Supplementary Material for Yang et al., Generation of oligodendroglial cells by direct lineage conversion
|
|
|
- Grant Hood
- 5 years ago
- Views:
Transcription
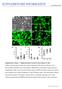
1 Supplementary Material for Yang et al., Generation of oligodendroglial cells by direct lineage conversion Supplementary Figure 1. The presence of Plp::EGFP + /O4 + cells 21 days after transgene induction. Plp::GFP MEF was transduced with a pool of 10 lentiviruses containing candidate genes. 21 days after transduction, cells expressing Plp::EGFP and O4 were detected. Scale bar: 50 µm.
2 Supplementary Figure 2. Combinations of three transcription factors induce Plp::EGFP expression at different levels. FACS analyses for GFP expression in mouse fibroblasts were performed 1 week after transgene induction.
3 Supplementary Figure 3. O4 + cells proliferate in response to PDGF signal. (a) Fourteen days after transduction, thymidine analogue EdU was added into the media. Six hours after the addition of EdU, cells were fixed for immunostaining using O4 antibody and EdU was detected using the Click-iT EdU cell proliferation kit (Invitrogen). Scale bar: 50 µm. (b) 11.39% ± 6.87 SD of the O4 + cells incorporated EdU within 6 hours.
4 Supplementary Figure 4. Astrocytes but not neurons were generated through reprogramming. (a) GFAP+/O4- astrocytes were detected 20 days after the induction, which constituted 18.11±7.83% of the entire culture. (b) After the purification and differentiation of O4+ cells, GFAP+/Nestin+ astrocyte-looking cells were detected. Scale bar: 50 µm.
5 Supplementary Figure 5. Screen of antibodies recognizing oligodendroglial cells using fibroblasts, OPCs and oligodendrocytes. From the tested panel of antibodies, only S100, O4, Olig2, MBP, MOG, CC1 and CNPase were specifically detected in OPCs or oligodendrocytes.
6 Supplementary Figure 6. Expression of reprogramming factors in O4 + cells. (a) Map of the lentiviral vector used to introduce the reprogramming factors and the schematic drawing of the experimental setup. The forward primer was specific to the candidate gene and the reverse primer was specific to the woodchuck response elements (WRE). (b) Genomic DNA was extracted from O4-immunopanned cells and PCR analysis showed the integration of individual provirus that express specific factors into the genome of O4 + cells.
7 Supplementary Figure 7. The graft-derived MBP+ cells do not express markers of periphery lineage. (a and c) The graft-derived MBP+ cells do not express P0 or Peripherin. (b and d) Positive stainings of P0 and Peripherin in the mouse spinal cord. Scale bar: 50 µm.
8 Supplementary Figure 8. Identification and imaging of the graft-derived myelination in the shiverer mouse corpus callosum. (a) Immunofluorescence of MPB + cells in the shiverer mouse corpus callosum. The asterisks mark two vessels in the vibratome section. (b) The region containing MBP + cells was micro-dissected, post-fixed, epon-imbedded and cut into semi-thin (1 µm)
9 sections. The framed region in (a) was identified in the semi-thin section stained by Toluidine Blue. The asterisks indicate the same vessels observed in (a). The numbers in the red frame in (b) labels the landmarks seen in both (b) and (c), including the small vessels (1, 2 and 6) and neuron soma (3-5). (c) Electronic microscope images of the ultrathin (80-90 nm) sections focusing on the region framed in (b) using the same landmarks. Colored frames highlight the cells shown in (d,e,f). The cell in (e) is the same cell shown in Fig.5g in lower magnification. The numbered signatures correlated to the same ones in (b). (d,e,f) High-magnification TEM images showed the myelin sheath and inserts highlight the ultra-structures. C.C: corpus callosum. Scale bars: 500 nm.
