The protein kinase A anchoring protein makap coordinates two integrated camp effector pathways
|
|
|
- Wilfred McBride
- 5 years ago
- Views:
Transcription
1 Vol September 2005 doi: /nature03966 The protein kinase A anchoring protein makap coordinates two integrated camp effector pathways Kimberly L. Dodge-Kafka 1, Joseph Soughayer 1, Genevieve C. Pare 2, Jennifer J. Carlisle Michel 1, Lorene K. Langeberg 1, Michael S. Kapiloff 2 & John D. Scott 1 Cyclic adenosine 3 0,5 0 -monophosphate (camp) is a ubiquitous mediator of intracellular signalling events. It acts principally through stimulation of camp-dependent protein kinases (PKAs) 1,2 but also activates certain ion channels and guanine nucleotide exchange factors (Epacs) 3. Metabolism of camp is catalysed by phosphodiesterases (PDEs) 4,5. Here we identify a camp-responsive signalling complex maintained by the musclespecific A-kinase anchoring protein (makap) that includes PKA, PDE4D3 and Epac1. These intermolecular interactions facilitate the dissemination of distinct camp signals through each effector protein. Anchored PKA stimulates PDE4D3 to reduce local camp concentrations, whereas an makap-associated ERK5 kinase module suppresses PDE4D3. PDE4D3 also functions as an adaptor protein that recruits Epac1, an exchange factor for the small GTPase Rap1, to enable camp-dependent attenuation of ERK5. Pharmacological and molecular manipulations of the makap complex show that anchored ERK5 can induce cardiomyocyte hypertrophy. Thus, two coupled camp-dependent feedback loops are coordinated within the context of the makap complex, suggesting that local control of camp signalling by AKAP proteins is more intricate than previously appreciated. Spatiotemporal control of camp flux requires the concerted action of adenylyl cyclases that synthesize camp, and phosphodiesterases that locally metabolize it into 5 0 -AMP (ref. 4). PDE binding proteins target distinct isozymes to specific subcellular sites 5 7.In cardiomyocytes, makap assembles a negative feedback loop containing PDE4D3 and PKA at the perinuclear membrane 8. PKA phosphorylation of Ser 13 on PDE4D3 increases binding to makap 9, whereas phosphorylation (of PDE4D3 by PKA) of Ser 54 enhances camp catabolism 10. This configuration may generate local fluctuations in camp and pulses of compartmentalized PKA activity 11. Here we used fluorescent reporters of PKA activity to test this model in cultured human and rat cells 12. AKAR2, a reversible A-kinase activity reporter 31, is an improved version of AKAR1. This chimaeric protein consists of cyan fluorescent protein (CFP), a consensus PKA substrate sequence, a forkhead-associated (FHA) domain that binds phosphoamino acids, and the yellow fluorescent protein (YFP) citrine (Fig. 1a). PKA phosphorylation of the consensus site engages the FHA domain, permitting fluorescence resonance energy transfer (FRET) between the fluorescent moieties. Two modified AKAR2 reporters were generated: AKAR PKA, which recruits PKA by means of a consensus anchoring sequence (Fig. 1b), and AKAR PKA PDE, which recruits PKA and tethers PDE4D3 through a binding site derived from makap (Fig. 1c). HeLa cells expressing AKAP PKA showed a rapid and sustained elevation of FRETon camp stimulation (Fig. 1b; red trace in Fig. 1d (n ¼ 10); Supplementary Video 1). In contrast, cells expressing AKAR PKA PDE showed a transient and less robust FRET response (Fig. 1c; green trace in Fig. 1d (n ¼ 13); Supplementary Video 2). The PKA antagonist H89 blocked all camp-dependent FRET changes (Fig. 1d). PKA activity was unaffected by the PDE3 inhibitor milrinone (10 mm). However, suppression of PKA activity was prevented when the same cells received the PDE4 inhibitor rolipram (1 mm) (Fig. 1e, n ¼ 10; Supplementary Video 3). Thus, recruitment of a PDE4 terminates activation of anchored PKA in the context of the AKAR PKA PDE reporter. ERK kinases phosphorylate PDE4D3 on Ser 579 to suppress phosphodiesterase activity 13. We predicted that makap might incorporate an ERK kinase module to counterbalance the anchored PDE4D3. Accordingly, sustained elevation of FRETon camp stimulation was observed in HeLa cells expressing AKAR PKA PDE plus constitutively active MEK, a kinase upstream of ERKs (black trace in Fig. 1f, n ¼ 11). From this, we suggest that ERK activity suppresses anchored phosphodiesterase activity. Complementary studies were performed in cultured rat neonatal ventriculocytes (RNVs). Treatment with 10% fetal calf serum activated ERK before immunoprecipitation of the makap complex. The associated PDE activity was attenuated by 52 ^ 9% (n ¼ 4, P, ) compared to unstimulated controls (Fig. 1g, column 2). The MEK inhibitor PD98059 (20 mm) blocked this effect (Fig. 1g, column 3, P, 0.002). Equivalent amounts of PDE4D3 were co-purified with the makap anchoring protein under each experimental condition, suggesting that the reduction in phosphodiesterase activity is not a consequence of displacing the enzyme from the makap complex (Fig. 1g, bottom panel). ERK activity was also measured in makapimmunoprecipitated complexes (Fig. 1h, column 1), and an increase of 3.9 ^ 0.7-fold (n ¼ 3, P, ) over the IgG control (immunoprecipitation with IgG) was observed (data not shown). PD98059 (2 mm) blocked makap-associated ERK activity (Fig. 1h, column 2, P, ). The PKA inhibitor PKI 5 24 peptide (50 nm) had no significant effect on ERK activity (Fig. 1h, column 3). Thus, makap-associated ERK activity suppresses anchored PDE4D3 activity. To determine which ERK family members might suppress makap-associated PDE4D3, we performed western blot analysis on makap immunoprecipitates from heart extracts. An 80-kDa protein that co-purified with makap was detected with an antibody that recognizes all ERK family members and also with an ERK5- specific antibody (Fig. 1i, top and middle panels, lane 3). ERK5 was enriched in makap immune complexes, but the 45-kDa ERK1/2 kinase was not (Fig. 1i, top panel, lane 1). The upstream kinase MEK5 (refs 14, 15) also co-precipitated with makap (Fig. 1i, bottom panel). Immunofluorescence detection of makap and ERK5 at the perinuclear membranes of hypertrophic RNVs confirmed the in situ assembly of this signalling complex 16 (Supplementary Fig. S1). 1 Howard Hughes Medical Institute, Vollum Institute and 2 Department of Pediatrics, Oregon Health and Sciences University, Portland, Oregon 97239, USA. Present address: Calhoun Center for Cardiology, University of Connecticut Health Center, 263 Farmington Avenue, Farmington, Connecticut 06030, USA. 574
