Sam68 STARR Sam68 QUA1- KH. p(r ) [Å] [Å] TSTAR STAR. Sam68 QUA1-KH and. constructs are
|
|
|
- Dorothy Johnson
- 6 years ago
- Views:
Transcription
1 a b Sam68 STARR Sam68 QUA1- KH c d e ) p(r p(r ) r [Å] r [Å] Supplementary Figure 1: The QUA2 domain is not involved in i the overall conformation of the STAR domain (a) Overlay of T-STAR QUA1-KH in complex with UAAU and T-STAR STAR in complex with AUUAAA structures. (b) HSQC spectra of Sam68 STAR ( black) andd QUA1-KH (red) domains in complex with AUUAAA. (c) Experimental SAXS dataa for the T-STAR and Sam68 QUA1-KH and STAR domains free and in complex with AAAUAA RNA. (d,e) Comparison of distance distributions of SAM68 and TSTAR QUA1-KH domains (d) and of SAM68 and TSTAR STAR domains (e). The maximum of both curves is att the samee distance.. TSTAR constructs are slightly more compact than Sam68 constructs.
2 Supplementary Figure 2: Protein-RNA contacts observed in the X-ray structures of T- STAR-RNA complexes. Intermolecular contacts between T-STAR KH and AAAUAA (the 5 AAA motif is recognized by one KH and the 3 UAA motif by another KH) (a), KH-QUA2 and AAUAAU (b), QUA1-KH and UAAU (c), and STAR and AUUAAA (d). Black and blue lines indicate van der Waals contacts and hydrogen bonds, respectively.
3 a b L77 I97 K755 c T-STAR 2-EEKYLPELMAEKDSLDPSFTHALRLVNQEIEKFQKGEGK--DEEKYIDVVINKNMKLGQKVV Sam ENKYLPELMAEKDSLDPSFTHAMQLLTAEIEKIQKGDSKKDDEENYLDLFSHKNMKLKERV * *** ***** *** * *** *** T-STAR Sam68 61-LIPVKQFPKFNFVGKLLGPRGNSLKRLQEETLTKMSILGKGSMRDKAKEEELRKSGEAKYF 161-LIPVKQYPKFNFVGKILGPQGNTIKRLQEETGAKISVLGKGSMRDKAKEEELRKGGDPKYA * T-STAR Sam HLNDDLHVLIEVFAPPAEAYARMGHALEEIKKFLIPDYNDEIRQAQLQELTYLNGGSENAD 222-HLNMDLHVFIEVFGPPCEAYALMAHAMEEVKKFLVPDMMDDICQEQFLELSYLNGVPEPSR d e L177 D258 I197 K175 I184 Supplementary Figure 3: NMR derived RNA recognitionn by T-STAR KH domain and Sam68 STAR domain. (a) HSQC spectra of T-STAR KH free (black) and in complex with AAAUAA at protein:rna molar ratios of 1:0.25 (magenta), 1:0.5 (green), 1:1 ( blue) and 1:1.5 (red). (b) Amino acids that display a significant chemical shift perturbation or disappear upon RNA binding are colored on the surface representation of T-STAR KH-UAA structure. (c) Sequence alignment of T-STARR and Sam68 STAR domains. Amino acids corresponding to the QUA1, KH and QUA2 domains are boxed in green, red and blue, respectively. Amino acids that contact the RNA in the T-STAR-AAT AAUAA structure are marked with an asterisk. (d) HSQC spectra of Sam68 STAR freee (black) and in complex with AUUAAA at protein:rna molar ratios of 1:0.5 (magenta), 1:1 (green) and a 1:1.2 (red). (e) Amino acids that display a significant chemical shift perturbation or disappear upon RNA binding are colored on the surface representationn of Sam68 KH structural model, derived from T-STAR KH structure. The UAAA RNA is positioned based on T-STAR KH-AAAUAA structure.
4 3 54.3Å Å 3 Supplementary Figure 4: The KH dimerization brings two RNA binding elements on opposite sides of the dimer. Overview of T-STAR QUA1-KH structure inn complex with UAAU. The distance between the 3 -end of one RNA and the 5 -end of the other is indicated.
5 GFP-Sam68 WT GFP-Sam68 Y241E GFP-T-STAR WT GFP-T-STAR Y141E Supplementary Figure 5: Localization of GFP-Sam68 WT and Y241E, and GFP-T-STAR WT and Y141E, in HEK293 cells. GFP-fusion constructs were transfected into HEK293 cells, and 24 h after transfection the protein expression patterns were analyzed by fluorescence microscopy. The cell nuclei were stained with DAPI.
6 T-STAR KH dimer PCBP1 P KH1 dimer Linker -strand 2 -strandd 2 Linker -helix 3 -helix 3 Supplementary Figure 6: Comparison of T-STAR KH andd PCBP1 interfaces. KH1 dimerization Top, -sheet view of the structures. The dimer interface involves -strands s 2 of PCBP1 (right) but not of T-STAR (left). Bottom, -helix 3 view of the structure. The dimer interfacee involves a large hydrophobic interface of -helix 3 of T-TSTAR (left) but not n PCBP1 (right).
