Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like
|
|
|
- Esther Lane
- 5 years ago
- Views:
Transcription
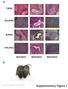
1 Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like morphology in co-culture with mouse embryonic feeder fibroblasts (Scale bars: left panel, 200 µm; right panel, 50 µm) (a), AP staining of hipscs (Scale bars of left panel, 200 µm) (b). c. As shown via immunostaining, hipsc clones express markers common to pluripotent cells, including OCT3/4, NANOG, SSEA3, SSEA4 and TRA , 6-Diamidino-2-phenylindole (DAPI) staining indicates cell nuclear. Scale bar, 30 µm.
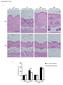
2 Supplementary Figure 2. Gene expression in hipscs is similar to hescs. qpcr assay for expression of OCT3/4(endo), SOX2(endo), NANOG, TERT, KLF4(endo) and REX1 in hescs, hipscs and parental fibroblasts (hfs). Individual PCR reactions were normalized against internal controls (β-actin). Data shown are mean ± SD of the expression from three different experiments.
3 Supplementary Figure 3. qpcr analysis of retroviral transgene expression in the hipsc clones. Transgene-specific PCR primers permit determination of the relative retrovirally expressed (transgene) genes (OCT3/4, SOX2 and KLF4) via qpcr. Tg represents transgene. Five different hipsc clones were tested. hf stands for human fibroblasts and hf/3f-4d stands for human fibroblasts 4 days after virally infection with OCT3/4, SOX2 and KLF4. Data shown are mean ± SD of the expression from three different experiments.
4 Supplementary Figure 4. Analysis of DNA methylation and histone modification in the OCT3/4 and NANOG promoters in the hipscs, hescs and hfs. a. Bisulfite genomic sequencing of the promoter regions of OCT3/4 and NANOG in 10 randomly selected hipsc and hesc clones, as well as human fibroblasts (hfs). Open circles indicate unmethylated CpG dinucleotides, whereas closed circles indicate methylated CpGs. b. Chromatin immunoprecipitation was performed using antibodies against dimethylated histone H3K4 (H3K4me2) and H3 acetylation (ach3). OCT3/4 and NANOG promoters showed enrichment for the active (H3K4me2 and ach3) mark in hipscs, similar to hescs. In hfs, OCT3/4 and NANOG promoters appeared in the inactive state. Data shown are mean ± SD of histone modification from three independent experiments.
5 Supplementary Figure 5. Pluripotent characterization of hipscs. hipscs were injected into immunodeficient mouse and generated well-differentiated teratomalike masses containing all three embryonic germ layers (endoderm, mesoderm and ectoderm). Scale bar, 50 µm.
6 Supplementary Figure 6. Flow cytometric analysis of positive cells for CD200 and ITGA6 during epithelial cell differentiation from hipscs. Flow cytometric analysis was performed using antibodies specific for CD200 and ITGA6. CD200 + /ITGA6 + cell percentage was monitored at 0, 11, 18, 25 and 30 days respectively.
7 Supplementary Figure 7. Flow cytometric analysis of positive cells for KRT15 during epithelial cell differentiation from hipscs. Flow cytometric analysis was performed using antibody specific for KRT15 and KRT15 + cell percentage was monitored at 0, 11, 18, 25 and 30 days respectively.
8 Supplementary Figure 8. Flow cytometric analysis of the percentage of CD200 + /KRT15 +, CD200 + /KRT14 +, ITGA6 + /KRT15 +, ITGA6 + /KRT14 + and KRT14 + /KRT15 + cell populations at day 18 druing the epithelial cell differentiation.
9 Supplementary Figure 9. Flow cytometric analysis of ITGB1 + or KRT5 + cell percentage in CD200 + /ITGA6 + cells. CD200 + /ITGA6 + cell population was gated out. Almost all the cells expressed ITGB1 and only about 50% cells expressed KRT15.
10 Supplementary Figure 10. Flow cytometric analysis of positive cells for KRT14 during epithelial cell differentiation from hipscs. Flow cytometric analysis was performed using antibody against KRT14 and KRT14 + cell percentage was monitored at 0, 11, 18, 25 and 30 days respectively.
11 Supplementary Figure 11. Flow cytometric analysis of positive cells for SSEA3 during epithelial cell differentiation from hipscs. Flow cytometric analysis was performed using antibody specific for SSEA3 and SSEA3 + cell percentage was monitored at 0, 11, 18, 25 and 30 days respectively.
12 Supplementary Figure 12. qpcr analysis of OCT3/4, NANOG, KRT5, KRT8, KRT14, KRT15, LamB3, Involucrin and Filaggrin in hipsc-derived cells at different stages of differentiation. Data shown are mean ± SD of gene expression from three independent experiments. * indicates p<0.05 and ** indicates p<0.01. One-Way ANOVA was used to calculate the p values. The expression of OCT3/4 and NANOG decreased significantly at day 11, 18, 25 and 30 compared with the expression at day 0. Reversely, the expression of KRT5, KRT8, KRT15, KRT18, KRT14, LamB3, involucrin and filaggrin increased significantly at day 11, 18, 25 and 30 compared with the expression at day 0. The expression of KRT15 decreased significantly at day 30 compared with the expression at day 18 (P<0.01). Student s Test was used to calculate the p value.
13 Supplementary Figure 13.Quantification of size and number of colonies from unfractionated cells, CD200 + /ITGA6 + cells (hipsc-cd200 + /ITGA6 + cells), CD200 + /ITGA6 - cells (hipsc- CD200 + /ITGA6 - cells ) and CD200 - /ITGA6 + cells (hipsc-cd200 - /ITGA6 + cells) derived from hipscs at day 18 differentiation; mature keratinocytes derived from hipsc (hipsckeratinocytes)and normal skin keratinocytes (passage 3). Data shown are mean ± SD of the colony numbers from three independent experiments. 3T3 feeder cells used as a negative control.
14 Supplementary Figure 14. qpcr analysis of known EpSC markers, including LGR5, LGR6, CD200, KRT15, ITGA6, TCF4, FZD2, DKK3, CTNNB1, LEF1 and LHX2 in KRT14 + /KRT15 + / ITGA6 + cells compared with control CD200 + /ITGA6 + cells and hepscs. Data shown are mean ± SD of the expression from three independent experiments.
15 Supplementary Figure 15. Microarray analysis of the expression of KRT1, KRT8, KRT10, KRT15, ITGA6, ITGB1 and ITGB4 in hipscs, hipsc-derived EpSCs and hepscs from hair follicles. KRT1, KRT8, KRT10, KRT15, ITGA6, ITGB1 and ITGB4 were selected and the expression data were shown from raw data of microarray.
