macrophage polarization (a, c) For M2->M1 cytokine profiles, -derived M2 macrophages at day 5 were either left in containing medium or exchanged for (1ng/ml) containing medium and after 24h subjected to LPS stimulation (1ng/ml). (b, d) For M1->M2 cytokine profiles, derived M1 macrophages at day 5 were either left in containing medium or exchanged for (1ng/ml) containing medium and after 24h subjected to LPS stimulation (1ng/ml). (a, b) The change in secretion of IL-12p7, IL-23 and IL-1 was determined by ELISA. Nature Immunology: doi:1.138/ni.199
3 (c, d) The change in protein expression was analysed by Western blotting followed by densitometric analysis using Quantity One software. The measurements were normalised to actin. Shown are the mean ± SEM from 4 independent experiments presented as % of increase (c) or decrease (d) in protein levels relative to the initial condition: p<.5 (One-way ANOVA with Dunnett's Multiple Comparison Post Test). (e) For M2->M1->M2 cytokine profiles, M2 macrophages at day 5 were either left in M- CSF containing medium, or exchanged for IFN- (5ng/ml) containing medium, or further reversed to containing medium (1ng/ml) and after 48h subjected to LPS stimulation (1ng/ml). The amount of secreted IL-12p7, IL-23 and IL-1 protein following 24h of LPS stimulation was determined by ELISA. Data shown are the mean ± SEM of 3 independent experiments each using macrophages derived from a different donor. Figure S3: defines the production of lineage specific cytokines in human macrophages (a) M2 macrophages were infected as in (Fig. 2 A) and left unstimulated or stimulated with LPS (1ng/ml) for 4, 8, 24, 32 and 48h. The amount of secreted IL-12p7 and IL- 23 protein was determined by ELISA. Data shown are the mean ± SD and are representative of 3 independent experiments each using macrophages derived from a different donor. (b) M2 macrophages were infected with adenoviral vectors encoding, or empty vector () and stimulated with LPS for 24h. The amount of secreted IL-1 and TNF protein was determined by ELISA. Data show the trend of cytokine secretion in 4-8 independent experiments each using M2 macrophages derived from a different donor: p<.1, p<.1 (One-way ANOVA with Dunnett's Multiple Comparison Post Test). (c) M1 macrophages were transfected with sirna targeting (si) or control sirna (sic). ~5% of protein was degraded estimated by serial dilutions of the sic control sample analysed by Western blotting. Figure S4: induces T cell proliferation and expression of T cell subset specific markers (a) M2 macrophages were infected with adenoviral vectors encoding or empty vector () and cultured with T lymphocytes from unmatched donors. After 4 days, cells were stimulated for 3h with PMA/ionomycine/Brefeldin A. The percentage of Nature Immunology: doi:1.138/ni.199
4 CD4+/IL-17+ or CD4+/IFN + cells was determined by ICC staining and representative FACS plots are shown. (b) M2 macrophages were infected with adenoviral vectors encoding, or empty vector () and cultured in triplicate for 72h with T lymphocytes from unmatched donors. Cultures were pulsed with thymidine for the last 16h to measure DNA synthesis. Control cultures contained macrophages or T-cells alone. Results are expressed as counts per minute (CPM) minus proliferation of macrophage-only cultures. Data are shown as the mean ± SEM of 6 independent experiments each using cells derived from a different donor: p<.1 (One-way ANOVA with Dunnett's Multiple Comparison Post Test). (c, e) M2 macrophages were infected with adenoviral vectors encoding, or empty vector () and cultured with T lymphocytes from unmatched donors. After 4 days, cells were stimulated for 3h with PMA/ionomycine/Brefeldin A and IFN- and IL-17 expression were determined by ICC staining. Data are shown as the percentage of IFN- +/IL-17- (c) or IFN- -/ IL-17+ (e) cells ± SEM of 8 independent experiments. (d, f) M2 macrophages were infected with adenoviral vectors encoding, or empty vector () and cultured with T lymphocytes from unmatched donors. IFN- (d) or IL-17A, IL-17F, IL-21, IL-22, IL-26, IL-23R (f) mrna expression was analysed after 2 days of co-culture. Data are shown as the mean ± SEM of 6-9 independent experiments each using cells derived from a different donor: p<.5, p<.1, p<.1 (One-way ANOVA with Dunnett's Multiple Comparison Post Test). Figure S5: drives expression of IL12p4 mrna and production of selected M1 and M2 cytokines (a) M2 macrophages were infected with adenoviral vectors encoding or empty vector () and left unstimulated or stimulated with LPS (1ng/ml) for 4, 8, 16 and 24h. IL12p4 mrna expression was compared to unstimulated control cells. Data shown are the mean ± SD and are representative of 3 independent experiments each using macrophages derived from a different donor. (b) M1 macrophages were transfected with sirna targeting (si) or control sirna (sic) and left unstimulated or stimulated with LPS (1ng/ml) for 2, 4, 8, 16 and 24h. IL-12p4 mrna expression was compared to control cells transfected with nontargeting sirna (sic). Data shown are the mean ± SD of representative experiments presented as a % of reduction in IL-12p4 mrna levels by si. Nature Immunology: doi:1.138/ni.199