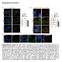
2 NATURE Vol September 2005 We wanted to determine whether anchored PKA and ERK5 facilitate bidirectional regulation of the makap-associated phosphodiesterase, hence controlling localized camp metabolism. Glutathione S-transferase (GST) pulldowns from heart extracts established the formation of a makap PDE4D3 ERK5 ternary complex (Supplementary Fig. S2). Cellular binding studies showed that PDE4D3 acts as an adaptor protein linking ERK5 to the makap complex. This was confirmed when a mutant PDE4D3 form unable to bind ERK kinases (the KIM/FQF double mutant, see Methods 17 ) could not recruit ERK5 to makap (Supplementary Figs S3 S5). Thus, ERK5 is well positioned to suppress PDE4D3 activity and influence localized accumulation of camp. camp modulates ERK signalling in a context-specific manner 18. camp activates ERK in neuronally derived cell lines, but it inhibits ERK in certain fibroblasts and kidney cells 19,20. We evaluated crosstalk between makap-anchored PKA and ERK5 in RNVs. When ERK was activated with 10% serum before immunoprecipitation of the makap complex, makap-associated ERK activity was increased 2.9 ^ 0.6-fold (n ¼ 3, P, ) over controls (Fig. 2a, column 2). However, pretreatment with forskolin (20 mm) to elevate intracellular camp resulted in an 89 ^ 8% reduction in the serum-dependent activation of the makap-associated ERK5 pool (n ¼ 3, P, ; Fig. 2a, column 3). To our surprise, the PKA antagonist H89 (10 mm) did not overcome the camp-dependent suppression of ERK5 activity (Fig. 2a, column 4). Similar results were obtained using two other well-characterized PKA inhibitors, KT5720 (1 mm) and Rp-cAMPs (1 mm; Fig. 2a, columns 5, 6). Thus, the camp-dependent block of ERK5 activity was independent of PKA. Control experiments show that this effect was not a consequence of displacing ERK5 from the makap complex (Fig. 2a, bottom). Therefore, another camp effector within the makap complex may function to suppress ERK5 activation. One such candidate is Epac1 (exchange protein directly activated by camp), a camp-dependent guanine nucleotide exchange factor for the small G-protein Rap1 (refs 21, 22). Epac1 co-purified with makap from heart extracts, but was not detected in an IgG control immunoprecipitation (Fig. 2b). Immunofluorescence techniques detected Epac1 at the perinuclear membranes of hypertrophic RNVs, where makap is also found (Fig. 2c e). The camp analogue 8-CPT-2 0 -O-Me-cAMP was used to assess whether Epac1 is the camp effector that inhibits makap-associated ERK5. This compound, also called 007, is a potent and selective activator of Epac proteins, with up to 300-fold selectivity over the R subunits of PKA 23. Treatment of RNVs with 007 alone resulted in the inhibition of ERK5 activity in makap immune complexes (Fig. 2f, column 3, n ¼ 3, P, ). KT5720 did not increase the effect of 007 (Fig. 2f, column 4), emphasizing that PKA does not have an additive effect on camp-dependent suppression of ERK5. Further biochemical experiments demonstrated that PDE4D3 recruits Epac1 to the makap complex (Supplementary Figs S6 S8). This permits firm control of camp concentrations in the vicinity of Epac1, thereby regulating its guanine nucleotide exchange activity. Epac1 is an effector protein of Rap1, a member of the Ras family of small G-proteins 21,22. Rap1 signalling is attenuated by the GTPase activating protein RapGAP 24. Adenovirus-mediated expression of constitutively active RapGAP in heart cells blocked Rap1 and, consequently, camp-mediated suppression of ERK5 activity (Fig. 2g, columns 1 3, n ¼ 4, P, 0.007). Treatment with the PKA inhibitor KT5720 did not reverse this effect (Fig. 2g, column 4). Figure 1 Bidirectional control of the makap-associated PDE4D3 activity. a, The modular composition and action of AKAR2. b, Schematic of AKAR2 PKA (top), and pseudo-coloured images of FRET changes in AKAR2 PKA-expressing HeLa cells stimulated with camp for 15 min (bottom). c, Schematic of AKAR2 PKA PDE (top), and pseudo-coloured images of FRET changes in AKAR2 PKA PDE-expressing HeLa cells stimulated with camp for 15 min (bottom). d, FRET measurements for AKAR PKA (red, n ¼ 10), AKAR PKA PDE (green, n ¼ 13), AKAR PKA þ H89 (blue, n ¼ 10) and AKAR PKA PDE þ H89 (black, n ¼ 7) for 15 min after camp stimulation with forskolin (arrow). e, FRET traces (n ¼ 10) using the AKAR PKA PDE reporter after application of the PDE3 inhibitor milrinone (0 12 min) and the PDE4 inhibitor rolipram (24 36 min). Stimulation with camp was at 0 and 24 min (arrows). f, FRET measurements from HeLa cells expressing the AKAR PKA PDE reporter in the presence (black, n ¼ 11) or absence (green, n ¼ 7) of a dominant active MEK5 for 15 min after camp stimulation with forskolin (arrow). g, Phosphodiesterase activity (n ¼ 4) in makap immune complexes isolated from RNVs. Treatment conditions are indicated. IB, immunoblot. h, ERK activity in makap complexes immunoprecipitated from heart extracts (n ¼ 3). Treatments with kinase inhibitors are indicated. Error bars in g, h show s.e.m. P values,0.01 are indicated by asterisks (black, relative to control; red, relative to sample). i, Co-precipitated ERK was detected by immunoblot of makap complexes from heart extracts using pan-erk (top), ERK5-specific (middle) and MEK5-specific (bottom) antibodies. 575
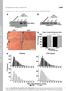