16 Supplementary Figure 16. Statistical analysis of the densities of hair follicles produced by patch assay and chamber assay using EpSCs derived from hipscs and neonatal mouse dermal fibroblasts. Data shown are mean ± SD of the hair follicle numbers from three independent experiments in the chamber assay. Data shown are mean ± SD of the hair follicle numbers from seven independent experiments in the patch assay.
17 Supplementary Figure 17. a. Flow cytometric analysis of the percentage of CD200 + and KRT15 + cells in the neonatal foreskin-derived keratinocytes and hipsc-derived keratinocytes. b. qpcr analysis of the expression of epithelial marekers among hipscs, hepscs, hipsc-epscs and neonatal foreskin-derived keratinocytes (Foreskinkeratinocytes). Data shown are mean ± SD of the expression from three independent experiments.
18 Supplementary Figure 18. Skin permeability assay Human-like skin in the chamber graft was treated with toluidine blue dissolved in DMSO (up panel) and H 2 O (down panel) respectively. Human-like skin was impermeable to toluidine blue dissolved in H 2 O. Toluidine blue dissolved DMSO permeated into skin and skin was dyed in blue. Scale bar indicates 2mm.
19 Supplementary Figure 19. Original immunostaing results of AE13 (a), AE15 (b) and K75 (c) in the hair follicle derived from hipscs, which are illustrated in Figure 4c, 4d and 4e. Scale bar indicates 50 um (a and b) and 200 um(c).
20 Supplementary Figure 20. Oil red staining of sebocyte and qpcr analysis of sebocyte markers. a. Sebocytes derived from ipsc-epscs were stained with Oil and showed Oil red staining. Scale bar: left panel, 100 um; right panel, 20 um. b. qpcr analysis of sebocyte markers, including KRT7, PPAR-α and LPL in hipsc-epscs, hepscs, hipsc-sebocytes and HaCaT- Sebocytes. Data shown are mean ± SD of the expression from three independent experiments.
21 Supplementary Figure 21. Flow cytometric analysis of cells positive for Lrig1 + /ITGA6 +, Lrg6 + /ITGA6 + or MTS24 + /ITGA6 + during epithelial cell differentiation from hipscs at day 18. Flow cytometric analysis was performed using antibodies specific for Lrig1, Lrg6, MTS24 and ITGA6. Positive cell percentage was monitored at day 18.
22 Supplementary Figure 22. Flow cytometric analysis of positive cells for SSEA3 when only BMP4 was added during the differentiation. Flow cytometric analysis was performed using antibody specific for SSEA3. SSEA + cell percentage was monitored at 11, 18 and 25 days respectively
23 Supplementary Tables Supplementary Table 1 Primer sequences Gene Forward sequence Reverse sequence Applications ACTB TGAAGTGTGACGTGGACATC GGAGGAGCAATGATCTTGAT RT-PCR ITGB4 CTGTACCCGTATTGCGACT AGGCCATAGCAGACCTCGTA RT-PCR ITGA6 GCTGGTTATAATCCTTCAATATCAATTGT TTGGGCTCAGAACCTTGGTTT RT-PCR COL7A1 GATGACCCACGGACAGAGTT ACTTCCCGTCTGTGATCAGG RT-PCR LAMB3 GACAGGACTGGAGAAGCGTGTG CCATTGGCTCAGGCTCAGCT RT-PCR KRT5 ATCTCTGAGATGAACCGGATGATC CAGATTGGCGCACTGTTTCTT RT-PCR KRT14 GGCCTGCTGAGATCAAAGACTAC CACTGTGGCTGTGAGAATCTTGTT RT-PCR KRT8 GATCGCCACCTACAGGAAGCT ACTCATGTTCTGCATCCCAGACT RT-PCR KRT18 GAGTATGAGGCCCTGCTGAACATCA GCGGGTGGTGGTCTTTTGGAT RT-PCR OCT4(endo) CCTCACTTCACTGCACTGTA CAGGTTTTCTTTCCFTAGCT RT-PCR NANOG TGAACCTCAGCTACAAACAG TGGTGGTAGGAAGAGTAAAG RT-PCR KLF4(endo) GATGAACTGACCAGGCACTA GTGGGTCATATCCACTGTCT RT-PCR SOX2(endo) CCCAGCAGACTTCACATGT CCTCCCATTTCCCTCGTTTT RT-PCR REX1 TCGCTGAGCTGAAACAAATG CCCTTCTTGAAGGTTTACAC RT-PCR htert TGTGCACCAACATCTACAAG GCGTTCTTGGCTTTCAGGAT RT-PCR OCT4 transgene CCCCAGGGCCCCATTTTGGTACC CAACAACCGAAAATGCACCAGCCCCAG RT-PCR KLF4 transgene ACGATCGTGGCCCCGGAAAAGGACC CAACAACCGAAAATGCACCAGCCCCAG RT-PCR SOX2 transgene GGCACCCCTGGCATGGCTCTTGGCTC CAACAACCGAAAATGCACCAGCCCCAG RT-PCR Involucrin CTCCTCCAGTCAATACCCATCAG ACATTCTTGCTCAGGCAGTCC RT-PCR Flaggrin TTCGGCAAATCCTGAAGAATC CTTGAGCCAACTTGAATACCATC RT-PCR LGR5 CCTTCCAACCTCAGCGTCTTC GGGAATGTATGTCAGAGCGTTTC RT-PCR LGR6 CACACCCCAGTGTCCAGTGTAG CCACAGGAAATGCCAGTCAAG RT-PCR TCF4 CAGCAAGCACTGCCGACTAC CCAGGCTGATTCATCCCACT RT-PCR CD200 TGACTCTGTCTCACCCAAATG GCTTAGCAATAGCGGAACTG RT-PCR FZD2 GGACATCGCCTACAACCAGAC GCGTACATGGAGCACAGGAAG RT-PCR DKK3 ACCGAGAAATTCACAAGATAACC CTGGCAGGTGTACTGGAAGC RT-PCR LEF1 CCATCACGGGTGGATTCAG TCTGTTCATGCTGAGGCTTCAC RT-PCR LHX2 ATACCCGAGCAGCCAGAAGAC GGAGGACCCGCTTGGTGAG RT-PCR KRT7 GGACATCGAGATCGCCACCT TGCCACCGCCACTGCTACT RT-PCR KRT19 AACCAAGTTTGAGACGGAACAG GAGCGGAATCCACCTCCAC RT-PCR LPL TCATTCCCGGAGTAGCAGAGT GGCCACAAGTTTTGGCACC RT-PCR PPARA TTCGCAATCCATCGGCGAG CCACAGGATAAGTCACCGAGG RT-PCR meoct4 GAGGTTGGAGTAGAAGGATTGTTTTGGTTT CCCCCCTAACCCATCACCTCCACCACCTAA menanog TGGTTAGGTTGGTTTTAAATTTTT G AACCCACCCTTATAAATTCTCAATTA Bisulfite sequencing Bisulfite sequencing
24 Supplementary Table 2 Antibody Information Antibody Name Applications Dilution Company CD200 Flow Cytomentry 1:20 Ebioscience ITGA6 Flow Cytomentry 1:100 Ebioscience ITGB4 Flow Cytomentry 1:100 Ebioscience ITGB1 Flow Cytomentry 1:100 Ebioscience ITGB4 Immunofluorescence 1:200 Ebioscience ITGB1 Immunofluorescence 1:200 Ebioscience Krt14 Immunofluorescence 1:1000 Lab Vision p67 Immunofluorescence 1:500 Lab Vision Krt15 Immunofluorescence 1:500 Lab Vision E-CAD Immunofluorescence 1:40 Dako Cytokeratin Immunofluorescence 1:150 Dako SSEA3 Immunofluorescence 1:20 DSHB SSEA4 Immunofluorescence 1:20 DSHB TR-1-60 Immunofluorescence 1:100 Santa Cruz OCT4t Immunofluorescence 1:100 Santa Cruz Nanog Immunofluorescence 1:100 Millipore AE13 Immunochemistry 1:100 Abcam AE15 Immunochemistry 1:100 Abcam K75 Immunochemistry 1:100 Sigma-Aldrich