10 Supplementary Table 1 Primers used in this study for qrt-pcr analysis Gene Symbol Forward Primer Reverse Primer Amplicon Size Accession Number mbp taccctggctaaagcagagc gggagccgtagtgggtagtt 111nt NM_ ENSRNOT00000 nkx2-2 gcagcgacaacccctaca acttggagctcgagtcttgg 105nt sox6 caaaggacgaaaggaggaaa ggatttccagcgagatccta 89nt NM_ sox2 attacccgcagcaaaatgac tttttgcgttaatttggatgg 65nt NM_ plp1 ctgccagtctattgccttcc agcattccatgggagaacac 94nt NM_ tubb4 gtcttccgaggacggatgt ttgctctgcacacttagcatc 62nt NM_ col5a1 gaattcaagcgtgggaaact ggcagaggcactcagcag 99nt NM_ col1a1 tgcttgaagacctatgtgggta aaaggcagcatttggggtat 71nt NM_ beta-actin cccgcgagtacaaccttct cgtcatccatggcgaact 72nt NM_
11 Supplementary Table 2 List of genes with higher expression level in the iopcs compared to fibroblasts, OPCs and differentiated OPCs. Gene Title Gene Symbol (iopc vs. Fibroblasts) Fold-Change (iopc vs. day3 OL) (iopc vs. day6 OL) (iopc vs. OPC) myelin protein zero Mpz region containing similar to NGF-binding Ig light chain LOC tyrosinase-related protein 1 Tyrp cadherin 1 Cdh desert hedgehog homolog (Drosophila) Dhh hydroxy-3-methylglutaryl-Coenzyme A synthase 2 Hmgcs olfactomedin-like 2A Olfml2a laminin, alpha 2 Lama CUE domain containing 2 Cuedc prepronociceptin Pnoc neurofilament, light polypeptide Nefl sodium channel, voltage-gated, type VII, alpha Scn7a hypothetical protein LOC LOC sulfotransferase family, cytosolic, 1B, member 1 Sult1b reelin Reln hypothetical protein LOC LOC myelin protein zero Mpz neuropeptide Y Npy melanoma inhibitory activity Mia superoxide dismutase 3, extracellular Sod fibrinogen beta chain Fgb calpain 6 Capn family with sequence similarity 180, member A Fam180a decorin Dcn
12 Supplementary Table 3 GO analyses of genes with higher expression level in the iopcs compared to fibroblasts, OPCs and differentiated OPCs. GO enrichment P-Value extracellular structure organization 6.83E-05 extracellular matrix organization 3.88E-04 transmission of nerve impulse response to organic substance thyroid hormone metabolic process protein polymerization
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplementary Information
 Supplementary Information Supplementary Figures Supplementary Figure 1. qrt-pcr analysis of the expression of candidate factors including SOX2, MITF, PAX3, DLX5, FOXD3, LEF1, MSX1, PAX6, SOX2 and SOX9
Supplementary Information Supplementary Figures Supplementary Figure 1. qrt-pcr analysis of the expression of candidate factors including SOX2, MITF, PAX3, DLX5, FOXD3, LEF1, MSX1, PAX6, SOX2 and SOX9
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Journal: Nature Neuroscience
 Journal: Nature Neuroscience Article Title: Corresponding Author: Yy1 as molecular link between neuregulin and transcriptional modulation of peripheral myelination. Patrizia Casaccia Supplementary Item
Journal: Nature Neuroscience Article Title: Corresponding Author: Yy1 as molecular link between neuregulin and transcriptional modulation of peripheral myelination. Patrizia Casaccia Supplementary Item
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Mass spectrometry characterization of epoxide reactions.
 Supplementary Figure 1 Mass spectrometry characterization of epoxide reactions. (a) Degree of amine reactivity of bovine serum albumin (BSA) with epoxide molecules having different numbers of epoxide groups:
Supplementary Figure 1 Mass spectrometry characterization of epoxide reactions. (a) Degree of amine reactivity of bovine serum albumin (BSA) with epoxide molecules having different numbers of epoxide groups:
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
Supplementary Figure 1.
 Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
SUPPLEMENTARY INFORMATION
 a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
SUPPLEMENTARY INFORMATION
 Supplementary Figure a 3 factors 4 factors Nanog-GFP phase b c.5 Endogenous Oct3/4 2 Total Oct3/4.5 Relative expression (ES = ).5.5 6 3 Endogenous Sox2 Nanog Relative expression (ES = ) 5 25 2 Total Sox2
Supplementary Figure a 3 factors 4 factors Nanog-GFP phase b c.5 Endogenous Oct3/4 2 Total Oct3/4.5 Relative expression (ES = ).5.5 6 3 Endogenous Sox2 Nanog Relative expression (ES = ) 5 25 2 Total Sox2
Changing fibroblasts into neurons: Direct lineage conversion. Journal Club Uli Herrmann
 Changing fibroblasts into neurons: Direct lineage conversion Journal Club Uli Herrmann 25.6.13 Outline Epigenetic landscape model Reprogramming cells Induced pluripotent stem cells Three papers: Direct
Changing fibroblasts into neurons: Direct lineage conversion Journal Club Uli Herrmann 25.6.13 Outline Epigenetic landscape model Reprogramming cells Induced pluripotent stem cells Three papers: Direct
Supplementary Information
 Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
PrimePCR Assay Validation Report
 Gene Information Gene Name integrin, alpha 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID ITGA1 Human This gene encodes the alpha 1 subunit of integrin
Gene Information Gene Name integrin, alpha 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID ITGA1 Human This gene encodes the alpha 1 subunit of integrin
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and
 Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10202 Supplementary Figure 1: Rapid generation of neurons from human ES cells. a, Phase contrast image of feeder-free culture of human ES cells infected with Brn2, Ascl1, Myt1l (BAM)
doi:10.1038/nature10202 Supplementary Figure 1: Rapid generation of neurons from human ES cells. a, Phase contrast image of feeder-free culture of human ES cells infected with Brn2, Ascl1, Myt1l (BAM)
Supplemental Figure 1.
 Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Information
 Cell Stem Cell, Volume 12 Supplemental Information Human ipsc-derived Oligodendrocyte Progenitor Cells Can Myelinate and Rescue a Mouse Model of Congenital Hypomyelination Su Wang, Janna Bates, Xiaojie
Cell Stem Cell, Volume 12 Supplemental Information Human ipsc-derived Oligodendrocyte Progenitor Cells Can Myelinate and Rescue a Mouse Model of Congenital Hypomyelination Su Wang, Janna Bates, Xiaojie
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix
 Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix Authors Kerim B. Kaylan, 1,2 Viktoriya Ermilova, 1,2 Ravi Chandra Yada,
Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix Authors Kerim B. Kaylan, 1,2 Viktoriya Ermilova, 1,2 Ravi Chandra Yada,
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Data. Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells
 Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Functional assays of hepatocyte-like cells induced from H1 hescs (a) P450 and AAT staining, PAS staining, LDL uptake assay, and ICG uptake and release assay
Supplementary Figure 1 Supplementary Figure 1. Functional assays of hepatocyte-like cells induced from H1 hescs (a) P450 and AAT staining, PAS staining, LDL uptake assay, and ICG uptake and release assay
Supplementary Figure 1 Characterization of OKSM-iNSCs
 Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State
 Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction)
 Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
Supplementary Data. Supplementary Table S1.
 Supplementary Data Supplementary Table S1. Primer Sequences for Real-Time Reverse Transcription-Polymerase Chain Reaction Gene Forward 5?3 Reverse 3?5 Housekeeping gene 36B4 TCCAGGCTTTGGGCATCA CTTTATCAGCTGCACATCACTCAGA
Supplementary Data Supplementary Table S1. Primer Sequences for Real-Time Reverse Transcription-Polymerase Chain Reaction Gene Forward 5?3 Reverse 3?5 Housekeeping gene 36B4 TCCAGGCTTTGGGCATCA CTTTATCAGCTGCACATCACTCAGA
Supplementary Table-1: List of genes that were identically matched between the ST2 and