5 (c, d, e) M2 macrophages were infected with adenoviral vectors encoding or empty vector () and stimulated with LPS for 24h. The amount of secreted CCL5 (c); CCL2, CCL13 (d) or CCL22, CXCL1 (e) protein was determined by ELISA. The amount of CD4 (c) or CD163 (c) surface expression was determined by FACS and expressed as MFI. Data are shown as the mean ± SEM of 4-6 independent experiments each using M2 macrophages derived from a different donor: p<.1, p<.5 (Student's t-test). Figure S6: activates transcription of the human IL-12p35 gene HEK-293-TLR4/MD2 cells were co-transfected with IL-12p35 wild type (IL-12p35-Luc wt) reporter plasmid or the IL-12p35 plasmid in which site-specific mutation was introduced into the ISRE site as described in Ref 37 and constructs coding for (black bars), DNA-binding mutant ( DBD) (grey bars) or empty vector () (white bars). Luciferase activity was measured 24h post-infection. Data are presented as the mean ± SD from a representative out of 3 independent experiments. Figure S7: Impaired production of M1 cytokines in Irf5-/- mice Littermate wild type (n = 1) and Irf5 -/- (n = 1) mice were intraperitoneally injected with LPS (2ug/ml). Mice were sacrificed after 3h and serum concentrations of Il-1, Il-6 and Tnf were measured by BD TM cytrometric bead assay. Data are shown as the mean ± SEM of 8-1 serum samples from 3 independent experiments: p<.1, p<.5 (Student's t-test). Nature Immunology: doi:1.138/ni.199
6 a IL-12p7 IL-23 IL-1 IL-12p7, pg/ml IL-23, pg/ml IL-1, ng/ml 5 n=4 n=4 n=5 b protein level, normalised (%) Mono n=3 c protein level, normalised (%) untreated IFN-γ LPS+IFN-γ n=6 d LPS: Mono p5 actin M2 M1 e p5 protein level, normalised (%) untreated IFN-γ LPS+IFN-γ n=3 Nature Immunology: doi:1.138/ni.199 Figure S1
7 M2->M1 a IL-12p7, pg/ml IL-12p7 IL-23, pg/ml IL-23 IL-1, pg/ml IL-1 c protein level, normalized (%) M2 M1 n=4 M1->M2 b IL-12p7, pg/ml IL-12p IL-23, pg/ml IL-23 IL-1, pg/ml IL-1 d protein level, normalized (%) M1 M2 n=4 e M2->M1->M2 4 IL-12p7 IL-12p7, pg/ml Nature Immunology: doi:1.138/ni.199 IFN-γ Figure S2
8 a IL-12p7 1 IL-23 IL-12p7, ng/ml 1 5 IL-23, pg/ml time after LPS, hours time after LPS, hours b 25 2 IL-1β 4 TNF IL-1β, pg/ml TNF, ng/ml n=8 n=4 c 1% sic 5% 25% si actin Nature Immunology: doi:1.138/ni.199 Figure S3
9 a IL-17 IFN-γ CD4 b T cell proliferation c CD4+/IFN-γ+ d CPM % positive cells IFN-g mrna (AU) IFN-γ mrna n=6 n=8 n=9 Nature Immunology: doi:1.138/ni.199 Figure S4
10 % positive cells e CD4+/IL n=8 f IL-17 mrna (AU) IL-17A mrna IL-17F mrna (AU) IL-17F mrna n=8 n=5 IL-21 mrna (AU) IL-21 mrna IL-22 mrna (AU) IL-22 mrna IL-26 mrna (AU) IL-26 mrna IL-23R mrna (AU) IL-23R mrna n=6 n=6 n=6 n=6 Nature Immunology: doi:1.138/ni.199 Figure S4
11 a IL-12p4 mrna (AU) IL-12p4 mrna time after LPS, hours b IL-12p4 mrna, % inhibition IL-12p4 mrna time after LPS, hours sic si c M1 d M2 CCL5, pg/ml CCL5 CD4, MFI CD4 CCL2 CCL13 CD CCL2, ng/ml 4 2 CCL13, pg/ml 5 CD163, MFIx1 1 n=5 n=5 n=4 n=6 n=5 e CCL22 CXCL CCL22, ng/ml CXCL1, ng/ml 1 5 n=4 Nature Immunology: doi:1.138/ni.199 n=6 Figure S5
12 3 IL-12p35-luc luciferase activity (AU) 2 1 wt ISREmut wt ISREmut wt ISREmut DDBD Nature Immunology: doi:1.138/ni.199 Figure S6
13 i 125 IL-1β 8 TNF 5 IL-6 IL-1β, pg/ml TNF, pg/ml ILl -6, ng/ml wt n=8 wt n=1 wt Irf5-/- Irf5-/- Irf5-/- n=1 Nature Immunology: doi:1.138/ni.199 Figure S7
14 Supplementary Table S1: Genes up-regulated by ectopic Gene symbol Entrez Gene ID Fold change CXCR CXCR CXCR CXCR EBI TNFSF TNFSF LTA LTB IFN CCL CCL CXCL IL IL Genes down-regulated by ectopic Gene symbol Entrez Gene ID Fold change CSF1R IL-1R IL-1RA TGF Nature Immunology: doi:1.138/ni.199