3 NATURE Vol September 2005 Other control experiments demonstrated that adenoviral expression of b-galactosidase did not affect camp-dependent suppression of anchored ERK5 activity (Fig. 2h, n ¼ 3, P, 0.009). Collectively, the data in Fig. 2 suggest that camp-dependent activation of Epac1 mobilizes a Rap1 signalling pathway, leading to inhibition of ERK5 in the makap complex. Cytokines, including leukaemia inhibitory factor (LIF), activate ERK5 in RNVs to induce eccentric cardiac hypertrophy, an increase in cell size and length 25. LIF treatment for 24 h enhanced RNV cell size by 1.6-fold over controls (Fig. 3a c, n ¼ 11 independent experiments). This effect was blocked when ERK kinases were inhibited by PD98059 or if Epac1 was activated by 007, and was partially blocked on PDE4 inhibition with rolipram (Fig. 3c, columns 3 5). Plasmid-based RNA interference of makap also suppressed LIF-mediated changes in RNV size compared with controls (Fig. 3d and Supplementary Fig. S9). The LIF response was rescued in RNVs Figure 2 Epac1 suppresses makap-associated ERK5 activity. a, Serumstimulated makap-associated ERK5 activity (n ¼ 3) in parallel cultures pretreated with forskolin to elevate camp (columns 2, 3) or in the presence of the PKA inhibitors H89, KT5720 or Rp-cAMPs (columns 4 6). Bottom panel shows the amount of ERK5 detected in each sample. b, Immunoblot detection of Epac1 in makap immune complexes from rat heart extracts. c e, Fluorescent staining of hypertrophic RNVs with an anti-epac1 antibody (c) and Alexa 568 phalliodin to reveal the actin cytoskeleton (d). Composite image (e) shows the distribution of Epac1 (green) and actin (red). Scale bar, 20 mm. f, The makap complex was immunoprecipitated from cultured RNVs following serum stimulation to activate ERK. Before serum stimulation, parallel cultures were pretreated with either the Epac-selective activator 007 or KT5720 (n ¼ 3). g, h, Serum-stimulated makap-associated ERK activity in cells expressing constitutively active RapGAP (g, n ¼ 4) or control b-galactosidase (h, n ¼ 3). Stimulation of intracellular camp or treatment with the kinase inhibitor KT5720 is indicated. P values,0.01 are indicated by asterisks (black, relative to control; red, relative to sample). Error bars in f h show s.e.m. 576 Figure 3 The makap complex facilitates cytokine-induced cardiac hypertrophy. a c, Leukaemia inhibitory factor (LIF) induces changes in RNV size. The outline of a rat neonatal ventriculocyte from control (a) and LIF-treated (b) samples is presented. Scale bar, 20 mm. c, Quantification of cell size in control RNVs (lane 1, n ¼ 11) and LIF-treated RNVs (lane 2 (n ¼ 11) and lanes 3 5). Pharmacological manipulation of ERK5 (PD98095, n ¼ 4), Epac1 (007, n ¼ 4) or PDE4 (rolipram, n ¼ 7) activity is indicated. d, Expression of makap was suppressed by RNA interference. Quantification of LIF-induced hypertrophy (cell size ^ LIF) in control RNVs (lane 1, n ¼ 12), makap-silenced RNVs (lane 2, n ¼ 19) and RNVs rescued with a variant of makap resistant to the shrna (lane 3, n ¼ 13). e, Displacement of makap from the nuclear membrane was achieved by overexpression of an makap targeting domain fragment using the TET OFF inducible promoter. Quantification of cell size from control (lane 1, n ¼ 3), from LIF-stimulated RNV controls (lane 2, n ¼ 4), and from LIFstimulated cells expressing the makap fragment (lane 3, n ¼ 4). P values are indicated by asterisks (black, relative to control; red, relative to sample): single asterisk, P, 0.05; two asterisks, P, Error bars in c e show s.e.m. f i, Schematics highlighting the main findings of this study. f, ERK5 phosphorylation of PDE4D3 shuts down camp metabolism. g, PKA phosphorylation of PDE4D3 enhances camp metabolism. h, Activation of Epac mobilizes Rap1 to suppress ERK5 activation. i, Low camp represses the Epac-mediated block of ERK5, allowing cardiac hypertrophy.
4 NATURE Vol September 2005 expressing a variant of makap resistant to the short hairpin (sh)rna (Fig. 3d, column 3). Finally, displacement of makap from the perinuclear membrane with an inducible competing makap fragment (residues ) blocked LIF-induced RNV growth (Fig. 3e and Supplementary Fig. S10). Collectively, these data suggest that makap organizes a camp-responsive network containing PDE4D3, Epac1 and ERK5 that modulates cardiomyocyte size. Similar results were observed when atrial natriuretic factor (ANF) expression was monitored as an independent index of hypertrophy (Supplementary Figs S11 and S12). b-adrenergic agonists that elevate intracellular camp control the strength, duration and frequency of heart contraction. Cell imaging experiments in cardiomyocytes have revealed transient microdomains of camp where PKA becomes active 26,27. Our FRET data show that co-localization of PKA and a phosphodiesterase generate localized pulses of camp. A variety of camp-responsive events that have different durations and are responsive to different thresholds of camp can emanate from the same microdomain. This is particularly relevant for makap signalling complexes, which contain three functionally distinct camp-dependent enzymes (PKA, PDE4D3 and Epac1). PKA responds to nanomolar concentrations of camp, and would become active early in a second messenger response. PDE4D3 (K m 1 4 mm) and Epac1 (K d 4 mm) would only become activated once camp concentrations reached micromolar levels 4. Conversely, inactivation of PDE4D3 and Epac1 would precede PKA holoenzyme reformation as camp levels decline. Furthermore, Epac1 suppresses makap-associated ERK5 activity in a PKAindependent manner. This is consistent with the idea that Epac proteins provide the camp-dependent link to ERK signalling events through Rap1 22,23,28. In conclusion, makap and PDE4D3 create an integrated and internally regulated signalling network. makap anchors PKA and directs the cellular localization of the complex, while PDE4D3 serves as the adaptor protein for Epac1 and ERK5. The key regulatory enzyme is PDE4D3. ERK phosphorylation of PDE4D3 decreases the phosphodiesterase activity, thereby favouring local accumulation of camp and subsequent PKA activation (Fig. 3f) and Epac1 activation (Fig. 3h). Conversely, PKA phosphorylation of PDE4D3 increases its affinity for makap 9 and its V max for camp 10, decreasing the local camp concentration (Fig. 3g). This ultimately sustains ERK5 activity by repressing Epac1-mediated inhibition of ERK5 (Fig. 3i). This configuration creates a complex in which transduction enzymes from distinct pathways are spatially organized to exploit the temporal constraints of camp action. METHODS Antibodies. Primary antibodies used were monoclonal anti-pan-erk (Transduction Labs), monoclonal anti-mek5 (Transduction Labs), monoclonal antipan-pde4d (ICOS), rabbit anti-erk5 (StressGen), monoclonal anti-9e10-myc (Upstate), rabbit anti-makap (VO54), rabbit anti-epac1 (Santa Cruz), mouse anti-a-actinin (Sigma) and rabbit anti-rat ANF (USB). Donkey HRP-conjugated (Jackson ImmunoResearch) and Alexa-conjugated (Molecular Probes) secondary antibodies were used. AKAR PKA and AKAR PKA PDE. Plasmids (pcdna4) encoding AKAR PKA or AKAR PKA PDE were derived from the original AKAR2 construct provided by R. Tsien. A DNA fragment encoding the consensus RII binding peptide from AKAPis (A-kinase anchoring protein-in silico: a 22-amino-acid peptide