 Supplementary data Supplementary Table-1: List of genes that were identically matched between the ST2 and ST3. Supplementary Table-2: List of genes that were differentially expressed in GD2 + cells compared
Supplementary data Supplementary Table-1: List of genes that were identically matched between the ST2 and ST3. Supplementary Table-2: List of genes that were differentially expressed in GD2 + cells compared
Gene name forward primer 5->3 reverse primer 5->3 product GCCCATA. Nfatc1 AGGTGCAGCCCAAGTCTCAC GTGGCCATCTGGAGCCTTC CTGT GATGC GCT ACCAA
 Supplementary able 1 is presenting the PCR primer list used. ene name forward primer 5->3 reverse primer 5->3 product size cdna VE-Cadherin AACCCAAAAA CACCCCAAAAC 260bp CCA CCCAA PECAM-1 CACCACAA CCCCCACC
Supplementary able 1 is presenting the PCR primer list used. ene name forward primer 5->3 reverse primer 5->3 product size cdna VE-Cadherin AACCCAAAAA CACCCCAAAAC 260bp CCA CCCAA PECAM-1 CACCACAA CCCCCACC
Supplemental Data. Sethi et al. (2014). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplementary Data. Flvcr1a TCTAAGGCCCAGTAGGACCC GGCCTCAACTGCCTGGGAGC AGAGGGCAACCTCGGTGTCC
 Supplementary Data Supplementary Materials and Methods Measurement of reactive oxygen species accumulation in fresh intestinal rings Accumulation of reactive oxygen species in fresh intestinal rings was
Supplementary Data Supplementary Materials and Methods Measurement of reactive oxygen species accumulation in fresh intestinal rings Accumulation of reactive oxygen species in fresh intestinal rings was
A) B) Ladder. Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical
 A) B) Ladder C) r4 r4 Nt- -Ct 78 kda Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical calpains is shown. The protease core consists of
A) B) Ladder C) r4 r4 Nt- -Ct 78 kda Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical calpains is shown. The protease core consists of
Supplementary Figures and supplementary figure legends
 Supplementary Figures and supplementary figure legends Figure S1. Effect of different percentage of FGF signaling knockdown on TGF signaling and EndMT marker gene expression. HUVECs were subjected to different
Supplementary Figures and supplementary figure legends Figure S1. Effect of different percentage of FGF signaling knockdown on TGF signaling and EndMT marker gene expression. HUVECs were subjected to different
Supplementary Information. A novel human endogenous retroviral protein inhibits cell-cell fusion. Supplementary Figures:
 Supplementary Information A novel human endogenous retroviral protein inhibits cell-cell fusion Jun Sugimoto, Makiko Sugimoto, Helene Bernstein, Yoshihiro Jinno and Danny J. Schust Supplementary Figures:
Supplementary Information A novel human endogenous retroviral protein inhibits cell-cell fusion Jun Sugimoto, Makiko Sugimoto, Helene Bernstein, Yoshihiro Jinno and Danny J. Schust Supplementary Figures:
SUPPLEMENTARY FIG. S9. Morphology and DNA staining of cipsc-differentiated trophoblast cell-like cell. Scale bar: 100 mm. SUPPLEMENTARY FIG. S10.
 Supplementary Data SUPPLEMENTARY FIG. S1. Lentivirus-infected canine fibroblasts show YFP expression. (A, B) Uninfected canine skin fibroblasts (CSFs); (C, D) infected CSFs; (E, F) uninfected CTFs; (G,
Supplementary Data SUPPLEMENTARY FIG. S1. Lentivirus-infected canine fibroblasts show YFP expression. (A, B) Uninfected canine skin fibroblasts (CSFs); (C, D) infected CSFs; (E, F) uninfected CTFs; (G,
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplement Figure S1:
 Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in
 Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Development 142: doi: /dev : Supplementary Material
 Development 142: doi:1.42/dev.11687: Supplementary Material Figure S1 - Relative expression of lineage markers in single ES cell and cardiomyocyte by real-time quantitative PCR analysis. Single ES cell
Development 142: doi:1.42/dev.11687: Supplementary Material Figure S1 - Relative expression of lineage markers in single ES cell and cardiomyocyte by real-time quantitative PCR analysis. Single ES cell
NEURONAL CELL CULTURE MATRIX FOR BETTER MAINTENANCE AND SURVIVAL OF NEURONAL CELL CULTURES IN TISSUE CULTURE.