15 Supplementary Table S2: putative binding sites in -2/+2 nt relative to the TSSs of selected genes Table S2.1 genes up-regulated by Gene Number of IRF sites Sequence Strand Core sim Matrix sim TNFSF4 9 aatgtactttacatttcccac cacaaactttctcttttaagt tgcctcatttccattttttct agatctttttctttttctttg agaccagttccactttcccat taaaatatttccatttttctt atttttctttcactttattct attattttttcttattcagta cacctccaatgaaaccagaat EBi3 8 ctctgtgtttctctttctgtt gtttctctttctgtttccatc gtatctctgtcactttctctg ctgtcactttctctgtcatct cctttggtttctttttggttt tttttggttttgttttttgag ccaggaattcgagaccagcct gcaacatagtgaaaccggacc TNFSF7 14 ctgcctcattcagtttctgtt cattcagtttctgtttctgtt gtttctgtttctgttttcaca aggggaataggaagattgaat catggaaatggaagatgactc ccaggaaaacgattcgggaaa aaaataaaatgaaataaaatc gagggaaacggagagggggag agaagaaggggaaagaaagaa cggagaaagagaaaaaagaca aagaagaaaggaaaagaaaaa aaaagaaaaagaaagaaagga aaaggaaaaagaaagaagaaa agaagaaagagaaaaaaagaa TNFSF9 6 tcaacactgtccctttcttgc gagacaaagagagactaaaga cagagataacggagccagaga agggagaaaggaacctggagc gccggaaacggaaaggagagc gtacccctctccctttcaaga CCL1 6 ttcatgatttcaatgcctaga aacaaaaagggaaaattcccc aatagaaatggcaaatatcta gtgtgaatatgaatttgggta actctactttctctatcagtg actggaaagagcaagggaacc Nature Immunology: doi:1.138/ni.199
16 CCL3 12 gctttcatttctttttctact caaagaaatgggaaatcaaga ccattgaacagaaacttcagc ttcagaaaaagaaaaaaataa ctcatgctttctattcctcca cccccagattccatttcccca gcccccaagagaaaagagaac cttggtctttctctttaagac cagagaaacagagaacccact agaggaaagggacaggaagaa aatttattttcgatttcacag agtttggttttgttttcctgg IL-2Ra 5 ggagggttttctttttgttaa aattgaacttgaaaaaaaaaa caatgaatttccttttattct tgcaaattttaaatttcattc ccaagaacaggaaaatcttga CCR7 9 tcaagaaagtgaaaagatgat aaaaaaaaaagaaaaaagaaa tcaacaatttcacttctaggt agctaaaagggaaaacagccc ttcagaataggaaaatctata aaaggaaaaggaagggagggg accccagactaggtttagggg gggagggtttctgttacacaa tacacaaaatgaaaactccca CXCR5 8 ttggtgatttcactttttttt ttttttttttcttttagagac aagttgatttcatttttgtct tgaggaaaatgaaggtttgga gtggtggtttcattacaagtt gaaagaagctgaaatgcttga caaaaaaacagaaaagaccca aatgcaaaatgaaaacatggg IL23a 6 accaggaagtgaaacaaagag gggtagatttccatttttttt gtgatgaaatcggtgtcagtg ccatgaaaccaggaccatcca ctgagaaaaagaagcccgttt ttgggaaagagaaatcgatgg TNF 1 agccaagactgaaaccagcat gggtcagaatgaaagaagaag agaagaaaccgagacagaagg caggcaggttctcttcctctc ccctggaaaggacaccatgag catgagcactgaaagcatgat ttctgggtttgggtttggggg gggggaaatttaaagttttgg Nature Immunology: doi:1.138/ni.199
17 IL12p35 9 gctctcatttctttttctttc atgtaaattagaaactgtgtc gcgaacatttcgctttcattt atttcgctttcattttgggcc atccgaaagcgccgcaagccc gaaggagacagaaagcaagag tcgtagaggagaaactgaggc cacctggtctgggtttccctg tgtctccagagaaagcaagag IL12p4 8 gttacagttttttttttttaa cccgggtttcccatttccccc gagggtatttcactttctgct aagtcagtttctagtttaagt tttctagtttaagtttccatc tgtacagtgtccattttaaaa gttaaaaaatgaaaagctatt actggaatctgaaattgtatg Table S2.2 genes down-regulated by Gene name Number of IRF sites Sequence Strand Core sim Matrix sim CD163 1 ggatgaaactggaaaccatca ttgctaatttttgtttcacca gtatgaaatggaacctcagct gtagccttttcattttcatga tcatgaaagtgaagtgatttt gatgttgtttccattttccag gccctcttttctttttcacag caaaggaggagaaacttcaga agataagtttcagtctagcgt ctagtcttttcatcttcataa IL-1 7 cttgttatttcaacttcttcc acaactaaaagaaactctaag acgcgaatgagaacccacagc tgcaaaaattgaaaactaagt cagggaaatttaaattgcctc cttctgctttcccttcaaaat ttgctcatttctctttgagca MS4A6A 12 aaagacaagagaaaggagaat agccaaaatggaaaaaaaaag cgctgagaactaatccagcct tgactggctctggtttccttg tggggaattagaaaagcaaga ttagaaaactgaagcttcaag aatttaacttgaaactccttg gaaggagtatctgtttttaac ccgtgaaaagggatccaagct tccatactatcagtttctttc Nature Immunology: doi:1.138/ni.199
18 ctatcagtttctttctctaat gactgagttactgtttttgga CXCL1 9 caactaaaataaaactgtcac tttgcctttccggtttcccac ctttttttttctttttctttg caacctgtttcccttctgtct atgatgttttcattcagggac tataagacgtgaaacttgttt tttggaaagtgaaacctaatt catgcagagtgaaacttaaat ttaggaaacggcaatcttggg CLEC4a 5 gacttggtgtgggtttcagaa gaaagacaatgaaagcaggtt gagagaaatccactccagttc tagagtacaagaaactatggg actatagttacgctttctaaa IL-1R2 isoform1 13 ggcatggttttgcttcctctc ccaaatatttcaccttctaat gtaagaaaatgaagatctgca ctctgaaaacaaaacaaaaca gaaaaatagggaaacttatgc cagagaaacagagacagaaag cagagaaacagacagagatag gacagagacagagaccaagac gctctcgggtggttttctggg ctcagggtctccatttccacc ctctctgtctctgtttctctc ctctatgtctctgtttctccc taattgcattcccttttgggg IL-1R2 isoform2 1 ttcactcttccagtttctcac tttgctctctccctttcctgg gaacaaaatttaaactgttct acgatggcttcacttacatgg ttataagacagaaagcaaaat tttagaaactgaagctgtatc tgaacactttctttttgcagc gggagaatttgaagcctgtgg ttgaatgagcgaaaacatgag ccatctgtatcagtttctgcc IL-1RA isoform1 9 ggaagaaatccaatctatttc aatctagtttctgattcttta agaggaaattgaaggccctta attctgatttcattatatata ctctaattttaagtttctaat taaataaaatgaaataaaata agaggaaatggatatagagag actcggactggaaactggaag Nature Immunology: doi:1.138/ni.199
19 actggaaactggaagggtgag IL-1RA isoform4 9 tacaaaaaatgaaaatgaact cacacagtttgaattcctggg tgggaaaactgaatctcaaaa ctcagaaaaggaagctggttt ggaggaaaatgcaaattgaaa aatgcaaattgaaaagttgct ccttgcttttccctttgaatg aagaggaataggaactgcacc cctcttccttcagtttcagct Nature Immunology: doi:1.138/ni.199
TRIM31 is recruited to mitochondria after infection with SeV.
 Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern
 Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern blot. Northern blot analysis of mir-302b expression following infection with PAO1, PAK and Kp in (A) lung
Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern blot. Northern blot analysis of mir-302b expression following infection with PAO1, PAK and Kp in (A) lung
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators.
 Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Nature Medicine doi: /nm.3554
 SUPPLEMENTARY FIGURES LEGENDS Supplementary Figure 1: Generation, purification and characterization of recombinant mouse IL-35 (ril-35). High-Five insect cells expressing high levels of the bicistronic
SUPPLEMENTARY FIGURES LEGENDS Supplementary Figure 1: Generation, purification and characterization of recombinant mouse IL-35 (ril-35). High-Five insect cells expressing high levels of the bicistronic
Supplementary Figure 1
 Supplementary Figure 1 Virus infection induces RNF128 expression. (a,b) RT-PCR analysis of Rnf128 (RNF128) mrna expression in mouse peritoneal macrophages (a) and THP-1 cells (b) upon stimulation with
Supplementary Figure 1 Virus infection induces RNF128 expression. (a,b) RT-PCR analysis of Rnf128 (RNF128) mrna expression in mouse peritoneal macrophages (a) and THP-1 cells (b) upon stimulation with
Supplementary Methods Plasmid constructs
 Supplementary Methods Plasmid constructs. Mouse cdna encoding SHP-1, amplified from mrna of RAW264.7 macrophages with primer 5'cgtgcctgcccagacaaactgt3' and 5'cggaattcagacgaatgcccagatcacttcc3', was cloned
Supplementary Methods Plasmid constructs. Mouse cdna encoding SHP-1, amplified from mrna of RAW264.7 macrophages with primer 5'cgtgcctgcccagacaaactgt3' and 5'cggaattcagacgaatgcccagatcacttcc3', was cloned
Mouse expression data were normalized using the robust multiarray algorithm (1) using
 Supplementary Information Bioinformatics statistical analysis of microarray data Mouse expression data were normalized using the robust multiarray algorithm (1) using a custom probe set definition that
Supplementary Information Bioinformatics statistical analysis of microarray data Mouse expression data were normalized using the robust multiarray algorithm (1) using a custom probe set definition that
Supplemental Figure 1
 Supplemental Figure 1 A IL-12p7 (pg/ml) 7 6 4 3 2 1 Medium then TLR ligands MDP then TLR ligands Medium then TLR ligands + MDP MDP then TLR ligands + MDP B IL-12p4 (ng/ml) 1.2 1..8.6.4.2. Medium MDP Medium
Supplemental Figure 1 A IL-12p7 (pg/ml) 7 6 4 3 2 1 Medium then TLR ligands MDP then TLR ligands Medium then TLR ligands + MDP MDP then TLR ligands + MDP B IL-12p4 (ng/ml) 1.2 1..8.6.4.2. Medium MDP Medium
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
SUPPLEMENTARY INFORMATION
 doi:.8/nature85 Supplementary Methods Plasmid construction Murine RIG-I, HMGB and Rab5 cdnas were obtained by polymerase chain reaction with reverse transcription (RT-PCR) on total RNA from, and then cloned
doi:.8/nature85 Supplementary Methods Plasmid construction Murine RIG-I, HMGB and Rab5 cdnas were obtained by polymerase chain reaction with reverse transcription (RT-PCR) on total RNA from, and then cloned
Supplementary Figure 1. Reconstitution of human-acquired lymphoid system in
 Supplementary Figure 1. Reconstitution of human-acquired lymphoid system in mouse NOD/SCID/Jak3 null mice were transplanted with human CD34 + hematopoietic stem cells. (Top) Four weeks after the transplantation
Supplementary Figure 1. Reconstitution of human-acquired lymphoid system in mouse NOD/SCID/Jak3 null mice were transplanted with human CD34 + hematopoietic stem cells. (Top) Four weeks after the transplantation
Figure S1: GM-CSF upregulates CCL17 expression in in vitro-derived and ex vivo murine macrophage populations
 Figure S1 A B C Figure S1: GM-CSF upregulates CCL17 expression in in vitro-derived and ex vivo murine macrophage populations (A-B) Murine bone cells were cultured in either M-CSF (5,000 U/ml) or GM-CSF
Figure S1 A B C Figure S1: GM-CSF upregulates CCL17 expression in in vitro-derived and ex vivo murine macrophage populations (A-B) Murine bone cells were cultured in either M-CSF (5,000 U/ml) or GM-CSF
Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative
 Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative RT-PCR For CD8-1 transcript, forward primer CD8-1F 5 -TAGTAACCAGAGGCCGCAAGA-3 reverse primer CD8-1R 5 -TCTACTAAGGTGTCCCATAGCATGAT-3
Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative RT-PCR For CD8-1 transcript, forward primer CD8-1F 5 -TAGTAACCAGAGGCCGCAAGA-3 reverse primer CD8-1R 5 -TCTACTAAGGTGTCCCATAGCATGAT-3
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1. Characterization of the POP2 transcriptional and post-transcriptional regulatory elements. (A) POP2 nucleotide sequence
 1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
Online Supplementary Information
 Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Supplementary Information
 Supplementary Information Supplementary Figure 1: Synthetic annealed poly(ra):poly(dt) hybrid induces type I interferon response. a) Bone marrow derived macrophages (BMDM) were transfected with or without
Supplementary Information Supplementary Figure 1: Synthetic annealed poly(ra):poly(dt) hybrid induces type I interferon response. a) Bone marrow derived macrophages (BMDM) were transfected with or without
Standard Data Analysis Report Agilent Gene Expression Service
 Standard Data Analysis Report Agilent Gene Expression Service Experiment: S534662 Date: 2011-01-01 Prepared for: Dr. Researcher Genomic Sciences Lab Prepared by S534662 Standard Data Analysis Report 2011-01-01
Standard Data Analysis Report Agilent Gene Expression Service Experiment: S534662 Date: 2011-01-01 Prepared for: Dr. Researcher Genomic Sciences Lab Prepared by S534662 Standard Data Analysis Report 2011-01-01
Supplemental Information. The TRAIL-Induced Cancer Secretome. Promotes a Tumor-Supportive Immune. Microenvironment via CCR2
 Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
DC enriched CD103. CD11b. CD11c. Spleen. DC enriched. CD11c. DC enriched. CD11c MLN
 CD CD11 + CD + CD11 d g CD CD11 + CD + CD11 CD + CD11 + Events (% of max) Events (% of max) CD + CD11 Events (% of max) Spleen CLN MLN Irf fl/fl e Irf fl/fl h Irf fl/fl DC enriched CD CD11 Spleen DC enriched
CD CD11 + CD + CD11 d g CD CD11 + CD + CD11 CD + CD11 + Events (% of max) Events (% of max) CD + CD11 Events (% of max) Spleen CLN MLN Irf fl/fl e Irf fl/fl h Irf fl/fl DC enriched CD CD11 Spleen DC enriched
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification.
 Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Supplementary Data Supplementary Figure 1. Knockdown of VentX with a different sirna sequence reduces terminal monocyte to macrophage
 Supplementary Data Supplementary Figure 1. Knockdown of VentX with a different sirna sequence reduces terminal monocyte to macrophage differentiation. Monocytes were transfected with either a scrambled
Supplementary Data Supplementary Figure 1. Knockdown of VentX with a different sirna sequence reduces terminal monocyte to macrophage differentiation. Monocytes were transfected with either a scrambled
Li et al., Supplemental Figures
 Li et al., Supplemental Figures Fig. S1. Suppressing TGM2 expression with TGM2 sirnas inhibits migration and invasion in A549-TR cells. A, A549-TR cells transfected with negative control sirna (NC sirna)
Li et al., Supplemental Figures Fig. S1. Suppressing TGM2 expression with TGM2 sirnas inhibits migration and invasion in A549-TR cells. A, A549-TR cells transfected with negative control sirna (NC sirna)
Human skin punch biopsies were obtained under informed consent from normal healthy
 SUPPLEMENTAL METHODS Acquisition of human skin specimens. Human skin punch biopsies were obtained under informed consent from normal healthy volunteers (n = 30) and psoriasis patients (n = 45) under a
SUPPLEMENTAL METHODS Acquisition of human skin specimens. Human skin punch biopsies were obtained under informed consent from normal healthy volunteers (n = 30) and psoriasis patients (n = 45) under a
Supplemental Table S1. RT-PCR primers used in this study
 Supplemental Table S1. RT-PCR primers used in this study -----------------------------------------------------------------------------------------------------------------------------------------------
Supplemental Table S1. RT-PCR primers used in this study -----------------------------------------------------------------------------------------------------------------------------------------------
Nature Immunology: doi: /ni Supplementary Figure 1. Zranb1 gene targeting.
 Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1. GST pull-down analysis of the interaction of GST-cIAP1 (A, B), GSTcIAP1
 Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Supplementary Material & Methods
 Supplementary Material & Methods Affymetrix micro-array Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, USA). RNA concentration and quality was determined using the ND-1000 spectrophotometer
Supplementary Material & Methods Affymetrix micro-array Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, USA). RNA concentration and quality was determined using the ND-1000 spectrophotometer
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
Supplementary Fig S1: Bone marrow macrophages and liver Kupffer cells increase IL-1 in response to adenosine signaling. (a) Murine bone marrow
 Supplementary Fig S1: Bone marrow macrophages and liver Kupffer cells increase IL-1 in response to adenosine signaling. (a) Murine bone marrow derived macrophages (BMDM) were primed with LPS for 16 hrs,
Supplementary Fig S1: Bone marrow macrophages and liver Kupffer cells increase IL-1 in response to adenosine signaling. (a) Murine bone marrow derived macrophages (BMDM) were primed with LPS for 16 hrs,
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr
 Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Figure S1- Targeting strategy for generating SIK1-T182A, SIK2-T175A and SIK3-T163A KI mice.
 Figure S1- Targeting strategy for generating SIK1-T12A, SIK2-T175A and SIK3-T163A mice. Left panel represents a depiction of the genomic loci of each gene, the targeting strategy and the final recombined
Figure S1- Targeting strategy for generating SIK1-T12A, SIK2-T175A and SIK3-T163A mice. Left panel represents a depiction of the genomic loci of each gene, the targeting strategy and the final recombined
Nature Biotechnology: doi: /nbt.4086
 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3
Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3
Supplemental Information. Inflammatory Ly6C high Monocytes Protect. against Candidiasis through IL-15-Driven. NK Cell/Neutrophil Activation
 Immunity, Volume 46 Supplemental Information Inflammatory Ly6C high Monocytes Protect against Candidiasis through IL-15-Driven NK Cell/Neutrophil Activation Jorge Domínguez-Andrés, Lidia Feo-Lucas, María
Immunity, Volume 46 Supplemental Information Inflammatory Ly6C high Monocytes Protect against Candidiasis through IL-15-Driven NK Cell/Neutrophil Activation Jorge Domínguez-Andrés, Lidia Feo-Lucas, María
No wash 2 Washes 2 Days ** ** IgG-bead phagocytosis (%)
 Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Cell were phenotyped using FITC-conjugated anti-human CD3 (Pharmingen, UK)
 SUPPLEMENTAL MATERIAL Supplemental Methods Flow cytometry Cell were phenotyped using FITC-conjugated anti-human CD3 (Pharmingen, UK) and anti-human CD68 (Dako, Denmark), anti-human smooth muscle cell α-actin
SUPPLEMENTAL MATERIAL Supplemental Methods Flow cytometry Cell were phenotyped using FITC-conjugated anti-human CD3 (Pharmingen, UK) and anti-human CD68 (Dako, Denmark), anti-human smooth muscle cell α-actin
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated
 Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Flowcytometry-based purity analysis of peritoneal macrophage culture.