that is a high-affinity PKA-anchoring anagonist developed using a bioinformatic approach) following a Kozak sequence was generated by polymerase chain reaction (PCR). This fragment was ligated at the 5 0 end of AKAR2 to generate AKAR PKA. The AKAR PKA PDE reporter was constructed by ligating a fragment encoding residues of makap to the 3 0 end of the AKAR PKA coding region. Cell culture. Preparation of primary RNVs and treatment to induce hypertrophy was as previously described 16. Live cell imaging. HeLa cells grown on glass coverslips were transfected with vectors encoding AKAR PKA or AKAR PKA PDE using Lipofectamine After incubation at 37 8C for h, cells were washed twice with Hank s balanced salt solution, mounted in a Ludin chamber (Life Imaging Services) and imaged using a Leica AS MDW workstation. Images were acquired using a Leica DM IRE2 microscope equipped with a CoolSNAP-HQ charge-coupled device camera (Roper Photometrics). This device was equipped with a Dual-view module (Optical Insights), D465/30 and HQ535/30 emission filters (Chroma) and a 505dcxr dichroic mirror (Chroma). CFP and YFP images were acquired simultaneously, aligned, background-corrected and analysed using FRET Applicator software (Leica). Exposure time was from 250 1,000 ms, and images were taken every 15 s. Treatment with the PKA inhibitor H89 or isoform-specific PDE inhibitors preceded camp stimulation with forskolin by 10 min. Expression constructs. Plamids encoding wild-type PDE4D3 and mutant PDE4D3 (K455A/K456A/F597A/Q598A/F599A; this is the KIM/FQF double mutant) forms were from M. Houslay. pcdna3 expression vectors encoding MYC-tagged ERK5 and Epac1, and a RAP-GAP adenovirus, were provided by P. Stork. For GST PDE4D3 and GST ERK5, complementary DNAs were subcloned into PGEX-4T2. shrna vectors incorporated double-stranded hairpin oligonucleotides based on rat makap messenger RNA sequence (NCBI GI: , base pairs ) under the direction of the U6 promoter: makap shrna (sense strand), 5 0 -GACGAACCTTCCTTCCGAATTCAAGA GATTCGGAAGGAAGGTTCGTCTTTTT-3 0 ; control shrna (sense strand) 5 0 -GACGAACCCCTGTTCCGAATTCAAGAGATTCGGAACAGGGGTTCG TCTTTTT-3 0. Bold base pairs in the control shrna are different from the wildtype makap shrna. Rescue experiments were performed with a full-length MYC His rat makap lacking the shrna recognition sequence that resides in the 3 0 untranslated region of the makap message. Enzyme assays. PDE activity was measured by the method in ref. 29, and PKA activity was measured as previously described 8. ERK assays were performed on immunoprecipitates from heart extracts and RNVs. Immunoprecipitates were washed three times in HSE buffer before the addition of inhibitors, 25 ml H 2 O and 25 ml ERK assay buffer (80 mm HEPES ph 7.4, 80 mm MgCl 2, 0.2 mm ATP, 2 mm sodium orthovanadate, 20 mm NaF, 1 mg tyrosine hydroxylase peptide (BioSource) and 10 mci g- 32 ATP). After 30 min incubation at 30 8C, the reaction mixture was spotted onto phosphocellulose strips and extensively washed in 75 mm phosphoric acid. Filters were washed in 95% ethanol, air-dried and counted by liquid scintillation. RNVs were serum-starved for 16 h before the addition of either 20 mm PD98059, the PKA inhibitors H 89 (10 mm), KT5720 (1 mm) or Rp-cAMPs (1 mm), for 1 h. Forskolin (20 mm) and 3-isobutyl-1-methylxanthine (IBMX, 75 mm) were then added for 20 min. For 8-CPT-2 0 -O-Me-cAMP treatment (50 mm, the Epac-selective camp analogue 007), cells were not exposed to forskolin. Cells were stimulated with 10% fetal calf serum for 10 min, lysed in 1 ml HSE buffer containing 50 mm NaF, 1 mm sodium orthovanadate, 0.1 mm okadaic acid and 0.1 mm cyclosporine, and immunoprecipitations were performed as described above. H9C2 heart cells were incubated with adenoviral constructs of Rap1GAP1 or b-galactosidase at a multiplicity of infection (MOI) of 50 for 48 h. The cells were treated as described above before the addition of forskolin/ibmx and before serum stimulation. Immunocytochemistry and microscopy. Fixation and staining of RNVs were as previously described 8. To determine the effect of shrna expression on cellular hypertrophy, myocytes were co-transfected with ptre-u6 makap shrna or control shrna plasmid, pegfpn3 green fluorescent protein expression plasmid (Clontech) and either pbluescript carrier DNA or pcdna3.1 MYC His makap expression vector for rescue experiments. Cells were stained with primary and Alexa-conjugated specific secondary antibodies. Anti-a-actinin stained sarcomeric Z-disks to distinguish myocytes from contaminating fibroblasts. Images were acquired using a Leica DMRA fluorescent microscope. Cell size was measured using the method in ref. 30. Data are presented as the average mean cell area ^ s.e.m. P-values were determined using a two-tailed Student s t-test. Received 5 May; accepted 27 June Walsh, D. A., Perkins, J. P. & Krebs, E. G. An adenosine 3 0,5 0 -monophosphatedependent protein kinase from rabbit skeletal muscle. J. Biol. Chem. 243, (1968). 2. Su, Y. et al. Regulatory subunit of protein kinase A: structure of deletion mutant with camp binding proteins. Science 269, (1995). 3. Bos, J. L. Epac: a new camp target and new avenues in camp research. Nature Rev. Mol. Cell Biol. 4, (2003). 4. Beavo, J. A. & Brunton, L. L. Cyclic nucleotide research still expanding after half a century. Nature Rev. Mol. Cell Biol. 3, (2002). 5. Houslay, M. D. & Adams, D. R. PDE4 camp phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 370, 1-18 (2003). 6. Perry, S. J. et al. Targeting of cyclic AMP degradation to b2-adrenergic receptors by b-arrestins. Science 298, (2002). 577
