SUPPLEMENTARY INFORMATION
|
|
|
- Pamela Tucker
- 6 years ago
- Views:
Transcription
1 a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm neural fold lateral ectoderm b amputation wound epidermis formation contribution of dermis-, muscle-, skeletonand Schwann-derived cells to the blastema Supplementary Figure 1. Blastema cells respect their developmental origin. a. Scheme summarizing our results. We found that epidermis-, lateral plate-, presomitic mesoderm- and neural fold- derived lineages (indicated by the respective coloured boxes) remain restricted to their developmental origin during regeneration. b. Scheme illustrating a revised view of blastema formation. Cells coming from different tissues occupy distinct spatial domains and largely remain in distinct lineages during regeneration, except dermis. As the tissue differentiates, dermally-derived cells at the tip of the blastema contribute to the hand cartilage and tendons. 1
2 ep c n c m ct Supplementary Figure 2. CAGGS-GFP expression is ubiquitous and does not get silenced during regeneration. Cross-section through a regenerated limb of a CAGGS-GFP transgenic animal, 8 weeks after amputation. Apart from blood cells (identified by autoflourescence) most DAPI stained nuclei were GFP positive (see Table S1). c= cartilage, ct= connective tissue, ep= epidermis, m= muscle, n=nerve. Arrowhead points to a nerve. Scale bar: 0.5 mm 2
3 doi: /nature08152 a stage 18 stage 18 2 cm GFP-Epidermis //Fibronectin b c pre-amputation regenerate Supplementary Figure 3. Epidermis only gives rise to epidermis during regeneration. a. Schematic to show the experimental design. b. Cross-section of a mature limb whose epidermis had been labelled as described in a. The basement membrane (visualized by Fibronectin staining, arrow heads) is located between the epidermal and dermal layer of the skin. The image demonstrates that only epidermal cells are GFP+. c. Cross section through the regenerated part of a limb whose epidermal cells had been labelled as described in a. GFP+ cells are only found in the epidermal region (see arrowheads in the inset that point to the Fibronectin-stained basement membrane). 3 limbs from 3 animals. Scale bars in b, c: 50 μm 3
4 aaa pre-amputation GFP-dermis MHCI b PAX7 c GFP-skeleton MHCI d PAX7 g e GFP-muscle MHCI Supplementary Figure 4. Characterization of fluorescent dermis and skeleton grafts before amputation Cross-sections of mature limbs. Graft type and antibody stainings are indicated. a. Fluorescent cells of a dermal graft do not react with an anti-mhci antibody. b. Fluorescent cells of a dermal graft do not react with an anti-pax7 antibody. c. Fluorescent cells of a cartilage graft do not react with an anti-mhci antibody. d. Fluorescent cells of a cartilage graft do not react with an anti-pax7 antibody. e. Fluorescent presomitic mesoderm grafts gave rise to MHCI+ muscle fibers in 8 cm animals. 5 limbs, 3 animals (a, b); 6 limbs, 4 animals (c, d); 3 limbs, 3 animals (e). Scale bars: 50 µm. 4
5 PAX7 Supplementary Figure 5. PAX7 expression in a 12-day midbud stage blastema. Longitudinal section of a midbud stage blastema stained for PAX7. Expression domains are highlighted by white rectangles, the amputation plane is depicted by a white line. Note that PAX7 expression is restricted to the periphery. Distal is to the top. Scale bar: 100μm. 5
6 a DAPI GFP PAX7 Merge b average intensity (A.U) 2,000 1,800 1,600 1,400 1,200 1, Dermis (CarAct) Satellite cells (CarAct) Satellite cells (CAGGS) Dermis (CAGGS) Muscle fibers (CAGGS) c average intensity (A.U) 2,000 1,800 1,600 1,400 1,200 1, Dermis GFP+ satellite cells GFP+ muscle fibers d DAPI GFP Autofluorescent erythrocytes Supplementary Figure 6. Fluorescent muscle grafts harbor GFP+ satellite cells a. Representative cross-section of a mature limb prior to amputation immunostained with an anti-pax7 antibody (3 limbs, 3 animals). PAX7+, GFP+ cells (white arrow head) and PAX7+, GFP- cells (red arrow head) were present. b. Comparison of the average GFP fluorescence in PAX7+ satellite cells in transgenic donor animals (CAGGS) to satellite cell and dermis cell fluorescence in a different transgenic line (as a negative control) where only the muscle fibres express GFP (CarAct). Error bars show standard deviation. n = for CAGGs::GFP, dermis 6; muscle fibers 6; satellite cells 4; for CarAct::GFP, dermis 5; muscle fibers 5; satellite cells 8. c. Quantification of average GFP-fluorescence signal found in PAX7+ satellite cells in the presomitic mesoderm (muscle) grafts compared to negative dermis cells. Error bars show standard deviation. n= dermis 11; muscle fibers 15; satellite cells 30. Similar results were obtained from another replicate. d. Some GFP+ cells outside muscle in limbs prior to and after regeneration exhibit blood vessel morphology and often surround autofluorescent erythrocytes. Scale bar: 50 μm. 6
7 a b HoxA13 MEIS Supplementary Figure 7. MEIS and HoxA13 expression in the blastema a. Longitudinal section of a 12- day blastema on which immunostaining using an anti-meis antibody had been performed. Note accumulation of nuclear MEIS in the cells in the proximal part of the blastema while cells in the distal domain of the blastema do not exhibit MEIS signal. b. Longitudinal section of a 12- day blastema on which in situ hybridization using a HoxA13 antisense probe was performed. Note that the signal concentrates in the distal domain. a,b. Distal is to the top. Scale bars: 100 µm. 7