 Liao et al., KLF4 regulates macrophage polarization Revision of Manuscript 45444-RG- Supplementary Figure Legends Figure S Flowcytometry-based purity analysis of peritoneal macrophage culture. Thioglycollate
Liao et al., KLF4 regulates macrophage polarization Revision of Manuscript 45444-RG- Supplementary Figure Legends Figure S Flowcytometry-based purity analysis of peritoneal macrophage culture. Thioglycollate
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas.
 Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Supplementary Information
 Supplementary Information Sam68 modulates the promoter specificity of NF-κB and mediates expression of CD25 in activated T cells Kai Fu 1, 6, Xin Sun 1, 6, Wenxin Zheng 1, 6, Eric M. Wier 1, Andrea Hodgson
Supplementary Information Sam68 modulates the promoter specificity of NF-κB and mediates expression of CD25 in activated T cells Kai Fu 1, 6, Xin Sun 1, 6, Wenxin Zheng 1, 6, Eric M. Wier 1, Andrea Hodgson
Nature Immunology: doi: /ni Supplementary Figure 1. Control experiments for Figure 1.
 Supplementary Figure 1 Control experiments for Figure 1. (a) Localization of SETX in untreated or PR8ΔNS1 virus infected A549 cells (4hours). Nuclear (DAPI) and Tubulin staining are shown. SETX antibody
Supplementary Figure 1 Control experiments for Figure 1. (a) Localization of SETX in untreated or PR8ΔNS1 virus infected A549 cells (4hours). Nuclear (DAPI) and Tubulin staining are shown. SETX antibody
Albumin. MMP-9 (tertiary granules) Lactoferrin. MPO (U/ml) ctrl PMN-sec
 Figure S1: Antibody cross-linking of CD18 induces release of primary, secondary, and tertiary granules as well as secretory vesicles. Freshly isolated PMN were incubated with primary anti-cd18 mab IB4
Figure S1: Antibody cross-linking of CD18 induces release of primary, secondary, and tertiary granules as well as secretory vesicles. Freshly isolated PMN were incubated with primary anti-cd18 mab IB4
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Supplemental Table 1 Primers used in study. Human. Mouse
 Supplemental Table 1 Primers used in study Human Forward primer region(5-3 ) Reverse primer region(5-3 ) RT-PCR GAPDH gagtcaacggatttggtcgt ttgattttggagggatctcg Raftlin atgggttgcggattgaacaagttaga ctgaggtataacaccaacgaatttcaggc
Supplemental Table 1 Primers used in study Human Forward primer region(5-3 ) Reverse primer region(5-3 ) RT-PCR GAPDH gagtcaacggatttggtcgt ttgattttggagggatctcg Raftlin atgggttgcggattgaacaagttaga ctgaggtataacaccaacgaatttcaggc
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Figure 1
 Supplementary Figure 1 PTEN promotes virus-induced expression of IFNB1 and its downstream genes. (a) Quantitative RT-PCR analysis of IFNB1 mrna (left) and ELISA of IFN-β (right) in HEK 293 cells (2 10
Supplementary Figure 1 PTEN promotes virus-induced expression of IFNB1 and its downstream genes. (a) Quantitative RT-PCR analysis of IFNB1 mrna (left) and ELISA of IFN-β (right) in HEK 293 cells (2 10
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex
 POSTER PRESENTATION Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex Hai-Chon Lee *, Je-In Youn, Kyungwha Lee, Hwanyul Yong, Seung-Yong
POSTER PRESENTATION Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex Hai-Chon Lee *, Je-In Youn, Kyungwha Lee, Hwanyul Yong, Seung-Yong
Anti-ERK1/2, anti-p-erk1/2, anti-p38 anti-p-msk1 (Thr581) and anti-p-msk2
 Supplementary methods Antibodies Anti-ERK1/2, anti-p-erk1/2, anti-p38 anti-p-msk1 (Thr581) and anti-p-msk2 polyclonal were from Cell Signalling and the anti-p-histone H3(Ser1) antibody was from Upstate.
Supplementary methods Antibodies Anti-ERK1/2, anti-p-erk1/2, anti-p38 anti-p-msk1 (Thr581) and anti-p-msk2 polyclonal were from Cell Signalling and the anti-p-histone H3(Ser1) antibody was from Upstate.
Strategies for Assessment of Immunotoxicology in Preclinical Drug Development
 Strategies for Assessment of Immunotoxicology in Preclinical Drug Development Rebecca Brunette, PhD Scientist, Analytical Biology SNBL USA Preclinical Immunotoxicology The study of evaluating adverse effects
Strategies for Assessment of Immunotoxicology in Preclinical Drug Development Rebecca Brunette, PhD Scientist, Analytical Biology SNBL USA Preclinical Immunotoxicology The study of evaluating adverse effects
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Kimura et al.,
 Supplemental material JCB Kimura et al., http://www.jcb.org/cgi/content/full/jcb.201503023/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. TRIMs regulate IFN-γ induced autophagy. (A and B) HC image analysis
Supplemental material JCB Kimura et al., http://www.jcb.org/cgi/content/full/jcb.201503023/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. TRIMs regulate IFN-γ induced autophagy. (A and B) HC image analysis
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Wnt16 smact merge VK/AB
 A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Nature Biotechnology: doi: /nbt Supplementary Figure 1. In vitro validation of OTC sgrnas and donor template.
 Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines
 Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
isolated from ctr and pictreated mice. Activation of effector CD4 +
 Supplementary Figure 1 Bystander inflammation conditioned T reg cells have normal functional suppressive activity and ex vivo phenotype. WT Balb/c mice were treated with polyi:c (pic) or PBS (ctr) via
Supplementary Figure 1 Bystander inflammation conditioned T reg cells have normal functional suppressive activity and ex vivo phenotype. WT Balb/c mice were treated with polyi:c (pic) or PBS (ctr) via
a Lamtor1 (gene) b Lamtor1 (mrna) c WT Lamtor1 Lamtor1 flox Lamtor2 A.U. p = LysM-Cre Lamtor3 Lamtor4 Lamtor5 BMDMs: Φ WT Φ KO β-actin WT BMDMs
 a Lamtor (gene) b Lamtor (mrna) c BMDMs: Φ WT Φ KO Lamtor flox 8 bp LysM-Cre 93 bp..5 p =.4 WT BMDMs: Φ WT Φ KO Lamtor (protein) BMDMs: Φ WT Φ KO Lamtor 8 kda Lamtor 4 kda Lamtor3 4 kda Lamtor4 kda Lamtor5.5
a Lamtor (gene) b Lamtor (mrna) c BMDMs: Φ WT Φ KO Lamtor flox 8 bp LysM-Cre 93 bp..5 p =.4 WT BMDMs: Φ WT Φ KO Lamtor (protein) BMDMs: Φ WT Φ KO Lamtor 8 kda Lamtor 4 kda Lamtor3 4 kda Lamtor4 kda Lamtor5.5
A group A Streptococcus ADP-ribosyltransferase toxin stimulates a protective IL-1β-dependent macrophage immune response
 A group A Streptococcus ADP-ribosyltransferase toxin stimulates a protective IL-1β-dependent macrophage immune response Ann E. Lin, Federico C. Beasley, Nadia Keller, Andrew Hollands, Rodolfo Urbano, Emily
A group A Streptococcus ADP-ribosyltransferase toxin stimulates a protective IL-1β-dependent macrophage immune response Ann E. Lin, Federico C. Beasley, Nadia Keller, Andrew Hollands, Rodolfo Urbano, Emily
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
CD1a-autoreactive T cells are a normal component of the human αβ T cell repertoire
 CDa-autoreactive T cells are a normal component of the human αβ T cell repertoire Annemieke de Jong, Victor Peña-Cruz, Tan-Yun Cheng, Rachael A. Clark, Ildiko Van Rhijn,, D. Branch Moody Division of Rheumatology,
CDa-autoreactive T cells are a normal component of the human αβ T cell repertoire Annemieke de Jong, Victor Peña-Cruz, Tan-Yun Cheng, Rachael A. Clark, Ildiko Van Rhijn,, D. Branch Moody Division of Rheumatology,
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Nature Immunology: doi: /ni Supplementary Figure 1. Overexpression of Ym1 reduces eosinophil numbers in the lungs.
 Supplementary Figure 1 Overexpression of Ym1 reduces eosinophil numbers in the lungs. (a-d) Absolute numbers per mg of tissue of CD8 + T cells (a), CD4 + T cells (b), CD19 + CD11b - B cells (c) and SigF
Supplementary Figure 1 Overexpression of Ym1 reduces eosinophil numbers in the lungs. (a-d) Absolute numbers per mg of tissue of CD8 + T cells (a), CD4 + T cells (b), CD19 + CD11b - B cells (c) and SigF
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Analyses of ECTRs by C-circle and T-circle assays.
 Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/258/rs3/dc1 Supplementary Materials for A Genome-Wide sirna Screen Reveals Positive and Negative Regulators of the NOD2 and NF-κB Signaling Pathways Neil Warner,
www.sciencesignaling.org/cgi/content/full/6/258/rs3/dc1 Supplementary Materials for A Genome-Wide sirna Screen Reveals Positive and Negative Regulators of the NOD2 and NF-κB Signaling Pathways Neil Warner,
Site-Directed Mutagenesis. Mutations in four Smad4 sites of mouse Gat1 promoter
 Supplement Supporting Materials and Methods Site-Directed Mutagenesis. Mutations in four Smad4 sites of mouse Gat1 promoter were independently generated using a two-step PCR method. The Smad4 binding site
Supplement Supporting Materials and Methods Site-Directed Mutagenesis. Mutations in four Smad4 sites of mouse Gat1 promoter were independently generated using a two-step PCR method. The Smad4 binding site
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1
 Supplementary Information p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1 Bei Wang 1, Dandan Niu 1, Liyun Lai 1, & Ee Chee Ren 1,2 1 Singapore Immunology
Supplementary Information p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1 Bei Wang 1, Dandan Niu 1, Liyun Lai 1, & Ee Chee Ren 1,2 1 Singapore Immunology
8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and
 1 Supplemental information 2 3 Materials and Methods 4 Reagents and animals 5 8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and 6 Silencer Select Pre-designed sirna
1 Supplemental information 2 3 Materials and Methods 4 Reagents and animals 5 8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and 6 Silencer Select Pre-designed sirna
Microarray Informatics
 Microarray Informatics Donald Dunbar MSc Seminar 4 th February 2009 Aims To give a biologistʼs view of microarray experiments To explain the technologies involved To describe typical microarray experiments
Microarray Informatics Donald Dunbar MSc Seminar 4 th February 2009 Aims To give a biologistʼs view of microarray experiments To explain the technologies involved To describe typical microarray experiments
used at a final concentration of 5 ng/ml. Rabbit anti-bim and mouse anti-mkp2 antibodies were
 1 Supplemental Methods Reagents and chemicals: TGFβ was a generous gift from Genzyme Inc. (Cambridge, MA) and was used at a final concentration of 5 ng/ml. Rabbit anti-bim and mouse anti-mkp2 antibodies
1 Supplemental Methods Reagents and chemicals: TGFβ was a generous gift from Genzyme Inc. (Cambridge, MA) and was used at a final concentration of 5 ng/ml. Rabbit anti-bim and mouse anti-mkp2 antibodies
Functional analysis reveals that RBM10 mutations. contribute to lung adenocarcinoma pathogenesis by. deregulating splicing
 Supplementary Information Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing Jiawei Zhao 1,+, Yue Sun 2,3,+, Yin Huang 6, Fan Song
Supplementary Information Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing Jiawei Zhao 1,+, Yue Sun 2,3,+, Yin Huang 6, Fan Song
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
Motivation From Protein to Gene
 MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
ClustalW algorithm. The alignment statistics are 96.9% for both pairwise identity and percent identical sites.