5 NATURE Vol September Verde, I. et al. Myomegalin is a novel protein of the Golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J. Biol. Chem. 276, (2001). 8. Dodge, K. L. et al. makap assembles a protein kinase A/PDE4 phosphodiesterase camp signalling module. EMBO J. 20, (2001). 9. Carlisle Michel, J. J. et al. PKA phosphorylation of PDE4D3 facilitates recruitment of the makap signalling complex. Biochem. J. 381, (2004). 10. Sette, C. & Conti, M. Phosphorylation and activation of a camp-specific phosphodiesterase by the camp-dependent protein kinase. J. Biol. Chem. 271, (1996). 11. Wong, W. & Scott, J. D. AKAP signalling complexes: Focal points in space and time. Nature Rev. Mol. Cell Biol. 5, (2004). 12. Zhang, J., Ma, Y., Taylor, S. S. & Tsien, R. Y. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc. Natl Acad. Sci. USA 98, (2001). 13. Hoffmann, R., Baillie, G. S., MacKenzie, S. J., Yarwood, S. J. & Houslay, M. D. The MAP kinase ERK2 inhibits the cyclic AMP-specific phosphodiesterase HSPDE4D3 by phosphorylating it at Ser579. EMBO J. 18, (1999). 14. Zhou, G., Bao, Z. Q. & Dixon, J. E. Components of a new human protein kinase signal transduction pathway. J. Biol. Chem. 270, (1995). 15. English, J. M., Vanderbilt, C. A., Xu, S., Marcus, S. & Cobb, M. H. Isolation of MEK5 and differential expression of alternatively spliced forms. J. Biol. Chem. 270, (1995). 16. Kapiloff, M. S., Schillace, R. V., Westphal, A. M. & Scott, J. D. makap: an A- kinase anchoring protein targeted to the nuclear membrane of differentiated myocytes. J. Cell Sci. 112, (1999). 17. MacKenzie, S. J., Baillie, G. S., McPhee, I., Bolger, G. B. & Houslay, M. D. ERK2 mitogen-activated protein kinase binding, phosphorylation, and regulation of the PDE4D camp-specific phosphodiesterases. The involvement of COOHterminal docking sites and NH 2 -terminal UCR regions. J. Biol. Chem. 275, (2000). 18. Pearson, G. W. & Cobb, M. H. Cell condition-dependent regulation of ERK5 by camp. J. Biol. Chem. 277, (2002). 19. Cook, S. J. & McCormick, F. Inhibition by camp of Ras-dependent activation of Raf. Science 262, (1993). 20. Vaillancourt, R. R., Gardner, A. M. & Johnson, G. L. B-Raf-dependent regulation of the MEK-1/mitogen-activated protein kinase pathway in PC12 cells and regulation by cyclic AMP. Mol. Cell. Biol. 14, (1994). 21. de Rooij, J. et al. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396, (1998). 22. Kawasaki, H. et al. A family of camp-binding proteins that directly activate Rap1. Science 282, (1998). 23. Enserink, J. M. et al. A novel Epac-specific camp analogue demonstrates independent regulation of Rap1 and ERK. Nature Cell Biol. 4, (2002). 24. Rubinfeld, B. et al. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell 65, (1991). 25. Nicol, R. L. et al. Activated MEK5 induces serial assembly of sarcomeres and eccentric cardiac hypertrophy. EMBO J. 20, (2001). 26. Zaccolo, M. & Pozzan, T. Discrete microdomains with high concentration of camp in stimulated rat neonatal cardiac myocytes. Science 295, (2002). 27. Mongillo, M. et al. Fluorescence resonance energy transfer-based analysis of camp dynamics in live neonatal rat cardiac myocytes reveals distinct functions of compartmentalized phosphodiesterases. Circ. Res. 95, (2004). 28. Laroche-Joubert, N., Marsy, S., Michelet, S., Imbert-Teboul, M. & Doucet, A. Protein kinase A-independent activation of ERK and H,K-ATPase by camp in native kidney cells: role of Epac I. J. Biol. Chem. 277, (2002). 29. Beavo, J. A., Bechtel, P. J. & Krebs, E. G. Preparation of homogeneous cyclic AMP-dependent protein kinase(s) and its subunits from rabbit skeletal muscle. Methods Enzymol. 38, (1974). 30. Kodama, H. et al. Significance of ERK cascade compared with JAK/STAT and PI3-K pathway in gp130-mediated cardiac hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 279, H1635 -H1644 (2000). 31. Zhang, J., Hupfeld, C. J., Taylor, S. S., Olefsky, J. M. & Tsien, R. Y. Insulin disrupts b-adrenergic signalling to protein kinase A in adipocytes. Nature doi: /nature04140 (this issue). Supplementary Information is linked to the online version of the paper at Acknowledgements This work was supported by grants from the National Institutes of Health (to J.D.S. and M.S.K.) and the American Heart Association (to K.L.D.-K.). The authors wish to thank N. Mayer, D. Bleckinger and R. Mouton for technical assistance, and R. Tsien, M. Houslay, J. E. Dixon, J. L. Bos and P. Stork for reagents. Author Information Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to J.D.S. (scott@ohsu.edu). 578
Supplemental Online Material. The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/-
 #1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
#1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
PKA-phosphorylation of PDE4D3 facilitates recruitment of the makap signalling complex
 Biochem. J. (2004) 381, 587 592 (Printed in Great Britain) 587 ACCELERATED PUBLICATION PKA-phosphorylation of PDE4D3 facilitates recruitment of the makap signalling complex Jennifer J. CARLISLE MICHEL
Biochem. J. (2004) 381, 587 592 (Printed in Great Britain) 587 ACCELERATED PUBLICATION PKA-phosphorylation of PDE4D3 facilitates recruitment of the makap signalling complex Jennifer J. CARLISLE MICHEL
CRE/CREB Reporter Assay Kit camp/pka Cell Signaling Pathway Catalog #: 60611
 Data Sheet CRE/CREB Reporter Assay Kit camp/pka Cell Signaling Pathway Catalog #: 60611 Background The main role of the camp response element, or CRE, is mediating the effects of Protein Kinase A (PKA)
Data Sheet CRE/CREB Reporter Assay Kit camp/pka Cell Signaling Pathway Catalog #: 60611 Background The main role of the camp response element, or CRE, is mediating the effects of Protein Kinase A (PKA)
Supplementary methods Shoc2 In Vitro Ubiquitination Assay
 Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration
 /, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
/, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
Data Sheet. CRE/CREB Reporter Assay Kit (camp/pka Cell Signaling Pathway) Catalog #: 60611
 Data Sheet CRE/CREB Reporter Assay Kit (camp/pka Cell Signaling Pathway) Catalog #: 60611 Background The main role of the camp response element, or CRE, is mediating the effects of Protein Kinase A (PKA)
Data Sheet CRE/CREB Reporter Assay Kit (camp/pka Cell Signaling Pathway) Catalog #: 60611 Background The main role of the camp response element, or CRE, is mediating the effects of Protein Kinase A (PKA)
Dynamic Regulation of camp Synthesis through Anchored PKA-Adenylyl Cyclase V/VI Complexes
 Molecular Cell 23, 925 931, September 15, 2006 ª2006 Elsevier Inc. DOI 10.1016/j.molcel.2006.07.025 Dynamic Regulation of camp Synthesis through Anchored PKA-Adenylyl Cyclase V/VI Complexes Short Article
Molecular Cell 23, 925 931, September 15, 2006 ª2006 Elsevier Inc. DOI 10.1016/j.molcel.2006.07.025 Dynamic Regulation of camp Synthesis through Anchored PKA-Adenylyl Cyclase V/VI Complexes Short Article
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous
 Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Chapter One. Construction of a Fluorescent α5 Subunit. Elucidation of the unique contribution of the α5 subunit is complicated by several factors