8 a skeleton-derived (distal region) MEIS GFP Merge b skeleton- derived (proximal region) MEIS GFP Merge c Schwann cell-derived (proximal region) MEIS GFP Merge d GFP+, Meis- GFP+, Meis skeleton Schwann cells dermis muscle Supplementary Figure 8. MEIS is expressed in skeleton-, but not in Schwann- derived blastema cells. a-c. MEIS immunofluorescence on 12-day upper arm blastema sections harbouring indicated GFP+ grafts. Scale bar: 50µm. d. Different fractions of blastema cells from different sources express MEIS. Number of cells/blastemas/animals examined- skeleton: 203/6/6, Schwann: 281/7/7, dermis: 234/10/7, muscle: 187/2/2. 8
9 a - reverse transcription cdna amplification dissociation FACSorting of single GFP+ cells - detection of genes using gene-specific primers b total number of sorted, blastema cells skeleton Schwann cells dermis muscle Rp4+, HoxA13- Rp4+, HoxA13+ Supplementary Figure 9. Single Cell PCR analysis of HoxA13 expression in blastema cells derived from GFP+ grafts a. 12-day blastemas harbouring fluorescent cells were dissociated and single GFP+ cells were FACSed into 96-well plates. After extraction, reverse transcription and cdna amplification, the generated cdnas were analysed by QPCR using gene specific primers. Only cells that displayed a low Ct value for ribosomal protein4 (Rp4) were considered for further analysis. b. Histogram showing the expression of the distal marker HoxA13 in GFP+ skeleton-, Schwann-, dermis-, and muscle-derived blastema cells. Number of cells/blastemas/ animals/ experiments analysed- skeleton: 152/8/8/4, Schwann cells: 402/6/6/6, dermis: 230/12/12/6, muscle: 184/6/6/3. 9
10 % of GFP+ cartilage cells found in the regenerated hand p= ïïï ïï ïïïïï ï ïïï ï ï ï ï ïï ïï ïï ï ïïï ï ï cartilage graft dermis graft 0 Tissue source 1of the GFP graft 2 Supplementary Figure 10. Dermal cells converting to skeletal fates preferentially contribute to the hand Histogram quantifying the fractional contribution of fluorescent skeleton- derived versus dermis- derived cells to the hand skeleton. Six fluorescent grafts of upper arm dermis or skeleton, respectively, were transplanted to the upper arm of normal hosts. Amputation was performed through the transplants. The fully regenerated limbs were sectioned, and fluorescent progeny in the skeletal elements were quantified. The graph shows the percentages of fluorescent cells found in the regenerated hand over the total number of fluorescent cells in lower arm and hand of the regenerate (n>1000 in each regenerate). Error bars indicate standard deviations. The p-value was determined using the Student s t-test (Welch; unpaired). 10
11 Supplementary Table 1. CAGGS-GFP transgene expression is not silenced after regeneration a. CAGGS-GFP transgenic animals express the transgene ubiquitously in the limb muscle, nerves and skeleton (4 limbs, 4 animals) Sample % GFP+ satellite cells (PAX7+) % GFP+ cells within the nerve % GFP+ muscle cells (MHCI+) % GFP+ cells in the skeleton % (n=62) 100% (n=91) 100% (n=456) 100% (n=137) 2 100% (n=80) 100% (n=93) 100% (n=95) 100% (n=93) % (n=102) 100% (n=74) 100% (n=112) 100% (n=150) 4 100% (n=54) 100% (n=73) 100% (n=326) 100% (n=150) b. Regenerated limb cells maintain CAGGS-GFP expression (4 limbs, 4 animals) Sample % GFP + satellite cells (PAX7 + ) % GFP + cells within the nerve % GFP + muscle cells (MHCI + ) % GFP + cells in the skeleton 1 100% (n=100) 100% (n=72) 100% (n=219) 100% (n=290) % (n=80) 100% (n=57) 100% (n=230) 98.9% (n=88) % (n=198) 100% (n=157) 99.8% (n=618) 100% (n=282) 4 100% (n=96) 100% (n=210) 100% (n=1265) 100% (n=110) GFP-expressing cells from different tissues were counted in mature, previously not amputated limbs of the cm long CAGGS-GFP transgenic animals (a) and in the 8 weeks old limb regenerates (b). Left and right arms of the same animal were used as experimental and control samples, respectively. There is no indication of silencing of the transgene during regeneration. n= number of cells counted. 11
12 Supplementary Table 2. Tracking of fluorescent dermal cells during regeneration a. Skin-derived cells do not express PAX7 in the blastema (5 blastemas from 3 animals) Sample GFP +, PAX7 - GFP -, PAX7 + GFP +, PAX b. Skin-derived cells do not express MHCI or PAX7 in the regenerated limb but contribute to skeletal cartilage (6 limbs from 5 animals) Sample GFP +, GFP -, GFP +, MHCI +, PAX7 - PAX7 + PAX7 + GFP + Contribution to skeleton? Yes Yes Yes Yes Yes Yes 12