 NEURONAL CELL CULTURE MATRIX FOR BETTER MAINTENANCE AND SURVIVAL OF NEURONAL CELL CULTURES IN TISSUE CULTURE. D. R. Aguirre, N. DiMassa, Chrystal Johnson, H. Eran, R. Perez, C.V.R. Sharma, M.V.R. Sharma,
NEURONAL CELL CULTURE MATRIX FOR BETTER MAINTENANCE AND SURVIVAL OF NEURONAL CELL CULTURES IN TISSUE CULTURE. D. R. Aguirre, N. DiMassa, Chrystal Johnson, H. Eran, R. Perez, C.V.R. Sharma, M.V.R. Sharma,
Chemically defined conditions for human ipsc derivation and culture
 Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Partial list of differentially expressed genes from cdna microarray, comparing MUC18-
 Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
To investigate the heredity of the WFP gene, we selected plants that were homozygous
 Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
NIA Aging Cell Repository
 Certificate of Analysis NIA Aging Cell Repository Human induced Pluripotent Stem Cell (ipsc) Line: AG25370*B Diagnosis Alzheimer Disease, Familial, Type 4 Mutation Reprogramming method Publication(s) describing
Certificate of Analysis NIA Aging Cell Repository Human induced Pluripotent Stem Cell (ipsc) Line: AG25370*B Diagnosis Alzheimer Disease, Familial, Type 4 Mutation Reprogramming method Publication(s) describing
Supplementary Figure 1
 number of cells, normalized number of cells, normalized number of cells, normalized Supplementary Figure CD CD53 Cd3e fluorescence intensity fluorescence intensity fluorescence intensity Supplementary
number of cells, normalized number of cells, normalized number of cells, normalized Supplementary Figure CD CD53 Cd3e fluorescence intensity fluorescence intensity fluorescence intensity Supplementary
Neurosphere Nestin SOX 2 Merge Nestin SOX 2 Merge
 Neurosphere Nestin SOX 2 Merge Nestin SOX 2 Merge A B Fig. S1. Generation and characterization of anscs in vitro. anscs were isolated and expanded from the SVZ region of adult naïve C57BL/6 mice and cultured
Neurosphere Nestin SOX 2 Merge Nestin SOX 2 Merge A B Fig. S1. Generation and characterization of anscs in vitro. anscs were isolated and expanded from the SVZ region of adult naïve C57BL/6 mice and cultured
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplement figure legends
 Supplement figure legends Figure S1. Alignments of PRDM16 amino acid sequences of mouse, human, dog, chicken, and Xenopus. Two putative CtBP-binding motifs are indicated in the boxes. Numbers represent
Supplement figure legends Figure S1. Alignments of PRDM16 amino acid sequences of mouse, human, dog, chicken, and Xenopus. Two putative CtBP-binding motifs are indicated in the boxes. Numbers represent
Supplementary figures (Zhang)
 Supplementary figures (Zhang) Supplementary figure 1. Characterization of hesc-derived astroglia. (a) Treatment of astroglia with LIF and CNTF increases the proportion of GFAP+ cells, whereas the percentage
Supplementary figures (Zhang) Supplementary figure 1. Characterization of hesc-derived astroglia. (a) Treatment of astroglia with LIF and CNTF increases the proportion of GFAP+ cells, whereas the percentage
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Figures
 Supplementary Figures Fig. S1. Specificity of perilipin antibody in SAT compared to various organs. (A) Images of SAT at E18.5 stained with perilipin and CD31 antibody. Scale bars: 100 μm. SAT stained
Supplementary Figures Fig. S1. Specificity of perilipin antibody in SAT compared to various organs. (A) Images of SAT at E18.5 stained with perilipin and CD31 antibody. Scale bars: 100 μm. SAT stained
SUPPLEMENTARY INFORMATION
 Supplementary Figure S1 Colony-forming efficiencies using transposition of inducible mouse factors. (a) Morphologically distinct growth foci appeared from rtta-mefs 6-8 days following PB-TET-mFx cocktail
Supplementary Figure S1 Colony-forming efficiencies using transposition of inducible mouse factors. (a) Morphologically distinct growth foci appeared from rtta-mefs 6-8 days following PB-TET-mFx cocktail
SUPPLEMENTARY INFORMATION
 doi:.38/nature08267 Figure S: Karyotype analysis of ips cell line One hundred individual chromosomal spreads were counted for each of the ips samples. Shown here is an spread at passage 5, predominantly