7 a SRSF1 intron 4 b Sgce exon 8 ~600 nt Exon 8 Exon 4 Exon 5 Exon 7 Exon 9 Supplementary Figure 7: Sam68 could regulate splicing by looping out regions of the pre-mrna. Structural models of Sam68 interaction with SRSF1 (a), and Sgce (b) pre-mrnas suggesting thatt Sam68 might function in alternative splicing control by looping out regions of the pre- mrna, to promote exon inclusion or exon skipping, respectively..
8 T-STAR STAR / AUUAAAA T-STAR QUA1-KH / UAAU T-STAR KH T-STAR KH / AAAUAA T-STAR KH-QUA2 / AAUAAU Supplementary Figure 8: Representa ative electron density maps of T-STAR structures. Representative stereo image of 2Fo-Fcc maps contoured at 1.5σ (bluee mesh).
9 Uncropped gel corresponding to Figuree 5a (CD444 exon v5) 850bp 650bp 500bp 400bp 300bp 200bp 100bp Uncropped gel corresponding to Figuree 5b (Neurexin 3 exon AS4) 850bp 650bp 500bp 400bp 300bp 200bp 100bp Uncropped gel corresponding to Figuree 5c (Neurexin 2 exon AS4) 850bp 650bp 500bp 400bp 300bp 200bp 100bp Supplementary Figure 9: Uncropped d gels corresponding to Figure 5 a-c
10 Supplementary Table 1: SAXS analysis of T-STAR and Sam68 R g [Å] D max [Å] V Porod [Å 3 ] MW SAXS [kda] (dimer) MW calc [kda] (dimer) T-STAR QUA1-KH T-STAR STAR Sam68 QUA1-KH Sam68 STAR Sam68 QUA1-KH (37) * 41.8 AAAUAA Sam68 STAR AAAUAA (42) * 46.9 * The density of the RNA is not considered.
11 Supplementary Table 2: Most enriched 6-mer motifs bound by T-STAR identified by HITS-CLIP k-mer k-mer k-mer frequency Corrected p-value sequence frequency in CLIP tags in genomic background tags frequency AAUUAA < 1.0E-300 AUAAAC < 1.0E-300 UUAAUU E-195 AAUAAU < 1.0E-300 AUUAAA E-197 AAAUAA E-238 UAAAAC E-246 UAAAUA < 1.0E-300 AAAAAA < 1.0E-300 AAAAUA E-150 UUAAAU E-01 AAAAAU E-104 AAUAAA E-119 AUAAAA E-239 AUAAAU < 1.0E-300
12 Supplementary Table 3: Fluorescence polarization affinity measurements of T-STAR and Sam68 QUA1-KH domains to 5-mer RNAs. Kd ( M) T-STAR QUA1-KH Sam68 QUA1-KH AUAAA UUAAA CUAAA AAAAA 6 13 ACAAA 55 >100 AUUAA >100 >100 AUCAA >100 >100 AUAUA >100 >100 AUACA >100 >100 AUAAU AUAAC 47 >100 CCCCC >100 >100 UUUUU >100 >100
13 Supplementary Table 4: list of primers used in this study Sam68: STAR construct: Forward: 5 -TACTTCCAATCCATGATGGAGCCAGAGAACAAGTACCTG-3 Reverse: 5 -TATCCACCTTTACTGTCAACCACGAGAGGGTTCAGGTAC-3 QUA1-KH construct: Forward: 5 -TACTTCCAATCCATGATGGAGCCAGAGAACAAGTACCTG-3 Reverse: 5 -TATCCACCTTTACTGTCACATCATATCCGGTACTAG-3 Y241E mutant: Forward: 5 -GTCTTTGGACCCCCATGTGAGGCTGAAGCTCTTATGGCCCATGCCATGGAG-3 Reverse: 5 -CTCCATGGCATGGGCCATAAGAGCTTCAGCCTCACATGGGGGTCCAAAGAC-3 T-STAR: STAR construct: Forward: 5 -TACTTCCAATCCATGATGGAGGAGAAGTACCTGCCC-3 Reverse: 5 -TATCCACCTTTACTGTCAAACATCTGCATTTTCTGAACC-3 QUA1-KH construct: Forward: 5 -TACTTCCAATCCATGATGGAGGAGAAGTACCTGCCC-3 Reverse: 5 -TATCCACCTTTACTGTCAATTATAATCAGGGATGAGGAACTT-3 KH-QUA2 construct: Forward: 5 -TACTTCCAATCCATGATTAATAAGAACATGAAGCTGGGA-3 Reverse: 5 -TATCCACCTTTACTGTCAAACATCTGCATTTTCTGAACC-3 KH construct: Forward: 5 -TACTTCCAATCCATGATTAATAAGAACATGAAGCTGGGA-3 Reverse: 5 -TATCCACCTTTACTGTCAATTATAATCAGGGATGAGGAACTT-3 Y141E mutant: Forward: 5 -GTGTTTGCCCCACCTGCAGAAGCTGAAGCCAGGATGGGACATGCTTTGGAA-3 Reverse: 5 - TTCCAAAGCATGTCCCATCCTGGCTTCAGCTTCTGCAGGTGGGGCAAACAC-3
SUPPLEMENTARY INFORMATION. Supplementary Figures 1-8
 SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain
 Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain architecture. Various C-terminal fragments were cloned and
Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain architecture. Various C-terminal fragments were cloned and
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
HEK293T. Fig. 1 in the
 Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid
 Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein.
 Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein. MR766 NS5 sequence 1 ACAGAGAACAGATTGGTGGTGGAGGTGGGACGGGAGAGACTCTGGGAGAGAAGTGGAAAG 61 CTCGTCTGAATCAGATGTCGGCCCTGGAGTTCTACTCTTATAAAAAGTCAGGTATCACTG
Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein. MR766 NS5 sequence 1 ACAGAGAACAGATTGGTGGTGGAGGTGGGACGGGAGAGACTCTGGGAGAGAAGTGGAAAG 61 CTCGTCTGAATCAGATGTCGGCCCTGGAGTTCTACTCTTATAAAAAGTCAGGTATCACTG
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Nature Structural & Molecular Biology: doi: /nsmb.3428
 Supplementary Figure 1 Biochemical characterization of the monou and oligou activity switch of TUT4(7). (a) Mouse TUT4 and human TUT7 were assayed for monou and Lin28-dependent oligou addition activities
Supplementary Figure 1 Biochemical characterization of the monou and oligou activity switch of TUT4(7). (a) Mouse TUT4 and human TUT7 were assayed for monou and Lin28-dependent oligou addition activities
Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a)
 Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a) Representative images of 2D class averages of gb-698glyc bound to 1G2 Fab. Top views of the complex were underrepresented
Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a) Representative images of 2D class averages of gb-698glyc bound to 1G2 Fab. Top views of the complex were underrepresented
Supplemental Information Molecular Cell, Volume 41
 Supplemental Information Molecular Cell, Volume 41 Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and Its Regulation by Upf2 Sutapa Chakrabarti, Uma Jayachandran, Fabien Bonneau, Francesca
Supplemental Information Molecular Cell, Volume 41 Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and Its Regulation by Upf2 Sutapa Chakrabarti, Uma Jayachandran, Fabien Bonneau, Francesca
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Engineering splicing factors with designed specificities
 nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Sequence Determinants of a Conformational Switch in a Protein
 Sequence Determinants of a Conformational Switch in a Protein Thomas A. Anderson, Matthew H. J. Cordes, and Robert T. Sauer Kyle Skalenko 4/17/09 Introduction Protein folding is guided by the following
Sequence Determinants of a Conformational Switch in a Protein Thomas A. Anderson, Matthew H. J. Cordes, and Robert T. Sauer Kyle Skalenko 4/17/09 Introduction Protein folding is guided by the following
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
PHF20 is an effector protein of p53 double lysine methylation
 SUPPLEMENTARY INFORMATION PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53 Gaofeng Cui 1, Sungman Park 2, Aimee I Badeaux 3, Donghwa Kim 2, Joseph Lee 1,
SUPPLEMENTARY INFORMATION PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53 Gaofeng Cui 1, Sungman Park 2, Aimee I Badeaux 3, Donghwa Kim 2, Joseph Lee 1,
Supplementary Figure 1
 Supplementary Figure 1 TPM2 331- - n 2 (PASS reads for pa identification) 3- n < 2, LAP near pa ( 24 nt) CU 5 T 45 RPM - 21- - 11- - 1- - 46- - n < 2, LAP not near pa (> 24 nt) n 2 (PASS reads for pa identification)
Supplementary Figure 1 TPM2 331- - n 2 (PASS reads for pa identification) 3- n < 2, LAP near pa ( 24 nt) CU 5 T 45 RPM - 21- - 11- - 1- - 46- - n < 2, LAP not near pa (> 24 nt) n 2 (PASS reads for pa identification)
Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade
 Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade Supplementary Materials Supplementary methods Table S1-S Figure S1-S 1 1 1 1 1 1 1 1 1 0 1 0 Supplementary Methods Competitive
Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade Supplementary Materials Supplementary methods Table S1-S Figure S1-S 1 1 1 1 1 1 1 1 1 0 1 0 Supplementary Methods Competitive
SUPPLEMENTARY INFORMATION. Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly
 SUPPLEMENTARY INFORMATION Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly Dustin J. E. Huard, Kathleen M. Kane and F. Akif Tezcan* Department of Chemistry and Biochemistry,
SUPPLEMENTARY INFORMATION Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly Dustin J. E. Huard, Kathleen M. Kane and F. Akif Tezcan* Department of Chemistry and Biochemistry,
Supplementary Figure 1 Two distinct conformational states of the HNH domain in crystal structures. a
 Supplementary Figure 1 Two distinct conformational states of the HNH domain in crystal structures. a HNH-state 1 in PDB 4OO8, in which the distance from the C atom of the HNH catalytic residue 840 to the
Supplementary Figure 1 Two distinct conformational states of the HNH domain in crystal structures. a HNH-state 1 in PDB 4OO8, in which the distance from the C atom of the HNH catalytic residue 840 to the
RNP purification, components and activity.