13 c. Dermis- derived cells do not express MHCI, PAX7 or MBP in the regenerated limb but contribute to skeletal cartilage (3 limbs from 3 animals). Sample GFP +, GFP +, GFP +, MHCI +, PAX7 - PAX7 + MBP + GFP + Contribution to skeleton? Yes Yes Yes a. Limb blastemas harbouring fluorescent skin-derived cells were sectioned longitudinally 12 days post amputation. 16 µm sections were stained with an anti-pax7 antibody. Note that we did not find GFP, PAX7 double positive cells. b. Labelling of dermis was achieved by transplanting fluorescent full-thickness skin from GFP-transgenic adult animals (8 cm snout-tail) to limbs of normal hosts. Fully regenerated limbs harbouring fluorescent dermis-derived cells were sectioned longitudinally 30 days post amputation. 16 µm sections were stained with antibodies directed against PAX7 or MHCI. Contribution of fluorescent cells to skeletal elements was tested by morphological observation. Note that we did not find GFP, PAX7 or GFP, MHCI double positive cells, however, in each case we found fluorescent cells being present in the skeleton. c. Labelling of dermis was achieved by transplanting lateral plate mesoderm from GFPtransgenic embryos to normal host embryos. Operated embryos were allowed to grow to a size of 8 cm. They were then used as donors to transplant full-thickness skin harbouring GFP + dermal cells to normal hosts. Fully regenerated limbs harbouring fluorescent dermis- derived cells were sectioned longitudinally 30 days post amputation. 16µm sections were stained with antibodies directed against PAX7, MHCI or MBP. Contribution of fluorescent cells to skeletal elements was tested by morphological observation. Note that we did not find GFP,PAX7; GFP, MHCI or GFP, MBP double positive cells, however, in each case we found fluorescent cells being present in the cartilage. To determine total numbers of values in a- c, every section containing GFP + cells was included into our analysis. 13
14 Supplementary Table 3. Tracking of cartilage- derived cells a. Cartilage- derived cells do not express PAX7 in the blastema (2 blastemas from 2 animals). Sample GFP +, PAX7 - GFP -, PAX7 + GFP +, PAX b. Skeleton- derived cells mainly give rise to skeleton (3 limbs from 3 animals). Sample Cells in Cells outside PAX7 +, GFP + GFP +, MHCI + skeletal skeletal structures structures (likely tendon and dermis) a. Limb blastemas harbouring fluorescent cartilage-derived cells were sectioned longitudinally 12 days post amputation. 16µm sections were stained with an antibody against PAX7. b. Fully regenerated limbs harbouring fluorescent cartilage-derived cells were sectioned longitudinally 30 days post amputation. 16µm sections were stained with antibodies directed against PAX7 and MHCI. Contribution of fluorescent cells to skeletal elements was tested by morphological observation. To analyze all GFP + cells of the regenerate, every section containing fluorescent cells was counted. Note that we did not find GFP, MHCI or GFP, PAX7 double positive cells. 14
15 Supplementary Table 4. Tracking of muscle- derived cells a. Fluorescent muscle grafts harbour GFP + satellite cells (3 limbs from 3 animals) Sample % GFP + satellite cells at amputation plane before regeneration 1 50% (n=38) 2 70% (n=43) % (n=56) b. Muscle derived cells express PAX7 in the blastema (2 blastemas from 2 animals) Sample GFP +, PAX7 - GFP -, PAX7 + GFP +, PAX c. Muscle- derived cells contribute to muscle, but not to cartilage or epidermis of the regenerated limb (6 limbs from 6 animals) Sample No. of nuclei GFP + cells in GFP + cells in GFP + cells in GFP + fibres epidermis/ total no. cartilage/ total no. outside muscle of epidermal cells of cartilage cells (blood vessels) / / / / / / / / / / / /
16 a. 5 sections (16 µm) next to the amputation plane were taken from the amputated limbs in samples 1-3 to assess GFP, PAX7 colocalization. Note that we detected GFP +,PAX7 + cells in all three samples. n= number of cells counted. b. Limb blastemas harbouring fluorescent muscle-derived cells were sectioned longitudinally 12 days post amputation. 16 µm sections were stained with an antibody directed against PAX7. For each sample, at least 3 representative sections were analyzed. Note that we found GFP, PAX7 double positive cells among the fluorescent progeny. c. Fully regenerated limbs harbouring fluorescent muscle-derived cells were sectioned longitudinally 30 days post amputation. 16 µm sections were stained with antibodies against PAX7 and MHCI. Contribution of fluorescent cells to epidermis and skeletal elements was determined by morphological observation. Here, every limb section was included in our analysis. Note that we did not detect GFP + cells in cartilage or epidermis. 16
17 Supplementary Table 5. Tracking of Schwann cells a. GFP-labelled Schwann cells are closely associated with MBP and/or BIII-tubulin after regeneration (4 limbs, 3 animals) Sample GFP +, cells next to MBP or BIII-tubulin GFP + cells not next to MBP or BIII-tubulin b. GFP-labelled Schwann cells remain lineage restricted when implanted into irradiated limbs. Sample GFP + cells GFP + cells outside GFP + Implant type / recipient next to MBP cartilage and not cells in the treatment or BIII- next to MBP or BIII- cartilage tubulin tubulin Schwann cell-gfp / P Schwann cell-gfp / P Schwann cell-gfp / P Schwann cell-gfp / X Schwann cell-gfp / X Schwann cell-gfp / X Schwann cell-gfp / X All-GFP / X All-GFP / X 17