doi:.38/nature08267 Figure S: Karyotype analysis of ips cell line One hundred individual chromosomal spreads were counted for each of the ips samples. Shown here is an spread at passage 5, predominantly
Supplementary Methods
 Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera
 SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of
 Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like
 Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like morphology in co-culture with mouse embryonic feeder fibroblasts
Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like morphology in co-culture with mouse embryonic feeder fibroblasts
Khaled_Fig. S MITF TYROSINASE R²= MITF PDE4D R²= Variance from mean mrna expression
 Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Uniquely Mapped. Total Reads. Non Uniquely mapped. Samples
 Supplementary Table S1: Mapping Statistics. Sequencing libraries were prepared using DP- seq method. The sequencing reads were mapped to the mouse transcriptome (RefSeq Database). BR refers to biological
Supplementary Table S1: Mapping Statistics. Sequencing libraries were prepared using DP- seq method. The sequencing reads were mapped to the mouse transcriptome (RefSeq Database). BR refers to biological
Detection of Histone Modifications at Specific Gene Loci in Single Cells in Histological Sections
 Detection of Histone Modifications at Specific Gene Loci in Single Cells in Histological Sections Delphine Gomez; Laura L Shankman; Anh T Nguyen; Gary K Owens. Supplementary Figures 1-13 Supplementary
Detection of Histone Modifications at Specific Gene Loci in Single Cells in Histological Sections Delphine Gomez; Laura L Shankman; Anh T Nguyen; Gary K Owens. Supplementary Figures 1-13 Supplementary
A population of Nestin expressing progenitors in the cerebellum exhibits increased tumorigenicity
 A population of Nestin expressing progenitors in the cerebellum exhibits increased tumorigenicity Peng Li 1,2, Fang Du 1, Larra W. Yuelling 1, Tiffany Lin 3, Renata E. Muradimova 1, Rossella Tricarico
A population of Nestin expressing progenitors in the cerebellum exhibits increased tumorigenicity Peng Li 1,2, Fang Du 1, Larra W. Yuelling 1, Tiffany Lin 3, Renata E. Muradimova 1, Rossella Tricarico
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
PrimePCR Assay Validation Report
 Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID Glyceraldehyde-3-phosphate dehydrogenase Gapdh Rat This gene encodes a member of
Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID Glyceraldehyde-3-phosphate dehydrogenase Gapdh Rat This gene encodes a member of
Supplementary Figure 1.
 Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
PrimePCR Assay Validation Report
 Gene Information Gene Name major histocompatibility complex, class II, DR beta 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID HLA-DRB1 Human HLA-DRB1 belongs
Gene Information Gene Name major histocompatibility complex, class II, DR beta 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID HLA-DRB1 Human HLA-DRB1 belongs
A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer s disease
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41593-018-0175-4 In the format provided by the authors and unedited. A 3D human triculture system modeling neurodegeneration and neuroinflammation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41593-018-0175-4 In the format provided by the authors and unedited. A 3D human triculture system modeling neurodegeneration and neuroinflammation
Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction
 Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Accelerating skin wound healing by M-CSF through generating SSEA-1 and -3 stem cells. in the injured sites
 Accelerating skin wound healing by M-CSF through generating SSEA-1 and -3 stem cells in the injured sites Yunyuan Li, Reza Baradar Jalili, Aziz Ghahary Department of Surgery, University of British Columbia,
Accelerating skin wound healing by M-CSF through generating SSEA-1 and -3 stem cells in the injured sites Yunyuan Li, Reza Baradar Jalili, Aziz Ghahary Department of Surgery, University of British Columbia,
Myofibroblasts in kidney fibrosis. LeBleu et al.