 4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Transcriptional regulation of BRCA1 expression by a metabolic switch: Di, Fernandez, De Siervi, Longo, and Gardner. H3K4Me3
 ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
SUPPLEMENTARY INFORMATION FILE
 SUPPLEMENTARY INFORMATION FILE Existence of a microrna pathway in anucleate platelets Patricia Landry, Isabelle Plante, Dominique L. Ouellet, Marjorie P. Perron, Guy Rousseau & Patrick Provost 1. SUPPLEMENTARY
SUPPLEMENTARY INFORMATION FILE Existence of a microrna pathway in anucleate platelets Patricia Landry, Isabelle Plante, Dominique L. Ouellet, Marjorie P. Perron, Guy Rousseau & Patrick Provost 1. SUPPLEMENTARY
Gene Forward (5 to 3 ) Reverse (5 to 3 ) Accession # PKA C- TCTGAGGAAATGGGAGAACC CGAGGGTTTTCTTCCTCTCAA NM_011100
 Supplementary Methods: Materials. BRL37344, insulin, 3-isobutyl-1-methylxanthine, dibutyryl camp (Bt2-cAMP) and 8-Bromoadenosine 3,5 -cyclic monophosphate sodium (8-br-cAMP), cilostamide, adenosine deaminase,
Supplementary Methods: Materials. BRL37344, insulin, 3-isobutyl-1-methylxanthine, dibutyryl camp (Bt2-cAMP) and 8-Bromoadenosine 3,5 -cyclic monophosphate sodium (8-br-cAMP), cilostamide, adenosine deaminase,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10016 Supplementary discussion on binding site density for protein complexes on the surface: The density of biotin sites on the chip is ~10 3 biotin-peg per µm 2. The biotin sites are
doi:10.1038/nature10016 Supplementary discussion on binding site density for protein complexes on the surface: The density of biotin sites on the chip is ~10 3 biotin-peg per µm 2. The biotin sites are
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
Cyclic AMP-Mediated Inhibition of Cell Growth Requires the Small G Protein Rap1
 MOLECULAR AND CELLULAR BIOLOGY, June 2001, p. 3671 3683 Vol. 21, No. 11 0270-7306/01/$04.00 0 DOI: 10.1128/MCB.21.11.3671 3683.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved.
MOLECULAR AND CELLULAR BIOLOGY, June 2001, p. 3671 3683 Vol. 21, No. 11 0270-7306/01/$04.00 0 DOI: 10.1128/MCB.21.11.3671 3683.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved.
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Materials and Methods
 Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
pt7ht vector and over-expressed in E. coli as inclusion bodies. Cells were lysed in 6 M
 Supplementary Methods MIG6 production, purification, inhibition, and kinase assays MIG6 segment 1 (30mer, residues 334 364) peptide was synthesized using standard solid-phase peptide synthesis as described
Supplementary Methods MIG6 production, purification, inhibition, and kinase assays MIG6 segment 1 (30mer, residues 334 364) peptide was synthesized using standard solid-phase peptide synthesis as described
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Development of an AlphaLISA MEK1 Kinase Assay Using Full-Length ERK2 Substrate
 TECHNICAL NOTE Development of an AlphaLISA MEK1 Kinase Assay Using Full-Length ERK2 Substrate AlphaLISA Technology Author: Jeanine Hinterneder, PhD PerkinElmer, Inc. Hopkinton, MA Introduction The mitogen-activated
TECHNICAL NOTE Development of an AlphaLISA MEK1 Kinase Assay Using Full-Length ERK2 Substrate AlphaLISA Technology Author: Jeanine Hinterneder, PhD PerkinElmer, Inc. Hopkinton, MA Introduction The mitogen-activated
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
AKAR2-AKAP12 fusion protein "biosenses" dynamic phosphorylation and localization of a GPCR-based scaffold
 RESEARCH ARTICLE Open Access Research article -AKAP12 fusion protein "biosenses" dynamic phosphorylation and localization of a GPCR-based scaffold Jiangchuan Tao 1, Hsien-yu Wang 2 and Craig C Malbon*
RESEARCH ARTICLE Open Access Research article -AKAP12 fusion protein "biosenses" dynamic phosphorylation and localization of a GPCR-based scaffold Jiangchuan Tao 1, Hsien-yu Wang 2 and Craig C Malbon*
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
Supplementary Material
 Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
camp ELISA Kit (Colorimetric)
 Product Manual camp ELISA Kit (Colorimetric) Catalog Numbers STA-500 STA-500-5 96 assays 5 x 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Adenosine 3,5 -cyclic monophosphate
Product Manual camp ELISA Kit (Colorimetric) Catalog Numbers STA-500 STA-500-5 96 assays 5 x 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Adenosine 3,5 -cyclic monophosphate
The MAP Kinase Family
 The MAP Kinase Family Extracellular stimuli Classical MAP kinases Atypical MAP kinases MAPKKK MLK1/2/3/7; LZK RAF-1/A/B TAK1; TPL2 c-mos MEKK1-4; DLK ASK1/2; MLTK TAO1/2 ASK1 TAK1 MEKK1-4 MEKK2/3 TPL2???
The MAP Kinase Family Extracellular stimuli Classical MAP kinases Atypical MAP kinases MAPKKK MLK1/2/3/7; LZK RAF-1/A/B TAK1; TPL2 c-mos MEKK1-4; DLK ASK1/2; MLTK TAO1/2 ASK1 TAK1 MEKK1-4 MEKK2/3 TPL2???
Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in
 Supplementary information Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer s disease Zhentao Zhang, Mingke Song, Xia Liu, Seong Su Kang, Il-Sun Kwon, Duc
Supplementary information Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer s disease Zhentao Zhang, Mingke Song, Xia Liu, Seong Su Kang, Il-Sun Kwon, Duc
Supplementary Information for. Regulation of Rev1 by the Fanconi Anemia Core Complex
 Supplementary Information for Regulation of Rev1 by the Fanconi Anemia Core Complex Hyungjin Kim, Kailin Yang, Donniphat Dejsuphong, Alan D. D Andrea* *Corresponding Author: Alan D. D Andrea, M.D. Alan_dandrea@dfci.harvard.edu
Supplementary Information for Regulation of Rev1 by the Fanconi Anemia Core Complex Hyungjin Kim, Kailin Yang, Donniphat Dejsuphong, Alan D. D Andrea* *Corresponding Author: Alan D. D Andrea, M.D. Alan_dandrea@dfci.harvard.edu
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/429/ra54/dc1 Supplementary Materials for Dephosphorylation of the adaptor LAT and phospholipase C by SHP-1 inhibits natural killer cell cytotoxicity Omri Matalon,
www.sciencesignaling.org/cgi/content/full/9/429/ra54/dc1 Supplementary Materials for Dephosphorylation of the adaptor LAT and phospholipase C by SHP-1 inhibits natural killer cell cytotoxicity Omri Matalon,
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the
 Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
Supplemental Information
 Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplemental Data Supplementary Figure Legends and Scheme Figure S1.