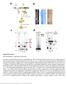 Supplementary Figure 1 RNP purification, components and activity. (a) Intein-mediated RNP production and purification. The Ll.LtrB intron RNA (red) (Exons E1 and E2 in green) and associated intron encoded
Supplementary Figure 1 RNP purification, components and activity. (a) Intein-mediated RNP production and purification. The Ll.LtrB intron RNA (red) (Exons E1 and E2 in green) and associated intron encoded
Conformational changes in IgE contribute to its. uniquely slow dissociation rate from receptor FcεRI
 Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI M.D. Holdom, A.M. Davies, J.E. Nettleship, S.C. Bagby, B. Dhaliwal, E. Girardi, J. Hunt, H.J. Gould,
Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI M.D. Holdom, A.M. Davies, J.E. Nettleship, S.C. Bagby, B. Dhaliwal, E. Girardi, J. Hunt, H.J. Gould,
Transcription & post transcriptional modification
 Transcription & post transcriptional modification Transcription The synthesis of RNA molecules using DNA strands as the templates so that the genetic information can be transferred from DNA to RNA Similarity
Transcription & post transcriptional modification Transcription The synthesis of RNA molecules using DNA strands as the templates so that the genetic information can be transferred from DNA to RNA Similarity
Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1
 Supplementary information Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1 Catherine A. Musselman 1, Nikita Avvakumov 2, Reiko Watanabe 3, Christopher G. Abraham 4, Marie-Eve Lalonde
Supplementary information Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1 Catherine A. Musselman 1, Nikita Avvakumov 2, Reiko Watanabe 3, Christopher G. Abraham 4, Marie-Eve Lalonde
human Cdc45 Figure 1c. (c)
 1 Details of the refined crystallographic model of human Cdc45 and comparison of its active-site region with that of bacterial RecJ. (a) Stereo view of a representative example of the final 2F o -F c electron
1 Details of the refined crystallographic model of human Cdc45 and comparison of its active-site region with that of bacterial RecJ. (a) Stereo view of a representative example of the final 2F o -F c electron
AD BD TOC1. Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between
 AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA.
 Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Make the protein through the genetic dogma process.
 Make the protein through the genetic dogma process. Coding Strand 5 AGCAATCATGGATTGGGTACATTTGTAACTGT 3 Template Strand mrna Protein Complete the table. DNA strand DNA s strand G mrna A C U G T A T Amino
Make the protein through the genetic dogma process. Coding Strand 5 AGCAATCATGGATTGGGTACATTTGTAACTGT 3 Template Strand mrna Protein Complete the table. DNA strand DNA s strand G mrna A C U G T A T Amino
Functional analysis reveals that RBM10 mutations. contribute to lung adenocarcinoma pathogenesis by. deregulating splicing
 Supplementary Information Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing Jiawei Zhao 1,+, Yue Sun 2,3,+, Yin Huang 6, Fan Song
Supplementary Information Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing Jiawei Zhao 1,+, Yue Sun 2,3,+, Yin Huang 6, Fan Song
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
MODULE 5: TRANSLATION
 MODULE 5: TRANSLATION Lesson Plan: CARINA ENDRES HOWELL, LEOCADIA PALIULIS Title Translation Objectives Determine the codons for specific amino acids and identify reading frames by looking at the Base
MODULE 5: TRANSLATION Lesson Plan: CARINA ENDRES HOWELL, LEOCADIA PALIULIS Title Translation Objectives Determine the codons for specific amino acids and identify reading frames by looking at the Base
Nature Methods: doi: /nmeth Supplementary Figure 1. DMS-MaPseq data are highly reproducible at elevated DMS concentrations.
 Supplementary Figure 1 DMS-MaPseq data are highly reproducible at elevated DMS concentrations. a, Correlation of Gini index for 202 yeast mrna regions with 15x coverage at 2.5% or 5% v/v DMS concentrations
Supplementary Figure 1 DMS-MaPseq data are highly reproducible at elevated DMS concentrations. a, Correlation of Gini index for 202 yeast mrna regions with 15x coverage at 2.5% or 5% v/v DMS concentrations
Nature Structural & Molecular Biology: doi: /nsmb.3018
 Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
Orange fluorescent proteins shift constructed from cyanobacteriochromes. chromophorylated with phycoerythrobilin. Kai-Hong Zhao 1 Ming Zhou 1, *
 Electronic Supplementary Material (ESI) for Photochemical. This journal is The Royal Society of Chemistry and Owner Societies 2014 Orange fluorescent proteins shift constructed from cyanobacteriochromes
Electronic Supplementary Material (ESI) for Photochemical. This journal is The Royal Society of Chemistry and Owner Societies 2014 Orange fluorescent proteins shift constructed from cyanobacteriochromes
Student Learning Outcomes (SLOS)
 Student Learning Outcomes (SLOS) KNOWLEDGE AND LEARNING SKILLS USE OF KNOWLEDGE AND LEARNING SKILLS - how to use Annhyb to save and manage sequences - how to use BLAST to compare sequences - how to get
Student Learning Outcomes (SLOS) KNOWLEDGE AND LEARNING SKILLS USE OF KNOWLEDGE AND LEARNING SKILLS - how to use Annhyb to save and manage sequences - how to use BLAST to compare sequences - how to get
The drawing of RNA and cdna is worth 3 points, if there is no second strand of cdna or no oligo dc or dg linker added, 1 point will be deducted.