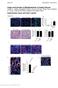 Origin and Function of Myofibroblasts in Kidney Fibrosis Valerie S. LeBleu, Gangadhar Taduri, Joyce O Connell, Yingqi Teng, Vesselina G. Cooke, Craig Woda, Hikaru Sugimoto and Raghu Kalluri Supplementary
Origin and Function of Myofibroblasts in Kidney Fibrosis Valerie S. LeBleu, Gangadhar Taduri, Joyce O Connell, Yingqi Teng, Vesselina G. Cooke, Craig Woda, Hikaru Sugimoto and Raghu Kalluri Supplementary
CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration
 /, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
/, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
Supplementary Figure S1. Alterations in Fzr1( / );Nestin-Cre brains. (a) P10 Cdh1-deficient brains display low levels of Myelin basic protein (MBP)
 Supplementary Figure S1. Alterations in Fzr1( / );Nestin-Cre brains. (a) P10 Cdh1-deficient brains display low levels of Myelin basic protein (MBP) in the cortex (area 1 as defined in Fig. 2a), and corpus
Supplementary Figure S1. Alterations in Fzr1( / );Nestin-Cre brains. (a) P10 Cdh1-deficient brains display low levels of Myelin basic protein (MBP) in the cortex (area 1 as defined in Fig. 2a), and corpus
Chromatin immunoprecipitation (ChIP) ES and FACS-sorted GFP+/Flk1+ cells were fixed in 1% formaldehyde and sonicated until fragments of an average
 Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted
Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted
PrimePCR Assay Validation Report
 Gene Information Gene Name SRY (sex determining region Y)-box 6 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID SOX6 Human This gene encodes a member of the
Gene Information Gene Name SRY (sex determining region Y)-box 6 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID SOX6 Human This gene encodes a member of the
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Human induced Pluripotent Stem Cell (ipsc) Line: GM23225*B
 Certificate of Analysis NIGMS Human tic Cell Repository Human induced Pluripotent Stem Cell (ipsc) Line: GM23225*B Diagnosis Mutation Reprogramming method Publication(s) describing ipsc line establishment
Certificate of Analysis NIGMS Human tic Cell Repository Human induced Pluripotent Stem Cell (ipsc) Line: GM23225*B Diagnosis Mutation Reprogramming method Publication(s) describing ipsc line establishment
PrimePCR Assay Validation Report
 Gene Information Gene Name laminin, beta 3 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID LAMB3 Human The product encoded by this gene is a laminin that
Gene Information Gene Name laminin, beta 3 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID LAMB3 Human The product encoded by this gene is a laminin that
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Artificial niche microarrays for probing single-stem-cell fate in high throughput
 Nature Methods Artificial niche microarrays for probing single-stem-cell fate in high throughput Samy Gobaa, Sylke Hoehnel, Marta Roccio, Andrea Negro, Stefan Kobel & Matthias P Lutolf Supplementary Figure
Nature Methods Artificial niche microarrays for probing single-stem-cell fate in high throughput Samy Gobaa, Sylke Hoehnel, Marta Roccio, Andrea Negro, Stefan Kobel & Matthias P Lutolf Supplementary Figure
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Protocol Using the Reprogramming Ecotropic Retrovirus Set: Mouse OSKM to Reprogram MEFs into ips Cells
 STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblast (MEF) cells into induced pluripotent stem (ips) cells in a 6-well format. Transduction
STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblast (MEF) cells into induced pluripotent stem (ips) cells in a 6-well format. Transduction
Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres
 Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres generated from the CD133-positive cells infected with
Supplementary Figure 1. Phenotype and genotype of cultured and transplanted S1 KCST (A) Brightfield and mcherry fluorescence images of the spheres generated from the CD133-positive cells infected with
A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase
 A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase tag insertion at the 3 end using CRISPR-Cas9 genome editing technique Sambuddha Basu 1, 3, Levi Adams 1, 3, Subhrangshu
A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase tag insertion at the 3 end using CRISPR-Cas9 genome editing technique Sambuddha Basu 1, 3, Levi Adams 1, 3, Subhrangshu
PrimePCR Assay Validation Report
 Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin)
Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin)
- Supplementary Information -
 TRIM32 dependent transcription in adult neural progenitor cells regulates neuronal differentiation. - Supplementary Information - Anna-Lena Hillje 1, 2#, Maria Angeliki S. Pavlou 1, 2#, Elisabeth Beckmann
TRIM32 dependent transcription in adult neural progenitor cells regulates neuronal differentiation. - Supplementary Information - Anna-Lena Hillje 1, 2#, Maria Angeliki S. Pavlou 1, 2#, Elisabeth Beckmann
PrimePCR Assay Validation Report
 Gene Information Gene Name collagen, type IV, alpha 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID COL4A1 Human This gene encodes the major type IV alpha
Gene Information Gene Name collagen, type IV, alpha 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID COL4A1 Human This gene encodes the major type IV alpha
AD BD TOC1. Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between
 AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