 Supplemental Data Supplementary Figure Legends and Scheme Figure S1. UTK1 inhibits the second EGF-induced wave of lamellipodia formation in TT cells. A and B, EGF-induced lamellipodia formation in TT cells,
Supplemental Data Supplementary Figure Legends and Scheme Figure S1. UTK1 inhibits the second EGF-induced wave of lamellipodia formation in TT cells. A and B, EGF-induced lamellipodia formation in TT cells,
Chapter 14 Regulation of Transcription
 Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
15 June 2011 Supplementary material Bagriantsev et al.
 Supplementary Figure S1 Characterization of K 2P 2.1 (TREK-1) GOF mutants A, Distribution of the positions of mutated nucleotides, represented by a red x, from a pool of 18 unselected K 2P 2.1 (KCNK2)
Supplementary Figure S1 Characterization of K 2P 2.1 (TREK-1) GOF mutants A, Distribution of the positions of mutated nucleotides, represented by a red x, from a pool of 18 unselected K 2P 2.1 (KCNK2)
Mechanism of action-1
 Mechanism of action-1 receptors: mediators of hormone action, membrane associated vs. intracellular receptors: measurements of receptor - ligand interaction, mechanism surface-receptors: kinases, phosphatases,
Mechanism of action-1 receptors: mediators of hormone action, membrane associated vs. intracellular receptors: measurements of receptor - ligand interaction, mechanism surface-receptors: kinases, phosphatases,
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
CAP BIOINFORMATICS Su-Shing Chen CISE. 10/5/2005 Su-Shing Chen, CISE 1
 CAP 5510-9 BIOINFORMATICS Su-Shing Chen CISE 10/5/2005 Su-Shing Chen, CISE 1 Basic BioTech Processes Hybridization PCR Southern blotting (spot or stain) 10/5/2005 Su-Shing Chen, CISE 2 10/5/2005 Su-Shing
CAP 5510-9 BIOINFORMATICS Su-Shing Chen CISE 10/5/2005 Su-Shing Chen, CISE 1 Basic BioTech Processes Hybridization PCR Southern blotting (spot or stain) 10/5/2005 Su-Shing Chen, CISE 2 10/5/2005 Su-Shing
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary information
 Supplementary information Table of Content: Supplementary Results... 2 Supplementary Figure S1: Experimental validation of AP-MS results by coimmunprecipitation Western blot analysis.... 3 Supplementary
Supplementary information Table of Content: Supplementary Results... 2 Supplementary Figure S1: Experimental validation of AP-MS results by coimmunprecipitation Western blot analysis.... 3 Supplementary
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Human/Mouse/Rat Phospho-CREB (S133) Immunoassay. An ELISA-based assay using fluorogenic substrates to measure phosphorylated CREB in whole cells.
 Cell-Based ELISA Human/Mouse/Rat Phospho-CREB (S133) Immunoassay Catalog Number KCB2510 An ELISA-based assay using fluorogenic substrates to measure phosphorylated CREB in whole cells. This package insert
Cell-Based ELISA Human/Mouse/Rat Phospho-CREB (S133) Immunoassay Catalog Number KCB2510 An ELISA-based assay using fluorogenic substrates to measure phosphorylated CREB in whole cells. This package insert
Supplementary Figure 1.
 Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Z -LYTE fluorescent kinase assay technology. Z -LYTE Kinase Assay Platform
 Z -LYTE fluorescent kinase assay technology Z -LYTE Kinase Assay Platform Z -LYTE Kinase Assay Platform Non-radioactive assay format for screening a diverse collection of over 185 kinase targets Assay
Z -LYTE fluorescent kinase assay technology Z -LYTE Kinase Assay Platform Z -LYTE Kinase Assay Platform Non-radioactive assay format for screening a diverse collection of over 185 kinase targets Assay
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells
 SUPPLEMENTARY FIGURES Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells Niklas Engels, Lars Morten König, Christina Heemann, Johannes
SUPPLEMENTARY FIGURES Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells Niklas Engels, Lars Morten König, Christina Heemann, Johannes
Supplementary Information
 Supplementary Information Supplementary Figure S1 (a) P-cRAF colocalizes with LC3 puncta. Immunofluorescence (IF) depicting colocalization of P-cRAF (green) and LC3 puncta (red) in NIH/3T3 cells treated
Supplementary Information Supplementary Figure S1 (a) P-cRAF colocalizes with LC3 puncta. Immunofluorescence (IF) depicting colocalization of P-cRAF (green) and LC3 puncta (red) in NIH/3T3 cells treated
Culture media, trypsin, penicillin and streptomycin were from Invitrogen (Breda, the Netherlands).
 Methods Materials Culture media, trypsin, penicillin and streptomycin were from Invitrogen (Breda, the Netherlands). Bovine fibroblast growth factor (BFGF), thrombin, forskolin, IBMX, H-89, BAPTA-AM and
Methods Materials Culture media, trypsin, penicillin and streptomycin were from Invitrogen (Breda, the Netherlands). Bovine fibroblast growth factor (BFGF), thrombin, forskolin, IBMX, H-89, BAPTA-AM and
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
No wash 2 Washes 2 Days ** ** IgG-bead phagocytosis (%)
 Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
SUPPLEMENTARY INFORMATION
 The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression
 Supplemental figures Supplemental Figure. 1. Silencing expression of Celsr3 by shrna. To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression plasmids for the shrna
Supplemental figures Supplemental Figure. 1. Silencing expression of Celsr3 by shrna. To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression plasmids for the shrna
Supplementary information
 Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
hnrnp C promotes APP translation by competing with FMRP for APP mrna recruitment to P bodies
 hnrnp C promotes APP translation by competing with for APP mrna recruitment to P bodies Eun Kyung Lee 1, Hyeon Ho Kim 1, Yuki Kuwano 1, Kotb Abdelmohsen 1, Subramanya Srikantan 1, Sarah S. Subaran 2, Marc
hnrnp C promotes APP translation by competing with for APP mrna recruitment to P bodies Eun Kyung Lee 1, Hyeon Ho Kim 1, Yuki Kuwano 1, Kotb Abdelmohsen 1, Subramanya Srikantan 1, Sarah S. Subaran 2, Marc
Cyclic AMP Controls mtor through Regulation of the Dynamic Interaction between Rheb and Phosphodiesterase 4D
 MOLECULAR AND CELLULAR BIOLOGY, Nov. 2010, p. 5406 5420 Vol. 30, No. 22 0270-7306/10/$12.00 doi:10.1128/mcb.00217-10 Copyright 2010, American Society for Microbiology. All Rights Reserved. Cyclic AMP Controls
MOLECULAR AND CELLULAR BIOLOGY, Nov. 2010, p. 5406 5420 Vol. 30, No. 22 0270-7306/10/$12.00 doi:10.1128/mcb.00217-10 Copyright 2010, American Society for Microbiology. All Rights Reserved. Cyclic AMP Controls
A guide to selecting control, diluent and blocking reagents