 1. a) The 3 end of mrna usually ends up copied in a cdna because the first strand of cdna is synthesized by reverse transcriptase using oligo dt as the primer. The enzyme will fall off after a while resulting
1. a) The 3 end of mrna usually ends up copied in a cdna because the first strand of cdna is synthesized by reverse transcriptase using oligo dt as the primer. The enzyme will fall off after a while resulting
SUPPLEMENTARY INFORMATION
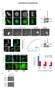 SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase
 A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase tag insertion at the 3 end using CRISPR-Cas9 genome editing technique Sambuddha Basu 1, 3, Levi Adams 1, 3, Subhrangshu
A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase tag insertion at the 3 end using CRISPR-Cas9 genome editing technique Sambuddha Basu 1, 3, Levi Adams 1, 3, Subhrangshu
Nature Structural & Molecular Biology: doi: /nsmb.2548
 Supplementary Figure 1. Structure of GltPhout. (a) Stereo view of a slice through a single GltPhout protomer shown in stick representation along with 2Fo-Fc and anomalous difference electron maps. The
Supplementary Figure 1. Structure of GltPhout. (a) Stereo view of a slice through a single GltPhout protomer shown in stick representation along with 2Fo-Fc and anomalous difference electron maps. The
Supplementary Information. Isl2b regulates anterior second heart field development in zebrafish
 Supplementary Information Isl2b regulates anterior second heart field development in zebrafish Hagen R. Witzel 1, Sirisha Cheedipudi 1, Rui Gao 1, Didier Y.R. Stainier 2 and Gergana D. Dobreva 1,3* 1 Origin
Supplementary Information Isl2b regulates anterior second heart field development in zebrafish Hagen R. Witzel 1, Sirisha Cheedipudi 1, Rui Gao 1, Didier Y.R. Stainier 2 and Gergana D. Dobreva 1,3* 1 Origin
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
embryos. Asterisk represents loss of or reduced expression. Brackets represent
 Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplementary Figure 1 PZA inhibits root hair formation as well as cell elongation in the maturation zone of eto1-2 roots. (A) The PI staining of the
 Supplementary Figure 1 PZA inhibits root hair formation as well as cell elongation in the maturation zone of eto1-2 roots. (A) The PI staining of the roots of three-day-old etiolated seedlings of Col-0
Supplementary Figure 1 PZA inhibits root hair formation as well as cell elongation in the maturation zone of eto1-2 roots. (A) The PI staining of the roots of three-day-old etiolated seedlings of Col-0
TALENs (Transcription Activator-Like Effector Nucleases)
 TALENs (Transcription Activator-Like Effector Nucleases) The fundamental rationale between TALENs and ZFNs is similar, namely, combine a sequencespecific DNA-binding peptide domain with a nuclease domain
TALENs (Transcription Activator-Like Effector Nucleases) The fundamental rationale between TALENs and ZFNs is similar, namely, combine a sequencespecific DNA-binding peptide domain with a nuclease domain
CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter
 CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter SUPPLEMENTARY DATA See Supplementary Sequence File: 1 Supplementary Figure S1: The total protein was extracted from
CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter SUPPLEMENTARY DATA See Supplementary Sequence File: 1 Supplementary Figure S1: The total protein was extracted from
Nature Structural & Molecular Biology: doi: /nsmb.2307
 A novel locally-closed conformation of a bacterial pentameric proton-gated ion channel Marie S. Prevost*, Ludovic Sauguet*, Hugues Nury, Catherine Van Renterghem, Christèle Huon, Frederic Poitevin, Marc
A novel locally-closed conformation of a bacterial pentameric proton-gated ion channel Marie S. Prevost*, Ludovic Sauguet*, Hugues Nury, Catherine Van Renterghem, Christèle Huon, Frederic Poitevin, Marc
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Supplementary information Bi-specific Aptamers mediating Tumour Cell Lysis
 Supplementary information Bi-specific Aptamers mediating Tumour Cell Lysis Figure S1 SELEX scheme of different CD16 DNA selection approaches. Nine rounds of filter SELEX were conducted in parallel, only
Supplementary information Bi-specific Aptamers mediating Tumour Cell Lysis Figure S1 SELEX scheme of different CD16 DNA selection approaches. Nine rounds of filter SELEX were conducted in parallel, only
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system
 SUPPLEMENTARY DATA Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system Daniel Gleditzsch 1, Hanna Müller-Esparza 1, Patrick Pausch 2,3, Kundan Sharma 4, Srivatsa Dwarakanath 1,
SUPPLEMENTARY DATA Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system Daniel Gleditzsch 1, Hanna Müller-Esparza 1, Patrick Pausch 2,3, Kundan Sharma 4, Srivatsa Dwarakanath 1,
Chapter 11. Transcription. The biochemistry and molecular biology department of CMU