Supplementary Information
 Supplementary Information Supplementary Figures Supplementary Figure 1. qrt-pcr analysis of the expression of candidate factors including SOX2, MITF, PAX3, DLX5, FOXD3, LEF1, MSX1, PAX6, SOX2 and SOX9
Supplementary Information Supplementary Figures Supplementary Figure 1. qrt-pcr analysis of the expression of candidate factors including SOX2, MITF, PAX3, DLX5, FOXD3, LEF1, MSX1, PAX6, SOX2 and SOX9
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Functional assays of hepatocyte-like cells induced from H1 hescs (a) P450 and AAT staining, PAS staining, LDL uptake assay, and ICG uptake and release assay
Supplementary Figure 1 Supplementary Figure 1. Functional assays of hepatocyte-like cells induced from H1 hescs (a) P450 and AAT staining, PAS staining, LDL uptake assay, and ICG uptake and release assay
hipscs were derived from human skin fibroblasts (CRL-2097) by ectopic
 Generation and characterization of hipscs hipscs were derived from human skin fibroblasts (CRL-2097) by ectopic expression of OCT4, SOX2, KLF4, and C-MYC as previously described 1. hipscs showed typical
Generation and characterization of hipscs hipscs were derived from human skin fibroblasts (CRL-2097) by ectopic expression of OCT4, SOX2, KLF4, and C-MYC as previously described 1. hipscs showed typical
T-iPSC. Gra-iPSC. B-iPSC. TTF-iPSC. Supplementary Figure 1. Nature Biotechnology: doi: /nbt.1667
 a T-iPSC Gra-iPSC B-iPSC TTF-iPSC Ectoderm Endoderm Mesoderm b Nature Biotechnology: doi:.38/nbt.667 Supplementary Figure Klf4 transgene expression Oct4 transgene expression.7 Fold GAPDH.6.5.4.3 Fold GAPDH
a T-iPSC Gra-iPSC B-iPSC TTF-iPSC Ectoderm Endoderm Mesoderm b Nature Biotechnology: doi:.38/nbt.667 Supplementary Figure Klf4 transgene expression Oct4 transgene expression.7 Fold GAPDH.6.5.4.3 Fold GAPDH
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplemental Figure 1 (Figure S1), related to Figure 1 Figure S1 provides evidence to demonstrate Nfatc1Cre is a mouse line that directed gene
 Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2239 Hepatocytes (Endoderm) Foreskin fibroblasts (Mesoderm) Melanocytes (Ectoderm) +Dox rtta TetO CMV O,S,M,K & N H1 hes H7 hes H9 hes H1 hes-derived EBs H7 hes-derived EBs H9 hes-derived
DOI: 10.1038/ncb2239 Hepatocytes (Endoderm) Foreskin fibroblasts (Mesoderm) Melanocytes (Ectoderm) +Dox rtta TetO CMV O,S,M,K & N H1 hes H7 hes H9 hes H1 hes-derived EBs H7 hes-derived EBs H9 hes-derived
Isolation of human ips cells using EOS lentiviral vectors to select for pluripotency
 nature methods Isolation of human ips cells using EOS lentiviral vectors to select for pluripotency Akitsu Hotta, Aaron Y L Cheung, Natalie Farra, Kausalia Vijayaragavan, Cheryle A Séguin, Jonathan S Draper,
nature methods Isolation of human ips cells using EOS lentiviral vectors to select for pluripotency Akitsu Hotta, Aaron Y L Cheung, Natalie Farra, Kausalia Vijayaragavan, Cheryle A Séguin, Jonathan S Draper,
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State
 Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
A Novel Platform to Enable the High-Throughput Derivation and Characterization of Feeder-Free Human ipscs
 Supplementary Information A Novel Platform to Enable the High-Throughput Derivation and Characterization of Feeder-Free Human ipscs Bahram Valamehr, Ramzey Abujarour, Megan Robinson, Thuy Le, David Robbins,
Supplementary Information A Novel Platform to Enable the High-Throughput Derivation and Characterization of Feeder-Free Human ipscs Bahram Valamehr, Ramzey Abujarour, Megan Robinson, Thuy Le, David Robbins,
SUPPLEMENTARY INFORMATION
 doi:.38/nature08267 Figure S: Karyotype analysis of ips cell line One hundred individual chromosomal spreads were counted for each of the ips samples. Shown here is an spread at passage 5, predominantly
doi:.38/nature08267 Figure S: Karyotype analysis of ips cell line One hundred individual chromosomal spreads were counted for each of the ips samples. Shown here is an spread at passage 5, predominantly
SUPPLEMENTARY FIG. S9. Morphology and DNA staining of cipsc-differentiated trophoblast cell-like cell. Scale bar: 100 mm. SUPPLEMENTARY FIG. S10.
 Supplementary Data SUPPLEMENTARY FIG. S1. Lentivirus-infected canine fibroblasts show YFP expression. (A, B) Uninfected canine skin fibroblasts (CSFs); (C, D) infected CSFs; (E, F) uninfected CTFs; (G,
Supplementary Data SUPPLEMENTARY FIG. S1. Lentivirus-infected canine fibroblasts show YFP expression. (A, B) Uninfected canine skin fibroblasts (CSFs); (C, D) infected CSFs; (E, F) uninfected CTFs; (G,
Biology Open (2015): doi: /bio : Supplementary information
 Fig. S1. Verification of optimal conditions for the generation of LIF-dependent inscs Representative images (from five repeat experiments) of human primary fibroblasts undergoing reprogramming for 21 days
Fig. S1. Verification of optimal conditions for the generation of LIF-dependent inscs Representative images (from five repeat experiments) of human primary fibroblasts undergoing reprogramming for 21 days
Supplementary Figure 1.
 Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Derivation of UCSFB lines from biopsied blastomeres of cleavage-stage human. Spare embryos were obtained through the UCSF IVF Tissue Bank from donors
 Supplementary Materials and Methods Derivation of UCSFB lines from biopsied blastomeres of cleavage-stage human embryos Spare embryos were obtained through the UCSF IVF Tissue Bank from donors undergoing
Supplementary Materials and Methods Derivation of UCSFB lines from biopsied blastomeres of cleavage-stage human embryos Spare embryos were obtained through the UCSF IVF Tissue Bank from donors undergoing
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Effect of timing of DE dissociation and RA concentration on lung field specification in hpscs. (a) Effect of duration of endoderm induction on expression of
Supplementary Figures Supplementary Figure 1. Effect of timing of DE dissociation and RA concentration on lung field specification in hpscs. (a) Effect of duration of endoderm induction on expression of
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb37 a mrna relative expression 1.3 1..8.5.3. CTR NGPS OCT4 SOX2 KLF4 c-myc b BANF1 mrna levels 1.2 1..8.6.4.2. plko.1 shbanf1 c Tra-1-6+ colonies 3 1 plko.1 shbanf1 d BANF1 locus Plasmid donor
DOI: 1.138/ncb37 a mrna relative expression 1.3 1..8.5.3. CTR NGPS OCT4 SOX2 KLF4 c-myc b BANF1 mrna levels 1.2 1..8.6.4.2. plko.1 shbanf1 c Tra-1-6+ colonies 3 1 plko.1 shbanf1 d BANF1 locus Plasmid donor
SUPPLEMENTAL METHODS Reverse transcriptase PCR (RT-PCR) and quantitative real-time PCR (Q-RT-PCR) RNA was isolated with TRIZOL (Invitrogen), and
 SUPPLEMENTAL METHODS Reverse transcriptase PCR (RT-PCR) and quantitative real-time PCR (Q-RT-PCR) RNA was isolated with TRIZOL (Invitrogen), and first strand cdna was synthesized using 1 μg of RNA and
SUPPLEMENTAL METHODS Reverse transcriptase PCR (RT-PCR) and quantitative real-time PCR (Q-RT-PCR) RNA was isolated with TRIZOL (Invitrogen), and first strand cdna was synthesized using 1 μg of RNA and
Supplementary Figure 1. Wnt10a mutation causes aberrant molar tooth development and formation of an ectopic M4 molar. (a,b) Smaller molar teeth with
 Supplementary Figure 1. Wnt10a mutation causes aberrant molar tooth development and formation of an ectopic M4 molar. (a,b) Smaller molar teeth with blunted cusp formation, and presence of an ectopic molar
Supplementary Figure 1. Wnt10a mutation causes aberrant molar tooth development and formation of an ectopic M4 molar. (a,b) Smaller molar teeth with blunted cusp formation, and presence of an ectopic molar
Supplementary Data. Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells
 Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
SUPPLEMENTARY INFORMATION
 Supplementary Figure S1 Colony-forming efficiencies using transposition of inducible mouse factors. (a) Morphologically distinct growth foci appeared from rtta-mefs 6-8 days following PB-TET-mFx cocktail
Supplementary Figure S1 Colony-forming efficiencies using transposition of inducible mouse factors. (a) Morphologically distinct growth foci appeared from rtta-mefs 6-8 days following PB-TET-mFx cocktail
NIA Aging Cell Repository
 Certificate of Analysis NIA Aging Cell Repository Human induced Pluripotent Stem Cell (ipsc) Line: AG25370*B Diagnosis Alzheimer Disease, Familial, Type 4 Mutation Reprogramming method Publication(s) describing
Certificate of Analysis NIA Aging Cell Repository Human induced Pluripotent Stem Cell (ipsc) Line: AG25370*B Diagnosis Alzheimer Disease, Familial, Type 4 Mutation Reprogramming method Publication(s) describing
Use of Induced Pluripotent Stem Cells in Dermatological Research Jason Dinella 1 3, Maranke I. Koster 1 3 and Peter J. Koch 1 4
 Use of Induced Pluripotent Stem Cells in Dermatological Research Jason Dinella 1 3, Maranke I. Koster 1 3 and Peter J. Koch 1 4 Journal of Investigative Dermatology (2014) 134, e23. doi:10.1038/jid.2014.238
Use of Induced Pluripotent Stem Cells in Dermatological Research Jason Dinella 1 3, Maranke I. Koster 1 3 and Peter J. Koch 1 4 Journal of Investigative Dermatology (2014) 134, e23. doi:10.1038/jid.2014.238
Supplementary Figure 1.
 Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Comparative study of human induced pluripotent stem cells derived from bone marrow cells, hair keratinocytes and skin fibroblasts
 ESC CONGRESS 2012, Munich Comparative study of human induced pluripotent stem cells derived from bone marrow cells, hair keratinocytes and skin fibroblasts I have no financial relationships to disclose
ESC CONGRESS 2012, Munich Comparative study of human induced pluripotent stem cells derived from bone marrow cells, hair keratinocytes and skin fibroblasts I have no financial relationships to disclose
Int. J. Mol. Sci. 2016, 17, 1259; doi: /ijms
 S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
Human induced Pluripotent Stem Cell (ipsc) Line: GM23225*B
 Certificate of Analysis NIGMS Human tic Cell Repository Human induced Pluripotent Stem Cell (ipsc) Line: GM23225*B Diagnosis Mutation Reprogramming method Publication(s) describing ipsc line establishment
Certificate of Analysis NIGMS Human tic Cell Repository Human induced Pluripotent Stem Cell (ipsc) Line: GM23225*B Diagnosis Mutation Reprogramming method Publication(s) describing ipsc line establishment
Supplementary Tables. Primers and probes. Target Primer / probe sequences Chemistry Human HBB F: AACTGTGTTCACTAGCAACCTCAAA
 Supplementary Tables Primers and probes Target Primer / probe sequences Chemistry H F: AACTGTGTTCACTAGCAACCTCAAA promoter R: ACAGGGCAGTAACGGCAGACT H Downstream Actb alpha (162) alpha (162.) (Pro) (16)
Supplementary Tables Primers and probes Target Primer / probe sequences Chemistry H F: AACTGTGTTCACTAGCAACCTCAAA promoter R: ACAGGGCAGTAACGGCAGACT H Downstream Actb alpha (162) alpha (162.) (Pro) (16)
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Supplemental Information
 Cell Stem Cell, Volume 15 Supplemental Information Generation of Naive Induced Pluripotent Stem Cells from Rhesus Monkey Fibroblasts Riguo Fang, Kang Liu, Yang Zhao, Haibo Li, Dicong Zhu, Yuanyuan Du,
Cell Stem Cell, Volume 15 Supplemental Information Generation of Naive Induced Pluripotent Stem Cells from Rhesus Monkey Fibroblasts Riguo Fang, Kang Liu, Yang Zhao, Haibo Li, Dicong Zhu, Yuanyuan Du,
Assuring Pluripotency of ESC and ipsc Lines
 Assuring Pluripotency of ESC and ipsc Lines Introduction Compared to other cell types, stem cells have the unique ability to self-renew their populations by dividing into identical daughter stem cells
Assuring Pluripotency of ESC and ipsc Lines Introduction Compared to other cell types, stem cells have the unique ability to self-renew their populations by dividing into identical daughter stem cells
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Disease Clinical features Genotype
 Disease Clinical features Genotype Alpha 1 Antitrypsin deficiency (A1ATD) Patient 1 65 year old caucasian male, liver transplant recipient, explant biopsy confirmed A1ATD induced liver cirrhosis Patient
Disease Clinical features Genotype Alpha 1 Antitrypsin deficiency (A1ATD) Patient 1 65 year old caucasian male, liver transplant recipient, explant biopsy confirmed A1ATD induced liver cirrhosis Patient
Supplementary Table S1
 Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Xeno-Free Systems for hesc & hipsc. Facilitating the shift from Stem Cell Research to Clinical Applications
 Xeno-Free Systems for hesc & hipsc Facilitating the shift from Stem Cell Research to Clinical Applications NutriStem Defined, xeno-free (XF), serum-free media (SFM) specially formulated for growth and
Xeno-Free Systems for hesc & hipsc Facilitating the shift from Stem Cell Research to Clinical Applications NutriStem Defined, xeno-free (XF), serum-free media (SFM) specially formulated for growth and
Directed Differentiation of Human Induced Pluripotent Stem Cells. into Fallopian Tube Epithelium
 Directed Differentiation of Human Induced Pluripotent Stem Cells into Fallopian Tube Epithelium Nur Yucer 1,2, Marie Holzapfel 2, Tilley Jenkins Vogel 2, Lindsay Lenaeus 1, Loren Ornelas 1, Anna Laury
Directed Differentiation of Human Induced Pluripotent Stem Cells into Fallopian Tube Epithelium Nur Yucer 1,2, Marie Holzapfel 2, Tilley Jenkins Vogel 2, Lindsay Lenaeus 1, Loren Ornelas 1, Anna Laury
Supplementary Figure 1 Characterization of OKSM-iNSCs
 Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
Human embryonic stem cells for in-vitro developmental toxicity testing
 Human embryonic stem cells for in-vitro developmental toxicity testing HESI workshop on alternatives assays for developmental toxicity Raimund Strehl Cellartis The company Founded in early 2001, University
Human embryonic stem cells for in-vitro developmental toxicity testing HESI workshop on alternatives assays for developmental toxicity Raimund Strehl Cellartis The company Founded in early 2001, University
Protocol Using the Reprogramming Ecotropic Retrovirus Set: Mouse OSKM to Reprogram MEFs into ips Cells
 STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblast (MEF) cells into induced pluripotent stem (ips) cells in a 6-well format. Transduction
STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblast (MEF) cells into induced pluripotent stem (ips) cells in a 6-well format. Transduction
- 1 - BIOalternatives SAS, 1 bis rue des Plantes, Gencay, France
 Manuscript clean - 1 - Human embryonic stem cells derivatives enable full reconstruction of the pluristratified epidermis. Hind Guenou 1, Xavier Nissan 1, Fernando Larcher 2, Jessica Feteira 1, Gilles
Manuscript clean - 1 - Human embryonic stem cells derivatives enable full reconstruction of the pluristratified epidermis. Hind Guenou 1, Xavier Nissan 1, Fernando Larcher 2, Jessica Feteira 1, Gilles
Table S1. Components for knockout serum replacer medium (KSR medium) (500 ml) Component Supplier Catalogue number
 Table S1. Components for knockout serum replacer medium (KSR medium) (500 ml) Component Supplier Catalogue number Stock Concentration Final concentration Volume used Advanced DMEM/F12 Invitrogen 12634010-78%
Table S1. Components for knockout serum replacer medium (KSR medium) (500 ml) Component Supplier Catalogue number Stock Concentration Final concentration Volume used Advanced DMEM/F12 Invitrogen 12634010-78%
WT Day 90 after injections
 Supplementary Figure 1 a Day 1 after injections Day 9 after injections Klf5 +/- Day 1 after injections Klf5 +/- Day 9 after injections BLM PBS b Day 1 after injections Dermal thickness (μm) 3 1 Day 9 after
Supplementary Figure 1 a Day 1 after injections Day 9 after injections Klf5 +/- Day 1 after injections Klf5 +/- Day 9 after injections BLM PBS b Day 1 after injections Dermal thickness (μm) 3 1 Day 9 after
Supplemental Materials. Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans
 Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and
 Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.
 LEGENDS TO SUPPLEMENTARY FIGURES Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.2) were stained with the antibodies oct3/4
LEGENDS TO SUPPLEMENTARY FIGURES Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.2) were stained with the antibodies oct3/4
produced in this study characterized using 3-8 % gradient SDS-PAGE under reducing
 Supplementary Figure 1 Characterisation of newly produced laminins. Human recombinant LN-521 produced in this study characterized using 3-8 % gradient SDS-PAGE under reducing 1 and non-reducing conditions.
Supplementary Figure 1 Characterisation of newly produced laminins. Human recombinant LN-521 produced in this study characterized using 3-8 % gradient SDS-PAGE under reducing 1 and non-reducing conditions.