 Specializing in Secondary Antibodies and Conjugates A guide to selecting control, diluent and blocking reagents Optimize your experimental protocols with Jackson ImmunoResearch Secondary antibodies and
Specializing in Secondary Antibodies and Conjugates A guide to selecting control, diluent and blocking reagents Optimize your experimental protocols with Jackson ImmunoResearch Secondary antibodies and
A guide to selecting control, diluent and blocking reagents
 Specializing in Secondary Antibodies and Conjugates A guide to selecting control, diluent and blocking reagents Optimize your experimental protocols with Jackson ImmunoResearch Secondary antibodies and
Specializing in Secondary Antibodies and Conjugates A guide to selecting control, diluent and blocking reagents Optimize your experimental protocols with Jackson ImmunoResearch Secondary antibodies and
Supplemental Table 1: Sequences of real time PCR primers. Primers were intronspanning
 Symbol Accession Number Sense-primer (5-3 ) Antisense-primer (5-3 ) T a C ACTB NM_001101.3 CCAGAGGCGTACAGGGATAG CCAACCGCGAGAAGATGA 57 HSD3B2 NM_000198.3 CTTGGACAAGGCCTTCAGAC TCAAGTACAGTCAGCTTGGTCCT 60
Symbol Accession Number Sense-primer (5-3 ) Antisense-primer (5-3 ) T a C ACTB NM_001101.3 CCAGAGGCGTACAGGGATAG CCAACCGCGAGAAGATGA 57 HSD3B2 NM_000198.3 CTTGGACAAGGCCTTCAGAC TCAAGTACAGTCAGCTTGGTCCT 60
Supplementary methods
 Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Supplementary Information
 Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
Supplemental Materials and Methods
 Supplemental Materials and Methods In situ hybridization In situ hybridization analysis of HFE2 and genin mrna in rat liver tissues was performed as previously described (1). Briefly, the digoxigenin-labeled
Supplemental Materials and Methods In situ hybridization In situ hybridization analysis of HFE2 and genin mrna in rat liver tissues was performed as previously described (1). Briefly, the digoxigenin-labeled
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1.
 Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
In vitro EGFP-CALI comprehensive analysis. Liang CAI. David Humphrey. Jessica Wilson. Ken Jacobson. Dept. of Cell and Developmental Biology
 In vitro EGFP-CALI comprehensive analysis Liang CAI David Humphrey Jessica Wilson Ken Jacobson Dept. of Cell and Developmental Biology 1/15 Liang s Rotation in Ken s Lab, Sep-Nov, 2003 1. CALI background
In vitro EGFP-CALI comprehensive analysis Liang CAI David Humphrey Jessica Wilson Ken Jacobson Dept. of Cell and Developmental Biology 1/15 Liang s Rotation in Ken s Lab, Sep-Nov, 2003 1. CALI background
HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid
 SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
Supplemental Materials and Methods
 Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
Control of Metabolic Processes
 Control of Metabolic Processes Harriet Wilson, Lecture Notes Bio. Sci. 4 - Microbiology Sierra College As described earlier, the metabolic processes occurring within living organisms (glycolysis, respiration,
Control of Metabolic Processes Harriet Wilson, Lecture Notes Bio. Sci. 4 - Microbiology Sierra College As described earlier, the metabolic processes occurring within living organisms (glycolysis, respiration,
Solution key. c) Consider the following perturbations in different components of this signaling pathway in cells.
 Solution key Question 1 During a summer hike you suddenly spy a grizzly bear. This triggers a fight or flight response activating the signaling pathway that is shown on last Page. a) Circle the best option.
Solution key Question 1 During a summer hike you suddenly spy a grizzly bear. This triggers a fight or flight response activating the signaling pathway that is shown on last Page. a) Circle the best option.
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Supplementary material for: Materials and Methods:
 Supplementary material for: Iron-responsive degradation of iron regulatory protein 1 does not require the Fe-S cluster: S.L. Clarke, et al. Materials and Methods: Fe-S Cluster Reconstitution: Cells treated
Supplementary material for: Iron-responsive degradation of iron regulatory protein 1 does not require the Fe-S cluster: S.L. Clarke, et al. Materials and Methods: Fe-S Cluster Reconstitution: Cells treated
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
TECHNICAL NOTE Phosphorylation Site Specific Aptamers for Cancer Biomarker Peptide Enrichment and Detection in Mass Spectrometry Based Proteomics
 TECHNICAL NOTE Phosphorylation Site Specific Aptamers for Cancer Biomarker Peptide Enrichment and Detection in Mass Spectrometry Based Proteomics INTRODUCTION The development of renewable capture reagents
TECHNICAL NOTE Phosphorylation Site Specific Aptamers for Cancer Biomarker Peptide Enrichment and Detection in Mass Spectrometry Based Proteomics INTRODUCTION The development of renewable capture reagents
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Regulation of autophagic activity by ζ proteins associated with class III. phosphatidylinositol-3 kinase. Mercedes Pozuelo Rubio
 ONLINE SUPPORTING INFORMATION Regulation of autophagic activity by 14-3-3ζ proteins associated with class III phosphatidylinositol-3 kinase Mercedes Pozuelo Rubio entro Andaluz de Biología Molecular y
ONLINE SUPPORTING INFORMATION Regulation of autophagic activity by 14-3-3ζ proteins associated with class III phosphatidylinositol-3 kinase Mercedes Pozuelo Rubio entro Andaluz de Biología Molecular y
Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and
 SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
Flag-Rac Vector V12 V12 N17 C40. Vector C40 pakt (T308) Akt1. Myc-DN-PAK1 (N-SP)
 a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
Supplementary Fig.1 Luton
 Supplementary Fig.1 Luton a 175 Brain Thymus Spleen Small Intestine Kidney Testis HeLa b 250 Lung Kidney MDCK c EFA6B si Control si Mismatch #637 #1564 #1770 83 62 47.5 175 IB: anti-efa6b #B1 130 66 Lysates
Supplementary Fig.1 Luton a 175 Brain Thymus Spleen Small Intestine Kidney Testis HeLa b 250 Lung Kidney MDCK c EFA6B si Control si Mismatch #637 #1564 #1770 83 62 47.5 175 IB: anti-efa6b #B1 130 66 Lysates
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Molecular Techniques. 3 Goals in Molecular Biology. Nucleic Acids: DNA and RNA. Disclaimer Nucleic Acids Proteins
 Molecular Techniques Disclaimer Nucleic Acids Proteins Houpt, CMN, 9-30-11 3 Goals in Molecular Biology Identify All nucleic acids (and proteins) are chemically identical in aggregate - need to identify
Molecular Techniques Disclaimer Nucleic Acids Proteins Houpt, CMN, 9-30-11 3 Goals in Molecular Biology Identify All nucleic acids (and proteins) are chemically identical in aggregate - need to identify
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