 Chapter 11 Transcription The biochemistry and molecular biology department of CMU Transcription The synthesis of RNA molecules using DNA strands as the templates so that the genetic information can be
Chapter 11 Transcription The biochemistry and molecular biology department of CMU Transcription The synthesis of RNA molecules using DNA strands as the templates so that the genetic information can be
Multiple choice questions (numbers in brackets indicate the number of correct answers)
 1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
STRUCTURAL BIOLOGY. α/β structures Closed barrels Open twisted sheets Horseshoe folds
 STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
SUPPLEMENTARY INFORMATION. Chemical modulation of Chaperone-mediated autophagy by novel
 SUPPLEMENTARY INFORMATION Chemical modulation of Chaperone-mediated autophagy by novel retinoic acid derivatives Jaime Anguiano 1, Thomas P Garner 2, Murugesan Mahalingam 1, Bhaskar C. Das 1, Evripidis
SUPPLEMENTARY INFORMATION Chemical modulation of Chaperone-mediated autophagy by novel retinoic acid derivatives Jaime Anguiano 1, Thomas P Garner 2, Murugesan Mahalingam 1, Bhaskar C. Das 1, Evripidis
Introduction to Cellular Biology and Bioinformatics. Farzaneh Salari
 Introduction to Cellular Biology and Bioinformatics Farzaneh Salari Outline Bioinformatics Cellular Biology A Bioinformatics Problem What is bioinformatics? Computer Science Statistics Bioinformatics Mathematics...
Introduction to Cellular Biology and Bioinformatics Farzaneh Salari Outline Bioinformatics Cellular Biology A Bioinformatics Problem What is bioinformatics? Computer Science Statistics Bioinformatics Mathematics...
Chapter One. Construction of a Fluorescent α5 Subunit. Elucidation of the unique contribution of the α5 subunit is complicated by several factors
 4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
30 Gene expression: Transcription
 30 Gene expression: Transcription Gene structure. o Exons coding region of DNA. o Introns non-coding region of DNA. o Introns are interspersed between exons of a single gene. o Promoter region helps enzymes
30 Gene expression: Transcription Gene structure. o Exons coding region of DNA. o Introns non-coding region of DNA. o Introns are interspersed between exons of a single gene. o Promoter region helps enzymes
Quiz Submissions Quiz 4
 Quiz Submissions Quiz 4 Attempt 1 Written: Nov 1, 2015 17:35 Nov 1, 2015 22:19 Submission View Released: Nov 4, 2015 20:24 Question 1 0 / 1 point Three RNA polymerases synthesize most of the RNA present
Quiz Submissions Quiz 4 Attempt 1 Written: Nov 1, 2015 17:35 Nov 1, 2015 22:19 Submission View Released: Nov 4, 2015 20:24 Question 1 0 / 1 point Three RNA polymerases synthesize most of the RNA present
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10258 Supplementary Figure 1 Reconstitution of the CENP-A nucleosome with recombinant human histones H2A, H2B, H4, and CENP-A. a, Purified recombinant human
SUPPLEMENTARY INFORMATION doi:10.1038/nature10258 Supplementary Figure 1 Reconstitution of the CENP-A nucleosome with recombinant human histones H2A, H2B, H4, and CENP-A. a, Purified recombinant human
If you wish to have extra practice with swiss pdb viewer or to familiarize yourself with how to use the program here is a tutorial:
 Name (s): Swiss PDB viewer assignment chapter 4. If you wish to have extra practice with swiss pdb viewer or to familiarize yourself with how to use the program here is a tutorial: http://spdbv.vital-it.ch/themolecularlevel/spvtut/index.html
Name (s): Swiss PDB viewer assignment chapter 4. If you wish to have extra practice with swiss pdb viewer or to familiarize yourself with how to use the program here is a tutorial: http://spdbv.vital-it.ch/themolecularlevel/spvtut/index.html
Exam 2 Key - Spring 2008 A#: Please see us if you have any questions!
 Page 1 of 5 Exam 2 Key - Spring 2008 A#: Please see us if you have any questions! 1. A mutation in which parts of two nonhomologous chromosomes change places is called a(n) A. translocation. B. transition.
Page 1 of 5 Exam 2 Key - Spring 2008 A#: Please see us if you have any questions! 1. A mutation in which parts of two nonhomologous chromosomes change places is called a(n) A. translocation. B. transition.
Supplementary Figure 1.
 Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
Differential Gene Expression
 Developmental Biology Biology 4361 Differential Gene Expression October 13, 2005 core transcription initiation site 5 promoter 3 TATAT +1 upstream downstream Basal transcription factors (eukaryotes) TFIID
Developmental Biology Biology 4361 Differential Gene Expression October 13, 2005 core transcription initiation site 5 promoter 3 TATAT +1 upstream downstream Basal transcription factors (eukaryotes) TFIID
Expanded View Figures
 EMO reports rystal structure of Mis18 Yippee-like domain Lakxmi Subramanian et al Expanded View Figures Figure EV1. Structural characterization of the N-terminal Yippee-like globular domain of spmis18.