18 a. Fate of Schwann cell derived GFP+ progenitors after regeneration was determined by staining sections of the regenerates with the antibodies against Myelin Basic Protein (MBP) and BIII-tubulin. The GFP+ cells next to BIII-tubulin or colocalizing with MBP were counted as being Schwann cells. 19 cells that scored as GFP +,MBP - /BIII-tubulin - were neither cartilage nor muscle cells and appeared to be immature glia with weak immunostaining. b. Regeneration of the X-rayed limbs was rescued by implanting non-irradiated GFPlabelled nerve. P=protected from X ray, X= X-rayed limbs. Samples 1-7: only Schwann cells in the implanted nerve were GFP+; samples 8,9: all cells in the implanted nerve were GFP + (All-GFP). The presence of GFP + cartilage in these samples but not in the Schwann cell-gfp samples shows that the cartilage derives from the GFP + nerve sheath but must come from a non-schwann cell source. Animal numbers used: Samples 1-3, N=3; 4-7, N=4; 8-9, N=2. N= number of animals 18
19 Supplementary Table 6. Proximo-distal location of fluorescent cartilage grafts from different limb levels Graft type No. of grafts Location of fluorescent cells Lower arm only Lower arm + hand Hand only Upper arm skeleton Finger skeleton Upper arm Schwann Cells Hand Schwann Cells Fluorescent tissues were grafted to the upper arm level of sibling host animals and allowed to heal. Amputation was performed through the transplants. The final location of fluorescent progeny in hand and lower arm of the fully regenerated limbs was determined. 19
Supplementary Figure 1
 Supplementary Figure 1 Preaxial dominance during salamander limb development. Panel (a, b) shows RNA in situ hybridisation on whole-mount limbs with Sox9 sense probe as a control. Note the absence of reactivity
Supplementary Figure 1 Preaxial dominance during salamander limb development. Panel (a, b) shows RNA in situ hybridisation on whole-mount limbs with Sox9 sense probe as a control. Note the absence of reactivity
Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and
 Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
supplementary information
 DOI: 10.1038/ncb2015 Figure S1 Confirmation of Sca-1 CD34 stain specificity using isotypematched control antibodies. Skeletal muscle preparations were stained and gated as described in the text (Hoechst
DOI: 10.1038/ncb2015 Figure S1 Confirmation of Sca-1 CD34 stain specificity using isotypematched control antibodies. Skeletal muscle preparations were stained and gated as described in the text (Hoechst
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplemental Information. Boundary Formation through a Direct. Threshold-Based Readout. of Mobile Small RNA Gradients
 Developmental Cell, Volume 43 Supplemental Information Boundary Formation through a Direct Threshold-Based Readout of Mobile Small RNA Gradients Damianos S. Skopelitis, Anna H. Benkovics, Aman Y. Husbands,
Developmental Cell, Volume 43 Supplemental Information Boundary Formation through a Direct Threshold-Based Readout of Mobile Small RNA Gradients Damianos S. Skopelitis, Anna H. Benkovics, Aman Y. Husbands,
Supplemental Figure 1 (Figure S1), related to Figure 1 Figure S1 provides evidence to demonstrate Nfatc1Cre is a mouse line that directed gene
 Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Neural Development. How does a single cell make a brain??? How are different brain regions specified??? Neural Development
 Neural Development How does a single cell make a brain??? How are different brain regions specified??? 1 Neural Development How do cells become neurons? Environmental factors Positional cues Genetic factors
Neural Development How does a single cell make a brain??? How are different brain regions specified??? 1 Neural Development How do cells become neurons? Environmental factors Positional cues Genetic factors
Supplementary Data. Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells
 Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Figure 1.
 Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
sides of the aleurone (Al) but it is excluded from the basal endosperm transfer layer
 Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
ANNOUNCEMENTS. HW2 is due Thursday 2/4 by 12:00 pm. Office hours: Monday 12:50 1:20 (ECCH 134)
 ANNOUNCEMENTS HW2 is due Thursday 2/4 by 12:00 pm. Office hours: Monday 12:50 1:20 (ECCH 134) Lectures 6 8 Outline Stem Cell Division - symmetric - asymmetric Stem cell lineage potential - pluripotent
ANNOUNCEMENTS HW2 is due Thursday 2/4 by 12:00 pm. Office hours: Monday 12:50 1:20 (ECCH 134) Lectures 6 8 Outline Stem Cell Division - symmetric - asymmetric Stem cell lineage potential - pluripotent
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
NPTEL Biotechnology Tissue Engineering. Stem cells
 Stem cells S. Swaminathan Director Centre for Nanotechnology & Advanced Biomaterials School of Chemical & Biotechnology SASTRA University Thanjavur 613 401 Tamil Nadu Joint Initiative of IITs and IISc
Stem cells S. Swaminathan Director Centre for Nanotechnology & Advanced Biomaterials School of Chemical & Biotechnology SASTRA University Thanjavur 613 401 Tamil Nadu Joint Initiative of IITs and IISc
Nature Structural & Molecular Biology: doi: /nsmb.3308
 Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
SUPPLEMENTARY INFORMATION
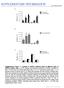 SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
Detection of Histone Modifications at Specific Gene Loci in Single Cells in Histological Sections
 Detection of Histone Modifications at Specific Gene Loci in Single Cells in Histological Sections Delphine Gomez; Laura L Shankman; Anh T Nguyen; Gary K Owens. Supplementary Figures 1-13 Supplementary