 Identity Human Mouse Supplemental Figure 1. Amino acid sequence alignment of -MyHC. The alignment was created using the ClustalW algorithm. The alignment statistics are 96.9% for both pairwise identity
Identity Human Mouse Supplemental Figure 1. Amino acid sequence alignment of -MyHC. The alignment was created using the ClustalW algorithm. The alignment statistics are 96.9% for both pairwise identity
Supplementary Table, Figures and Videos
 Supplementary Table, Figures and Videos Table S1. Oligonucleotides used for different approaches. (A) RT-qPCR study. (B) qpcr study after ChIP assay. (C) Probes used for EMSA. Figure S1. Notch activation
Supplementary Table, Figures and Videos Table S1. Oligonucleotides used for different approaches. (A) RT-qPCR study. (B) qpcr study after ChIP assay. (C) Probes used for EMSA. Figure S1. Notch activation
Microarray Informatics
 Microarray Informatics Donald Dunbar MSc Seminar 31 st January 2007 Aims To give a biologist s view of microarray experiments To explain the technologies involved To describe typical microarray experiments
Microarray Informatics Donald Dunbar MSc Seminar 31 st January 2007 Aims To give a biologist s view of microarray experiments To explain the technologies involved To describe typical microarray experiments
Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
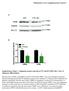 Merkestein et al. Supplementary Figure 1 A PLIN WT FTO KO 72 kda FABP4 17 kda HSC70 72 kda B Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
Merkestein et al. Supplementary Figure 1 A PLIN WT FTO KO 72 kda FABP4 17 kda HSC70 72 kda B Supplementary Figure 1. Adipogenic protein expression in WT and KO MEFs after 7 days of adipogenic differentiation.
Mutagenesis and generation of expression vectors Gene expression profiling Lentiviral production and infection of TF1 cells
 Mutagenesis and generation of expression vectors Human c-kit wild-type cdna encoding the short c-kit isoform was excised from vector pbshkitwt (kind gift from Dr. L Gros, France) by Sal I-Acc65 I digestion.
Mutagenesis and generation of expression vectors Human c-kit wild-type cdna encoding the short c-kit isoform was excised from vector pbshkitwt (kind gift from Dr. L Gros, France) by Sal I-Acc65 I digestion.
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various
 Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Supplementary Fig. 1 Kinetics of appearence of the faster migrating form of Bcl-10.
 α-cd3 + α-cd28: Time (min): + + + + + + + + + 0 5 15 30 60 120 180 240 300 360 360 n.s. Supplementary Fig. 1 Kinetics of appearence of the faster migrating form of. Immunoblot of lysates from Jurkat cells
α-cd3 + α-cd28: Time (min): + + + + + + + + + 0 5 15 30 60 120 180 240 300 360 360 n.s. Supplementary Fig. 1 Kinetics of appearence of the faster migrating form of. Immunoblot of lysates from Jurkat cells
Supplementary Figure 1
 Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Electrochemiluminescence-Based Immunoassays For Cytokines
 Electrochemiluminescence-Based Immunoassays George B. Sigal*, Rob Calamunci, and Jacob N. Wohlstadter Abstract Immunoassays for human cytokines are demonstrated using a new assay detection system from
Electrochemiluminescence-Based Immunoassays George B. Sigal*, Rob Calamunci, and Jacob N. Wohlstadter Abstract Immunoassays for human cytokines are demonstrated using a new assay detection system from
MEF-AOM MEF-DMH Symbol WT-AOM SKO-Mock SKO-AOM Symbol WT-DMH SKO-Mock SKO-DMH EG Rgs1 Ifit3 Ccl3 Usp18 Cxcl2 Rnf2 EG LOC Rgs1
 MEF-AOM Symol WT-AOM SKO-Mock SKO-AOM EG546166 2.097 2.571 2.850 Ifit3 2.064-2.984-3.111 Usp18 1.898-2.500-2.251 Rnf2 1.896 1.656 1.565 LOC667370 1.871-1.682-1.741 Ifit3 1.866-1.620-1.722 Usp18 1.860-8.756-6.861
MEF-AOM Symol WT-AOM SKO-Mock SKO-AOM EG546166 2.097 2.571 2.850 Ifit3 2.064-2.984-3.111 Usp18 1.898-2.500-2.251 Rnf2 1.896 1.656 1.565 LOC667370 1.871-1.682-1.741 Ifit3 1.866-1.620-1.722 Usp18 1.860-8.756-6.861
Supplementary figures
 Relative intensity Relative intensity Relative intensity Supplementary figures a None Caffeine None Caffeine c None Caffeine 6 6 6 ISG 6 6 6 UBE1L 6 6 6 UBCH8 6 6 6 EFP 1 1 DOX (h) 1 1 CPT (h) 1 1 UV (h)
Relative intensity Relative intensity Relative intensity Supplementary figures a None Caffeine None Caffeine c None Caffeine 6 6 6 ISG 6 6 6 UBE1L 6 6 6 UBCH8 6 6 6 EFP 1 1 DOX (h) 1 1 CPT (h) 1 1 UV (h)
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Analysis of Microarray Data
 Analysis of Microarray Data Lecture 3: Visualization and Functional Analysis George Bell, Ph.D. Bioinformatics Scientist Bioinformatics and Research Computing Whitehead Institute Outline Review Visualizing
Analysis of Microarray Data Lecture 3: Visualization and Functional Analysis George Bell, Ph.D. Bioinformatics Scientist Bioinformatics and Research Computing Whitehead Institute Outline Review Visualizing
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