EMO reports rystal structure of Mis18 Yippee-like domain Lakxmi Subramanian et al Expanded View Figures Figure EV1. Structural characterization of the N-terminal Yippee-like globular domain of spmis18.
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Differential Gene Expression
 Biology 4361 Developmental Biology Differential Gene Expression September 28, 2006 Chromatin Structure ~140 bp ~60 bp Transcriptional Regulation: 1. Packing prevents access CH 3 2. Acetylation ( C O )
Biology 4361 Developmental Biology Differential Gene Expression September 28, 2006 Chromatin Structure ~140 bp ~60 bp Transcriptional Regulation: 1. Packing prevents access CH 3 2. Acetylation ( C O )
RNA does not adopt the classic B-DNA helix conformation when it forms a self-complementary double helix
 Reason: RNA has ribose sugar ring, with a hydroxyl group (OH) If RNA in B-from conformation there would be unfavorable steric contact between the hydroxyl group, base, and phosphate backbone. RNA structure
Reason: RNA has ribose sugar ring, with a hydroxyl group (OH) If RNA in B-from conformation there would be unfavorable steric contact between the hydroxyl group, base, and phosphate backbone. RNA structure
Supplementary Figure 1. Ratiometric fluorescence visualization of DNA cleavage by
 Supplementary Figure 1. Ratiometric fluorescence visualization of DNA cleavage by Cas9:gRNA. (a) A labeled by Cy3 and Cy5 with an inter-probe distance of > 30 bp was tethered to a PEG-coated surface via
Supplementary Figure 1. Ratiometric fluorescence visualization of DNA cleavage by Cas9:gRNA. (a) A labeled by Cy3 and Cy5 with an inter-probe distance of > 30 bp was tethered to a PEG-coated surface via
Structure and Possible Mechanism of the CcbJ Methyltransferase from Streptomyces caelestis
 Supplemental material to accompany Structure and Possible Mechanism of the CcbJ Methyltransferase from Streptomyces caelestis Jacob Bauer, a Gabriela Ondrovičová, a Lucie Najmanová, b Vladimír Pevala,
Supplemental material to accompany Structure and Possible Mechanism of the CcbJ Methyltransferase from Streptomyces caelestis Jacob Bauer, a Gabriela Ondrovičová, a Lucie Najmanová, b Vladimír Pevala,
Genetic Parts in Bacterial Gene Expression
 Genetic Parts in Bacterial Gene Expression The Focus Promoters Operators Transcription Factors Transcriptional Terminators Ribosome Binding Sites An Abstract Annotation We ll Pay Particular Attention To:
Genetic Parts in Bacterial Gene Expression The Focus Promoters Operators Transcription Factors Transcriptional Terminators Ribosome Binding Sites An Abstract Annotation We ll Pay Particular Attention To:
BIOCHEMISTRY REVIEW. Overview of Biomolecules. Chapter 12 Transcription
 BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 12 Transcription 2 3 4 5 Are You Getting It?? Which are general characteristics of transcription? (multiple answers) a) An entire DNA molecule is transcribed
BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 12 Transcription 2 3 4 5 Are You Getting It?? Which are general characteristics of transcription? (multiple answers) a) An entire DNA molecule is transcribed
More on fluorescence
 More on fluorescence Last class Fluorescence Absorption emission Jablonski diagrams This class More on fluorescence Common fluorophores Jablonski diagrams to spectra Properties of fluorophores Excitation
More on fluorescence Last class Fluorescence Absorption emission Jablonski diagrams This class More on fluorescence Common fluorophores Jablonski diagrams to spectra Properties of fluorophores Excitation
Supplementary Information. A superfolding Spinach2 reveals the dynamic nature of. trinucleotide repeat RNA
 Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Fig Ch 17: From Gene to Protein
 Fig. 17-1 Ch 17: From Gene to Protein Basic Principles of Transcription and Translation RNA is the intermediate between genes and the proteins for which they code Transcription is the synthesis of RNA
Fig. 17-1 Ch 17: From Gene to Protein Basic Principles of Transcription and Translation RNA is the intermediate between genes and the proteins for which they code Transcription is the synthesis of RNA
Nucleic acids. How DNA works. DNA RNA Protein. DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology
 Nucleic acid chemistry and basic molecular theory Nucleic acids DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology Cell cycle DNA RNA Protein Transcription Translation
Nucleic acid chemistry and basic molecular theory Nucleic acids DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology Cell cycle DNA RNA Protein Transcription Translation
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence
 Supplementary Information Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Yoshinori Kousoku, Aya Ohkuni,
Supplementary Information Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Yoshinori Kousoku, Aya Ohkuni,
Molecular Techniques. 3 Goals in Molecular Biology. Nucleic Acids: DNA and RNA. Disclaimer Nucleic Acids Proteins
 Molecular Techniques Disclaimer Nucleic Acids Proteins Houpt, CMN, 9-30-11 3 Goals in Molecular Biology Identify All nucleic acids (and proteins) are chemically identical in aggregate - need to identify
Molecular Techniques Disclaimer Nucleic Acids Proteins Houpt, CMN, 9-30-11 3 Goals in Molecular Biology Identify All nucleic acids (and proteins) are chemically identical in aggregate - need to identify
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
Transcription and Post Transcript Modification
 Transcription and Post Transcript Modification You Should Be Able To 1. Describe transcription. 2. Compare and contrast eukaryotic + prokaryotic transcription. 3. Explain mrna processing in eukaryotes.