Detection of Histone Modifications at Specific Gene Loci in Single Cells in Histological Sections Delphine Gomez; Laura L Shankman; Anh T Nguyen; Gary K Owens. Supplementary Figures 1-13 Supplementary
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Stem cell: a cell capable of 1) tissue plasticity - make different cell types 2) infinite self renewal through asymmetric division
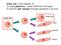 Stem cell: a cell capable of 1) tissue plasticity - make different cell types 2) infinite self renewal through asymmetric division stem cell stem cell skin muscle nerve Properties of STEM cells Plasticity
Stem cell: a cell capable of 1) tissue plasticity - make different cell types 2) infinite self renewal through asymmetric division stem cell stem cell skin muscle nerve Properties of STEM cells Plasticity
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2239 Hepatocytes (Endoderm) Foreskin fibroblasts (Mesoderm) Melanocytes (Ectoderm) +Dox rtta TetO CMV O,S,M,K & N H1 hes H7 hes H9 hes H1 hes-derived EBs H7 hes-derived EBs H9 hes-derived
DOI: 10.1038/ncb2239 Hepatocytes (Endoderm) Foreskin fibroblasts (Mesoderm) Melanocytes (Ectoderm) +Dox rtta TetO CMV O,S,M,K & N H1 hes H7 hes H9 hes H1 hes-derived EBs H7 hes-derived EBs H9 hes-derived
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
Quantitative real-time RT-PCR analysis of the expression levels of E-cadherin
 Supplementary Information 1 Quantitative real-time RT-PCR analysis of the expression levels of E-cadherin and ribosomal protein L19 (RPL19) mrna in cleft and bud epithelial cells of embryonic salivary
Supplementary Information 1 Quantitative real-time RT-PCR analysis of the expression levels of E-cadherin and ribosomal protein L19 (RPL19) mrna in cleft and bud epithelial cells of embryonic salivary
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2880 Supplementary Figure 1 Sequence alignment of Deup1 and Cep63. The protein sequence alignment was generated by the Clustal X 2.0 multiple sequence alignment program using default parameters.
DOI: 10.1038/ncb2880 Supplementary Figure 1 Sequence alignment of Deup1 and Cep63. The protein sequence alignment was generated by the Clustal X 2.0 multiple sequence alignment program using default parameters.
To assess the localization of Citrine fusion proteins, we performed antibody staining to
 Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking
 Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Supplementary Data. Supplementary Table S1.
 Supplementary Data Supplementary Table S1. Primer Sequences for Real-Time Reverse Transcription-Polymerase Chain Reaction Gene Forward 5?3 Reverse 3?5 Housekeeping gene 36B4 TCCAGGCTTTGGGCATCA CTTTATCAGCTGCACATCACTCAGA
Supplementary Data Supplementary Table S1. Primer Sequences for Real-Time Reverse Transcription-Polymerase Chain Reaction Gene Forward 5?3 Reverse 3?5 Housekeeping gene 36B4 TCCAGGCTTTGGGCATCA CTTTATCAGCTGCACATCACTCAGA
Pattern Formation via Small RNA Mobility SUPPLEMENTAL FIGURE 1 SUPPLEMENTAL INFORMATION. Daniel H. Chitwood et al.
 SUPPLEMENTAL INFORMATION Pattern Formation via Small RNA Mobility Daniel H. Chitwood et al. SUPPLEMENTAL FIGURE 1 Supplemental Figure 1. The ARF3 promoter drives expression throughout leaves. (A, B) Expression
SUPPLEMENTAL INFORMATION Pattern Formation via Small RNA Mobility Daniel H. Chitwood et al. SUPPLEMENTAL FIGURE 1 Supplemental Figure 1. The ARF3 promoter drives expression throughout leaves. (A, B) Expression
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and
 Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
initial single-cell analysis, with a pragmatic focus on surface markers with the highest potential for
 Supplementary Figure 1: Summary of the exclusionary approach to surface marker selection for initial single-cell analysis, with a pragmatic focus on surface markers with the highest potential for protein
Supplementary Figure 1: Summary of the exclusionary approach to surface marker selection for initial single-cell analysis, with a pragmatic focus on surface markers with the highest potential for protein
Supplemental Data. Borg et al. Plant Cell (2014) /tpc
 Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplementary Figure 1. Mouse genetic models used for identification of Axin2-
 Supplementary Figure 1. Mouse genetic models used for identification of Axin2- expressing cells and their derivatives. Schematic representations illustrate genetic cell labeling strategies to identify
Supplementary Figure 1. Mouse genetic models used for identification of Axin2- expressing cells and their derivatives. Schematic representations illustrate genetic cell labeling strategies to identify
Piscataway, NJ February 2007
 Final Progress Report for the New Jersey Commission on Spinal Cord Research Submitted by David P. Crockett, PhD for the late Ira B. Black, MD Department of Neuroscience and Cell Biology UMDNJ-Robert Wood
Final Progress Report for the New Jersey Commission on Spinal Cord Research Submitted by David P. Crockett, PhD for the late Ira B. Black, MD Department of Neuroscience and Cell Biology UMDNJ-Robert Wood
To investigate the heredity of the WFP gene, we selected plants that were homozygous
 Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary Table, Figures and Videos