Transcription and Post Transcript Modification You Should Be Able To 1. Describe transcription. 2. Compare and contrast eukaryotic + prokaryotic transcription. 3. Explain mrna processing in eukaryotes.
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Chapter 8. Microbial Genetics. Lectures prepared by Christine L. Case. Copyright 2010 Pearson Education, Inc.
 Chapter 8 Microbial Genetics Lectures prepared by Christine L. Case Structure and Function of Genetic Material Learning Objectives 8-1 Define genetics, genome, chromosome, gene, genetic code, genotype,
Chapter 8 Microbial Genetics Lectures prepared by Christine L. Case Structure and Function of Genetic Material Learning Objectives 8-1 Define genetics, genome, chromosome, gene, genetic code, genotype,
Introduction to Bioinformatics Online Course: IBT
 Introduction to Bioinformatics Online Course: IBT Multiple Sequence Alignment Building Multiple Sequence Alignment Lec5: Interpreting your MSA Using Logos Using Logos - Logos are a terrific way to generate
Introduction to Bioinformatics Online Course: IBT Multiple Sequence Alignment Building Multiple Sequence Alignment Lec5: Interpreting your MSA Using Logos Using Logos - Logos are a terrific way to generate
BETA STRAND Prof. Alejandro Hochkoeppler Department of Pharmaceutical Sciences and Biotechnology University of Bologna
 Prof. Alejandro Hochkoeppler Department of Pharmaceutical Sciences and Biotechnology University of Bologna E-mail: a.hochkoeppler@unibo.it C-ter NH and CO groups: right, left, right (plane of the slide)
Prof. Alejandro Hochkoeppler Department of Pharmaceutical Sciences and Biotechnology University of Bologna E-mail: a.hochkoeppler@unibo.it C-ter NH and CO groups: right, left, right (plane of the slide)
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Transcription is the first stage of gene expression
 Transcription is the first stage of gene expression RNA synthesis is catalyzed by RNA polymerase, which pries the DNA strands apart and hooks together the RNA nucleotides The RNA is complementary to the
Transcription is the first stage of gene expression RNA synthesis is catalyzed by RNA polymerase, which pries the DNA strands apart and hooks together the RNA nucleotides The RNA is complementary to the
PrimePCR Assay Validation Report
 Gene Information Gene Name fibroblast growth factor receptor 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID FGFR1 Human The protein encoded by this gene
Gene Information Gene Name fibroblast growth factor receptor 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID FGFR1 Human The protein encoded by this gene
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Supplementary Online Material. Structural mimicry in transcription regulation of human RNA polymerase II by the. DNA helicase RECQL5
 Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Nucleic Acids. Information specifying protein structure
 Nucleic Acids Nucleic acids represent the fourth major class of biomolecules (other major classes of biomolecules are proteins, carbohydrates, fats) Genome - the genetic information of an organism Information
Nucleic Acids Nucleic acids represent the fourth major class of biomolecules (other major classes of biomolecules are proteins, carbohydrates, fats) Genome - the genetic information of an organism Information
Information specifying protein structure. Chapter 19 Nucleic Acids Nucleotides Are the Building Blocks of Nucleic Acids
 Chapter 19 Nucleic Acids Information specifying protein structure Nucleic acids represent the fourth major class of biomolecules (other major classes of biomolecules are proteins, carbohydrates, fats)
Chapter 19 Nucleic Acids Information specifying protein structure Nucleic acids represent the fourth major class of biomolecules (other major classes of biomolecules are proteins, carbohydrates, fats)
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/11/e1601625/dc1 Supplementary Materials for A molecular mechanism of chaperone-client recognition This PDF file includes: Lichun He, Timothy Sharpe, Adam Mazur,
advances.sciencemag.org/cgi/content/full/2/11/e1601625/dc1 Supplementary Materials for A molecular mechanism of chaperone-client recognition This PDF file includes: Lichun He, Timothy Sharpe, Adam Mazur,
Delve AP Biology Lecture 7: 10/30/11 Melissa Ko and Anne Huang
 Today s Agenda: I. DNA Structure II. DNA Replication III. DNA Proofreading and Repair IV. The Central Dogma V. Transcription VI. Post-transcriptional Modifications Delve AP Biology Lecture 7: 10/30/11
Today s Agenda: I. DNA Structure II. DNA Replication III. DNA Proofreading and Repair IV. The Central Dogma V. Transcription VI. Post-transcriptional Modifications Delve AP Biology Lecture 7: 10/30/11