 Supplementary Table, Figures and Videos Table S1. Oligonucleotides used for different approaches. (A) RT-qPCR study. (B) qpcr study after ChIP assay. (C) Probes used for EMSA. Figure S1. Notch activation
Supplementary Table, Figures and Videos Table S1. Oligonucleotides used for different approaches. (A) RT-qPCR study. (B) qpcr study after ChIP assay. (C) Probes used for EMSA. Figure S1. Notch activation
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full//59/ra7/dc1 Supplementary Materials for Dok-7 ctivates the Muscle Receptor Kinase MuSK and Shapes Synapse Formation kane Inoue, Kiyoko Setoguchi, Yosuke Matsubara,
www.sciencesignaling.org/cgi/content/full//59/ra7/dc1 Supplementary Materials for Dok-7 ctivates the Muscle Receptor Kinase MuSK and Shapes Synapse Formation kane Inoue, Kiyoko Setoguchi, Yosuke Matsubara,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/331/6018/753/dc1 Supporting Online Material for Embryological Evidence Identifies Wing Digits in Birds as Digits 1, 2, and 3 Koji Tamura,* Naoki Nomura, Ryohei Seki,
www.sciencemag.org/cgi/content/full/331/6018/753/dc1 Supporting Online Material for Embryological Evidence Identifies Wing Digits in Birds as Digits 1, 2, and 3 Koji Tamura,* Naoki Nomura, Ryohei Seki,
Supplementary Fig. 5
 Supplementary Fig. 5 Supplemental Figures legends Supplementary Figure 1 (A) Additional dot plots from CyTOF analysis from untreated group. (B) Gating strategy for assessment of CD11c + NK cells frequency
Supplementary Fig. 5 Supplemental Figures legends Supplementary Figure 1 (A) Additional dot plots from CyTOF analysis from untreated group. (B) Gating strategy for assessment of CD11c + NK cells frequency
Supplemental Material
 Supplemental Material Figure S1.(A) Immuno-staining of freshly isolated Myf5-Cre:ROSA-YFP fiber (B) Satellite cell enumeration in WT and cko mice from resting hind limb muscles. Bulk hind limb muscles
Supplemental Material Figure S1.(A) Immuno-staining of freshly isolated Myf5-Cre:ROSA-YFP fiber (B) Satellite cell enumeration in WT and cko mice from resting hind limb muscles. Bulk hind limb muscles
Two classes of silencing RNAs move between Caenorhabditis elegans tissues.
 Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Apoptosis analyses and quantification. Activated caspase3 immunostaining on sections of E9.0 Tcof1 +/ -;p53 +/- (a) and Tcof1 +/ -;p53 -/- (b) littermate embryos
Supplementary Figures Supplementary Figure 1. Apoptosis analyses and quantification. Activated caspase3 immunostaining on sections of E9.0 Tcof1 +/ -;p53 +/- (a) and Tcof1 +/ -;p53 -/- (b) littermate embryos
Supplementary Materials
 Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
supplementary information
 Figure S1 Distribution of heterokaryons in vermis and hemispheres of the cerebellum. Schematic dorsal view of an adult cerebellum with the vermis in the middle (red) flanked by the two hemispheres (grey).
Figure S1 Distribution of heterokaryons in vermis and hemispheres of the cerebellum. Schematic dorsal view of an adult cerebellum with the vermis in the middle (red) flanked by the two hemispheres (grey).
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in
 Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
Irradiated cells and blastema formation in the adult newt, Notophthalmus viridescens
 /. Embryol. exp. Morph. Vol. 32, 2, pp. 405-416, 1974 405 Printed in Great Britain Irradiated cells and blastema formation in the adult newt, Notophthalmus viridescens By DAVID L.DESHA 1 From Tulane University,
/. Embryol. exp. Morph. Vol. 32, 2, pp. 405-416, 1974 405 Printed in Great Britain Irradiated cells and blastema formation in the adult newt, Notophthalmus viridescens By DAVID L.DESHA 1 From Tulane University,
A Survey of Genetic Methods
 IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
Figure S1 is related to Figure 1B, showing more details of outer segment of
 Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.
 LEGENDS TO SUPPLEMENTARY FIGURES Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.2) were stained with the antibodies oct3/4
LEGENDS TO SUPPLEMENTARY FIGURES Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.2) were stained with the antibodies oct3/4
Supplemental Figure 1
 12 1 8 Embryo 6 5 4 Silk 6 4 2 3 2 1 1 15 21 24 3 35 4 45 days after pollination 2 6 12 24 72 hours after pollination 35 6 3 25 2 15 1 5 Endosperm 8 12 21 22 3 35 4 45 days after pollination 5 4 3 2 1
12 1 8 Embryo 6 5 4 Silk 6 4 2 3 2 1 1 15 21 24 3 35 4 45 days after pollination 2 6 12 24 72 hours after pollination 35 6 3 25 2 15 1 5 Endosperm 8 12 21 22 3 35 4 45 days after pollination 5 4 3 2 1
Accelerating skin wound healing by M-CSF through generating SSEA-1 and -3 stem cells. in the injured sites
 Accelerating skin wound healing by M-CSF through generating SSEA-1 and -3 stem cells in the injured sites Yunyuan Li, Reza Baradar Jalili, Aziz Ghahary Department of Surgery, University of British Columbia,
Accelerating skin wound healing by M-CSF through generating SSEA-1 and -3 stem cells in the injured sites Yunyuan Li, Reza Baradar Jalili, Aziz Ghahary Department of Surgery, University of British Columbia,
Flow cytometry Stained cells were analyzed and sorted by SORP FACS Aria (BD Biosciences).
 Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Supplementary Figure 1. ERK signaling is not activated at early hypertension. a, Western blot analysis for the level of phospho-erk (perk) and total
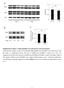 Supplementary Figure 1. ERK signaling is not activated at early hypertension. a, Western blot analysis for the level of phospho-erk (perk) and total ERK in the aortic tissue from the saline- or AngII-infused
Supplementary Figure 1. ERK signaling is not activated at early hypertension. a, Western blot analysis for the level of phospho-erk (perk) and total ERK in the aortic tissue from the saline- or AngII-infused
Notch1 (forward: 5 -TGCCTGTGCACACCATTCTGC-3, reverse: Notch2 (forward: 5 -ATGCACCATGACATCGTTCG-3, reverse:
 Supplementary Information Supplementary Methods. RT-PCR. For mouse ES cells, the primers used were: Notch1 (forward: 5 -TGCCTGTGCACACCATTCTGC-3, reverse: 5 -CAATCAGAGATGTTGGAATGC-3 ) 1, Notch2 (forward:
Supplementary Information Supplementary Methods. RT-PCR. For mouse ES cells, the primers used were: Notch1 (forward: 5 -TGCCTGTGCACACCATTCTGC-3, reverse: 5 -CAATCAGAGATGTTGGAATGC-3 ) 1, Notch2 (forward:
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
bronchial epithelial cells (I). Bronchi are outlined with dashed line. Scale bars = 25 µm, if not
 Supplemental Figure S1: ronchial epithelial cell polarity and integrity is maintained in bronchi. (A-E) Staining for selected markers of bronchial cell differentiation and intracellular compartments is
Supplemental Figure S1: ronchial epithelial cell polarity and integrity is maintained in bronchi. (A-E) Staining for selected markers of bronchial cell differentiation and intracellular compartments is
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum. Blood-stage Development
 Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12138 Supplementary Figure 1. Knockdown of KRAS leads to a reduction in macropinocytosis. (a) KRAS knockdown in MIA PaCa-2 cells expressing KRASspecific shrnas
SUPPLEMENTARY INFORMATION doi:10.1038/nature12138 Supplementary Figure 1. Knockdown of KRAS leads to a reduction in macropinocytosis. (a) KRAS knockdown in MIA PaCa-2 cells expressing KRASspecific shrnas
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Figure S1. Schematic of myeloid lineage reporter mouse model used in this study.
 Supplementary Figure Legends Figure S1. Schematic of myeloid lineage reporter mouse model used in this study. (A) The Lyzs-Cre mouse was crossed with Gt(ROSA)26Sor tm1(eyfp)cos/ J mice to label cells expressing
Supplementary Figure Legends Figure S1. Schematic of myeloid lineage reporter mouse model used in this study. (A) The Lyzs-Cre mouse was crossed with Gt(ROSA)26Sor tm1(eyfp)cos/ J mice to label cells expressing
Supplemental Table/Figure Legends
 MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
embryos. Asterisk represents loss of or reduced expression. Brackets represent
 Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplementary Information
 Supplementary Information Supplementary Figures Supplementary Figure 1. qrt-pcr analysis of the expression of candidate factors including SOX2, MITF, PAX3, DLX5, FOXD3, LEF1, MSX1, PAX6, SOX2 and SOX9
Supplementary Information Supplementary Figures Supplementary Figure 1. qrt-pcr analysis of the expression of candidate factors including SOX2, MITF, PAX3, DLX5, FOXD3, LEF1, MSX1, PAX6, SOX2 and SOX9
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles
 Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
(A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: WT; lower
 Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Q-PCR QUANTITATIVE-PCR 세포생물학및실험 2 박태식교수님
 Q-PCR QUANTITATIVE-PCR 세포생물학및실험 2 박태식교수님 Title : Study that the Drug A inhibits the accumulation of fat. Schedules 1. Mouse necropsy : take up the blood and major tissues(organs) ex) heart, liver, fat,
Q-PCR QUANTITATIVE-PCR 세포생물학및실험 2 박태식교수님 Title : Study that the Drug A inhibits the accumulation of fat. Schedules 1. Mouse necropsy : take up the blood and major tissues(organs) ex) heart, liver, fat,
Supplement Figure S1:
 Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Facts and theories of organ regeneration in adults
 Harvard-MIT Division of Health Sciences and Technology HST.535: Principles and Practice of Tissue Engineering Instructor: I. V. Yannas Facts and theories of organ regeneration in adults I.V.Yannas, PhD
Harvard-MIT Division of Health Sciences and Technology HST.535: Principles and Practice of Tissue Engineering Instructor: I. V. Yannas Facts and theories of organ regeneration in adults I.V.Yannas, PhD
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12783 Bone marrow transplantation experiments While we confirmed that CD45+ bone marrow derived cells contributed to the dermis during normal homeostasis and wound healing (Fig. E9d,
doi:10.1038/nature12783 Bone marrow transplantation experiments While we confirmed that CD45+ bone marrow derived cells contributed to the dermis during normal homeostasis and wound healing (Fig. E9d,
Stem cells in Development
 ANAT 2341 Embryology Lab 10 8 Oct 2009 Therapeutic Use of Stem Cells Practical Hurdles & Ethical Issues Stem cells in Development Blastocyst Cord blood Antonio Lee PhD Neuromuscular & Regenerative Medicine
ANAT 2341 Embryology Lab 10 8 Oct 2009 Therapeutic Use of Stem Cells Practical Hurdles & Ethical Issues Stem cells in Development Blastocyst Cord blood Antonio Lee PhD Neuromuscular & Regenerative Medicine
Stem cells in Development
 ANAT 2341 Embryology Lab 10 8 Oct 2009 Therapeutic Use of Stem Cells Practical Hurdles & Ethical Issues Stem cells in Development Blastocyst Cord blood Antonio Lee PhD Neuromuscular & Regenerative Medicine
ANAT 2341 Embryology Lab 10 8 Oct 2009 Therapeutic Use of Stem Cells Practical Hurdles & Ethical Issues Stem cells in Development Blastocyst Cord blood Antonio Lee PhD Neuromuscular & Regenerative Medicine
2.79J/2.79J/3.96J/BE.441J/HST.522J. A theory of induced regeneration in adults. Note: [C, S, R] data only cited; no kinetics
![2.79J/2.79J/3.96J/BE.441J/HST.522J. A theory of induced regeneration in adults. Note: [C, S, R] data only cited; no kinetics 2.79J/2.79J/3.96J/BE.441J/HST.522J. A theory of induced regeneration in adults. Note: [C, S, R] data only cited; no kinetics](/thumbs/87/96611421.jpg) 2.79J/2.79J/3.96J/BE.441J/HST.522J A theory of induced regeneration in adults. Note: [C, S, R] data only cited; no kinetics Outline 1. Irreversible injury 2. Regenerative and nonregenerative tissues 3.
2.79J/2.79J/3.96J/BE.441J/HST.522J A theory of induced regeneration in adults. Note: [C, S, R] data only cited; no kinetics Outline 1. Irreversible injury 2. Regenerative and nonregenerative tissues 3.
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2.
 Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Supplementary Figure S1. Hoechst. cells/field. Myogenin. % of Myogenin+ cells. 0 h chase CONTROL. BrdU. 72 h chase. CONTROL BrdU CONTROL BrdU
 Hoechst Myogenin amyhc 24 h 48 h 72 h b cells/field 50 40 30 20 10 0 24h 48h 72h h after plating (4 days growth+ t) c e CONTROL 0 h chase % of Myogenin+ cells h after plating (4 days growth+ t) CONTROL
Hoechst Myogenin amyhc 24 h 48 h 72 h b cells/field 50 40 30 20 10 0 24h 48h 72h h after plating (4 days growth+ t) c e CONTROL 0 h chase % of Myogenin+ cells h after plating (4 days growth+ t) CONTROL
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Mass spectrometry characterization of epoxide reactions.
 Supplementary Figure 1 Mass spectrometry characterization of epoxide reactions. (a) Degree of amine reactivity of bovine serum albumin (BSA) with epoxide molecules having different numbers of epoxide groups:
Supplementary Figure 1 Mass spectrometry characterization of epoxide reactions. (a) Degree of amine reactivity of bovine serum albumin (BSA) with epoxide molecules having different numbers of epoxide groups:
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Biology Open (2015): doi: /bio : Supplementary information
 Fig. S1. Verification of optimal conditions for the generation of LIF-dependent inscs Representative images (from five repeat experiments) of human primary fibroblasts undergoing reprogramming for 21 days
Fig. S1. Verification of optimal conditions for the generation of LIF-dependent inscs Representative images (from five repeat experiments) of human primary fibroblasts undergoing reprogramming for 21 days
Supplementary figures (Zhang)
 Supplementary figures (Zhang) Supplementary figure 1. Characterization of hesc-derived astroglia. (a) Treatment of astroglia with LIF and CNTF increases the proportion of GFAP+ cells, whereas the percentage
Supplementary figures (Zhang) Supplementary figure 1. Characterization of hesc-derived astroglia. (a) Treatment of astroglia with LIF and CNTF increases the proportion of GFAP+ cells, whereas the percentage
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Effect of timing of DE dissociation and RA concentration on lung field specification in hpscs. (a) Effect of duration of endoderm induction on expression of
Supplementary Figures Supplementary Figure 1. Effect of timing of DE dissociation and RA concentration on lung field specification in hpscs. (a) Effect of duration of endoderm induction on expression of
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1
 Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15
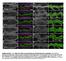 Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Supporting Information
 Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figure 1: List of primers/oligonucleotides used in this study. 1 Supplementary figure 2: Sequences and mirna-targets of i) mcherry expresses in transgenic fish used
SUPPLEMENTARY INFORMATION Supplementary figure 1: List of primers/oligonucleotides used in this study. 1 Supplementary figure 2: Sequences and mirna-targets of i) mcherry expresses in transgenic fish used
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
7.22 Example Problems for Exam 1 The exam will be of this format. It will consist of 2-3 sets scenarios.
 Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 10 7.22 Fall 2005 sample exam
Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 10 7.22 Fall 2005 sample exam
Xeno-Free Systems for hesc & hipsc. Facilitating the shift from Stem Cell Research to Clinical Applications
 Xeno-Free Systems for hesc & hipsc Facilitating the shift from Stem Cell Research to Clinical Applications NutriStem Defined, xeno-free (XF), serum-free media (SFM) specially formulated for growth and
Xeno-Free Systems for hesc & hipsc Facilitating the shift from Stem Cell Research to Clinical Applications NutriStem Defined, xeno-free (XF), serum-free media (SFM) specially formulated for growth and
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
7.22 Final Exam points
 Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 11 7.22 2004 FINAL FOR STUDY
Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 11 7.22 2004 FINAL FOR STUDY