HUMAN EMBRYONIC STEM CELL (hesc) ASSESSMENT BY REAL-TIME POLYMERASE CHAIN REACTION (PCR)
 HUMAN EMBRYONIC STEM CELL (hesc) ASSESSMENT BY REAL-TIME POLYMERASE CHAIN REACTION (PCR) OBJECTIVE: is performed to analyze gene expression of human Embryonic Stem Cells (hescs) in culture. Real-Time PCR
HUMAN EMBRYONIC STEM CELL (hesc) ASSESSMENT BY REAL-TIME POLYMERASE CHAIN REACTION (PCR) OBJECTIVE: is performed to analyze gene expression of human Embryonic Stem Cells (hescs) in culture. Real-Time PCR
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Gene name forward primer 5->3 reverse primer 5->3 product GCCCATA. Nfatc1 AGGTGCAGCCCAAGTCTCAC GTGGCCATCTGGAGCCTTC CTGT GATGC GCT ACCAA
 Supplementary able 1 is presenting the PCR primer list used. ene name forward primer 5->3 reverse primer 5->3 product size cdna VE-Cadherin AACCCAAAAA CACCCCAAAAC 260bp CCA CCCAA PECAM-1 CACCACAA CCCCCACC
Supplementary able 1 is presenting the PCR primer list used. ene name forward primer 5->3 reverse primer 5->3 product size cdna VE-Cadherin AACCCAAAAA CACCCCAAAAC 260bp CCA CCCAA PECAM-1 CACCACAA CCCCCACC
Chromatin immunoprecipitation (ChIP) ES and FACS-sorted GFP+/Flk1+ cells were fixed in 1% formaldehyde and sonicated until fragments of an average
 Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted
Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Chemically defined conditions for human ipsc derivation and culture
 Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Generation of NSCs from hpscs in SDC medium.
 Supplementary Figure 1 Generation of NSCs from hpscs in SDC medium. (a) Q-PCR of pluripotent markers (OCT4, NANOG), neural markers (SOX1, PAX6, N-Cadherin), markers for the other germ layers (T, EOMES,
Supplementary Figure 1 Generation of NSCs from hpscs in SDC medium. (a) Q-PCR of pluripotent markers (OCT4, NANOG), neural markers (SOX1, PAX6, N-Cadherin), markers for the other germ layers (T, EOMES,
Supplementary Fig. 1. A, Upper panel shows schematic molecular representation of mutation- and SNP-replacement with FANCA-c-HDAdV in FANCA-corrected
 Supplementary Fig. 1. A, Upper panel shows schematic molecular representation of mutation- and SNP-replacement with FANCA-c-HDAdV in FANCA-corrected FA-iPSC line (C-FA-iPSCs). Lower panels show the sequencing
Supplementary Fig. 1. A, Upper panel shows schematic molecular representation of mutation- and SNP-replacement with FANCA-c-HDAdV in FANCA-corrected FA-iPSC line (C-FA-iPSCs). Lower panels show the sequencing
 Supplemental Tables Supplemental Table 1. Phenotypic profile of different HSC lines. Marker d0 mhsc d7 mhsc 8B LX2 CD11b 3.61% 2.21% 2.16% 3.77% F4/80 4.54% 2.05% 0.127% 1.44% Elastin 3.90% 2.95% 4.23%
Supplemental Tables Supplemental Table 1. Phenotypic profile of different HSC lines. Marker d0 mhsc d7 mhsc 8B LX2 CD11b 3.61% 2.21% 2.16% 3.77% F4/80 4.54% 2.05% 0.127% 1.44% Elastin 3.90% 2.95% 4.23%
Development 142: doi: /dev : Supplementary Material
 Development 142: doi:1.42/dev.11687: Supplementary Material Figure S1 - Relative expression of lineage markers in single ES cell and cardiomyocyte by real-time quantitative PCR analysis. Single ES cell
Development 142: doi:1.42/dev.11687: Supplementary Material Figure S1 - Relative expression of lineage markers in single ES cell and cardiomyocyte by real-time quantitative PCR analysis. Single ES cell
Sundari Chetty, Felicia Walton Pagliuca, Christian Honore, Anastasie Kweudjeu, Alireza Rezania, and Douglas A. Melton
 A simple tool to improve pluripotent stem cell differentiation Sundari Chetty, Felicia Walton Pagliuca, Christian Honore, Anastasie Kweudjeu, Alireza Rezania, and Douglas A. Melton Supplementary Information
A simple tool to improve pluripotent stem cell differentiation Sundari Chetty, Felicia Walton Pagliuca, Christian Honore, Anastasie Kweudjeu, Alireza Rezania, and Douglas A. Melton Supplementary Information
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
hipsc-derived cardiomyocytes from Brugada Syndrome patients without identified mutations do not exhibit clear cellular electrophysiological
 hipsc-derived cardiomyocytes from Brugada Syndrome patients without identified mutations do not exhibit clear cellular electrophysiological abnormalities Authors: Christiaan C. Veerman, MD # 1 ; Isabella
hipsc-derived cardiomyocytes from Brugada Syndrome patients without identified mutations do not exhibit clear cellular electrophysiological abnormalities Authors: Christiaan C. Veerman, MD # 1 ; Isabella
Generation of ips-derived model cells for analyses of hair shaft differentiation
 Supplementary Material Generation of ips-derived model cells for analyses of hair shaft differentiation Takumi Kido, Tomoatsu Horigome, Minori Uda, Naoki Adachi, Yohei Hirai Department of Biomedical Chemistry,
Supplementary Material Generation of ips-derived model cells for analyses of hair shaft differentiation Takumi Kido, Tomoatsu Horigome, Minori Uda, Naoki Adachi, Yohei Hirai Department of Biomedical Chemistry,
Human Induced Pluripotent Stem Cells
 Applied StemCell, Inc. (866) 497-4180 www.appliedstemcell.com Datasheet Human Induced Pluripotent Stem Cells Product Information Catalog Number Description Tissue Age Sex Race Clinical information Quantity
Applied StemCell, Inc. (866) 497-4180 www.appliedstemcell.com Datasheet Human Induced Pluripotent Stem Cells Product Information Catalog Number Description Tissue Age Sex Race Clinical information Quantity
SUPPLEMENTARY INFORMATION
 Correction notice Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells Ángel Raya, Ignasi Rodríguez-Pizà, Guillermo Guenechea, Rita Vassena, Susana Navarro,
Correction notice Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells Ángel Raya, Ignasi Rodríguez-Pizà, Guillermo Guenechea, Rita Vassena, Susana Navarro,
bronchial epithelial cells (I). Bronchi are outlined with dashed line. Scale bars = 25 µm, if not
 Supplemental Figure S1: ronchial epithelial cell polarity and integrity is maintained in bronchi. (A-E) Staining for selected markers of bronchial cell differentiation and intracellular compartments is
Supplemental Figure S1: ronchial epithelial cell polarity and integrity is maintained in bronchi. (A-E) Staining for selected markers of bronchial cell differentiation and intracellular compartments is
TRIM31 is recruited to mitochondria after infection with SeV.
 Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
GENESDEV/2007/ Supplementary Figure 1 Elkabetz et al.,
 GENESDEV/2007/089581 Supplementary Figure 1 Elkabetz et al., GENESDEV/2007/089581 Supplementary Figure 2 Elkabetz et al., GENESDEV/2007/089581 Supplementary Figure 3 Elkabetz et al., GENESDEV/2007/089581
GENESDEV/2007/089581 Supplementary Figure 1 Elkabetz et al., GENESDEV/2007/089581 Supplementary Figure 2 Elkabetz et al., GENESDEV/2007/089581 Supplementary Figure 3 Elkabetz et al., GENESDEV/2007/089581
SUPPLEMENTARY INFORMATION
 Supplementary Discussion Interestingly, a recent study demonstrated that knockdown of Tet1 alone would lead to dysregulation of differentiation genes, but only minor defects in ES maintenance and no change
Supplementary Discussion Interestingly, a recent study demonstrated that knockdown of Tet1 alone would lead to dysregulation of differentiation genes, but only minor defects in ES maintenance and no change
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
Supplementary Figure 1. ips_3y+mir-302b. ips_3y+mir-372. ips_3y+mir-302b + mir-372. ips_4y. ips_4y+mir-302b. ips_4y + mir-372
 Supplementary Figure 1 ips_3y+mir-32b ips_3y+ ips_3y+mir-32b + ips_4y ips_4y+mir-32b ips_4y + Nature Biotechnology: doi:1.138/nb.t.1862 TRA-1-6 DAPI Oct3/4 Supplementary Figure 1: ips cells derived from
Supplementary Figure 1 ips_3y+mir-32b ips_3y+ ips_3y+mir-32b + ips_4y ips_4y+mir-32b ips_4y + Nature Biotechnology: doi:1.138/nb.t.1862 TRA-1-6 DAPI Oct3/4 Supplementary Figure 1: ips cells derived from
Nanog-Luc. R-Luc. 40 HA-Klf Relative protein level of Klf USP21. Vector USP2 USP21 *** *** *** ***
 :Flag Relative protein level of Relative protein level of : Flag Vec 21LV 21SV 2 : HA HA- HA- HA- HA-Klf4 Relative mrna expression Relative protein level of Relative protein level of Relative protein level
:Flag Relative protein level of Relative protein level of : Flag Vec 21LV 21SV 2 : HA HA- HA- HA- HA-Klf4 Relative mrna expression Relative protein level of Relative protein level of Relative protein level
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/11/e1701679/dc1 Supplementary Materials for Directed differentiation of human pluripotent stem cells to blood-brain barrier endothelial cells Tongcheng Qian,
advances.sciencemag.org/cgi/content/full/3/11/e1701679/dc1 Supplementary Materials for Directed differentiation of human pluripotent stem cells to blood-brain barrier endothelial cells Tongcheng Qian,
What we ll do today. Types of stem cells. Do engineered ips and ES cells have. What genes are special in stem cells?
 Do engineered ips and ES cells have similar molecular signatures? What we ll do today Research questions in stem cell biology Comparing expression and epigenetics in stem cells asuring gene expression
Do engineered ips and ES cells have similar molecular signatures? What we ll do today Research questions in stem cell biology Comparing expression and epigenetics in stem cells asuring gene expression
Do engineered ips and ES cells have similar molecular signatures?
 Do engineered ips and ES cells have similar molecular signatures? Comparing expression and epigenetics in stem cells George Bell, Ph.D. Bioinformatics and Research Computing 2012 Spring Lecture Series
Do engineered ips and ES cells have similar molecular signatures? Comparing expression and epigenetics in stem cells George Bell, Ph.D. Bioinformatics and Research Computing 2012 Spring Lecture Series
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector.
 Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11435 Supplementary Figures Figure S1. The scheme of androgenetic haploid embryonic stem (ahes) cells derivation. WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Figure S2.
doi:10.1038/nature11435 Supplementary Figures Figure S1. The scheme of androgenetic haploid embryonic stem (ahes) cells derivation. WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Figure S2.
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
gacgacgaggagaccaccgctttg aggcacattgaaggtctcaaacatg
 Supplementary information Supplementary table 1: primers for cloning and sequencing cloning for E- Ras ggg aat tcc ctt gag ctg ctg ggg aat ggc ttt gcc ggt cta gag tat aaa gga agc ttt gaa tcc Tpbp Oct3/4
Supplementary information Supplementary table 1: primers for cloning and sequencing cloning for E- Ras ggg aat tcc ctt gag ctg ctg ggg aat ggc ttt gcc ggt cta gag tat aaa gga agc ttt gaa tcc Tpbp Oct3/4
Supplementary Figure S1: (A) Schematic representation of the Jarid2 Flox/Flox
 Supplementary Figure S1: (A) Schematic representation of the Flox/Flox locus and the strategy used to visualize the wt and the Flox/Flox mrna by PCR from cdna. An example of the genotyping strategy is
Supplementary Figure S1: (A) Schematic representation of the Flox/Flox locus and the strategy used to visualize the wt and the Flox/Flox mrna by PCR from cdna. An example of the genotyping strategy is
Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on
 Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
H3K36me3 polyclonal antibody
 H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTRY INFORMTION DOI:.38/ncb Kdmb locus kb Long isoform Short isoform Long isoform Jmj XX PHD F-box LRR Short isoform XX PHD F-box LRR Target Vector 3xFlag Left H Neo Right H TK LoxP LoxP Kdmb Locus
SUPPLEMENTRY INFORMTION DOI:.38/ncb Kdmb locus kb Long isoform Short isoform Long isoform Jmj XX PHD F-box LRR Short isoform XX PHD F-box LRR Target Vector 3xFlag Left H Neo Right H TK LoxP LoxP Kdmb Locus
Supplementary Information
 Silva et al, 2006 Supplementary Information Nanog promotes transfer of pluripotency after cell fusion Supplementary Methods Antibodies RT-PCR analysis Trichostatin A and 5-azacytidine treatment Imaging
Silva et al, 2006 Supplementary Information Nanog promotes transfer of pluripotency after cell fusion Supplementary Methods Antibodies RT-PCR analysis Trichostatin A and 5-azacytidine treatment Imaging
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11272 Supplementary Figure 1. Experimental platform for epigenetic reversion of murine EpiSCs back to naïve ground state pluripotency. a, NGFP1 naïve-ipscs (40XY) expanded in 2i/LIF defined
doi:10.1038/nature11272 Supplementary Figure 1. Experimental platform for epigenetic reversion of murine EpiSCs back to naïve ground state pluripotency. a, NGFP1 naïve-ipscs (40XY) expanded in 2i/LIF defined
FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs
 FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs amount of standard analysed. Each point represents the
FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs amount of standard analysed. Each point represents the
Authors: Takuro Horii, Masamichi Yamamoto, Sumiyo Morita, Mika Kimura, Yasumitsu Nagao, Izuho Hatada *
 SUPPLEMENTARY INFORMATION Title: p53 Suppresses Tetraploid Development in Mice Authors: Takuro Horii, Masamichi Yamamoto, Sumiyo Morita, Mika Kimura, Yasumitsu Nagao, Izuho Hatada * Supplementary Methods
SUPPLEMENTARY INFORMATION Title: p53 Suppresses Tetraploid Development in Mice Authors: Takuro Horii, Masamichi Yamamoto, Sumiyo Morita, Mika Kimura, Yasumitsu Nagao, Izuho Hatada * Supplementary Methods
The p53-puma axis suppresses ipsc generation
 The p53-puma axis suppresses ipsc generation Yanxin Li, Haizhong Feng, Haihui Gu, Dale W. Lewis, Youzhong Yuan, Lei Zhang, Hui Yu, Peng Zhang, Haizi Cheng, Weimin Miao, Weiping Yuan, Shi-Yuan Cheng, Susanne
The p53-puma axis suppresses ipsc generation Yanxin Li, Haizhong Feng, Haihui Gu, Dale W. Lewis, Youzhong Yuan, Lei Zhang, Hui Yu, Peng Zhang, Haizi Cheng, Weimin Miao, Weiping Yuan, Shi-Yuan Cheng, Susanne
Supplementary Information
 Supplementary Information Supplementary Figure 1: Identification of new regulators of MuSC by a proteome-based shrna screen. (a) FACS plots of GFP + and GFP - cells from Pax7 ICN -Z/EG (upper panel) and
Supplementary Information Supplementary Figure 1: Identification of new regulators of MuSC by a proteome-based shrna screen. (a) FACS plots of GFP + and GFP - cells from Pax7 ICN -Z/EG (upper panel) and
Replacement of Oct4 by Tet1 during ipsc Induction Reveals an Important Role of DNA Methylation and Hydroxymethylation in Reprogramming
 Hydroxymethylation in Reprogramming, (2013), http://dx.doi.org/10.1016/j.stem.2013.02.005 Article Replacement of Oct4 by Tet1 during ipsc Induction Reveals an Important Role of DNA Methylation and Hydroxymethylation
Hydroxymethylation in Reprogramming, (2013), http://dx.doi.org/10.1016/j.stem.2013.02.005 Article Replacement of Oct4 by Tet1 during ipsc Induction Reveals an Important Role of DNA Methylation and Hydroxymethylation
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Protocol Reprogramming MEFs using the Dox Inducible Reprogramming Lentivirus Set: Mouse OKSM
 STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblasts (MEFs) into induced pluripotent stem (ips) cells in a 6-well format. Transduction
STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblasts (MEFs) into induced pluripotent stem (ips) cells in a 6-well format. Transduction
Proteasome activity is required for the initiation of precancerous pancreatic
 Proteasome activity is required for the initiation of precancerous pancreatic lesions Takaki Furuyama 1,2, Shinji Tanaka 1,2*, Shu Shimada 1, Yoshimitsu Akiyama 1, Satoshi Matsumura 2, Yusuke Mitsunori
Proteasome activity is required for the initiation of precancerous pancreatic lesions Takaki Furuyama 1,2, Shinji Tanaka 1,2*, Shu Shimada 1, Yoshimitsu Akiyama 1, Satoshi Matsumura 2, Yusuke Mitsunori
Nature Methods: doi: /nmeth Supplementary Figure 1. Extrinsic Signaling during Reprogramming.
 Supplementary Figure 1 Extrinsic Signaling during Reprogramming. a, In human pluripotent stem cells, exogenously- (medium) and endogenously-promoted (cell-secreted) FGF and TGF- pathways sustain the master
Supplementary Figure 1 Extrinsic Signaling during Reprogramming. a, In human pluripotent stem cells, exogenously- (medium) and endogenously-promoted (cell-secreted) FGF and TGF- pathways sustain the master
Supplemental Information. Tissue Mechanics Orchestrate Wnt-Dependent. Human Embryonic Stem Cell Differentiation
 Cell Stem Cell, Volume 19 Supplemental Information Tissue Mechanics Orchestrate Wnt-Dependent Human Embryonic Stem Cell Differentiation Laralynne Przybyla, Johnathon N. Lakins, and Valerie M. Weaver Supplemental
Cell Stem Cell, Volume 19 Supplemental Information Tissue Mechanics Orchestrate Wnt-Dependent Human Embryonic Stem Cell Differentiation Laralynne Przybyla, Johnathon N. Lakins, and Valerie M. Weaver Supplemental
0.9 5 H M L E R -C tr l in T w is t1 C M
 a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
Supplementary Table 1. Primary antibodies used in this study.
 Supplementary Table 1. Primary antibodies used in this study. Antibodies Mouse monoclonal Antibody(Ab) Working dilution Oct4 1:2 Pax6 1:1 Company Notes 1 Santa Cruz Biotechnology Use only Cy3 2nd antibody
Supplementary Table 1. Primary antibodies used in this study. Antibodies Mouse monoclonal Antibody(Ab) Working dilution Oct4 1:2 Pax6 1:1 Company Notes 1 Santa Cruz Biotechnology Use only Cy3 2nd antibody
Supplementary Figure 1 Schematic representation of ipsc line derivation and the samples assayed
 Supplementary igure 1 Schematic representation of ipsc line derivation and the samples assayed for DNA methylation. Most biological replicates or cell lines (i.e., samples ) in this study were represented
Supplementary igure 1 Schematic representation of ipsc line derivation and the samples assayed for DNA methylation. Most biological replicates or cell lines (i.e., samples ) in this study were represented
Characterization of new RNA polymerase III and RNA polymerase II transcriptional
 Characterization of new RNA polymerase III and RNA polymerase II transcriptional promoters in the Bovine Leukemia Virus genome Short title: New promoters in the BLV genome Benoit Van Driessche 1,#, Anthony
Characterization of new RNA polymerase III and RNA polymerase II transcriptional promoters in the Bovine Leukemia Virus genome Short title: New promoters in the BLV genome Benoit Van Driessche 1,#, Anthony
Protocol Reprogramming Human Fibroblasts into ips Cells using the Stemgent Reprogramming Lentivirus Set: Human OKSM
 STEMGENT Page 1 Reprogramming Lentivirus Set: Human OKSM OVERVIEW The following procedure describes the reprogramming of BJ Human Fibroblasts (BJ cells) to induced pluripotent stem (ips) cells using the
STEMGENT Page 1 Reprogramming Lentivirus Set: Human OKSM OVERVIEW The following procedure describes the reprogramming of BJ Human Fibroblasts (BJ cells) to induced pluripotent stem (ips) cells using the
Supplementary Figure 1. Western analysis of p-smad1/5/8 of differentiated hescs.
 kda 65 45 45 35 Supplementary Figure 1. Western analysis of p-smad1/5/8 of differentiated hescs. H9 hescs were differentiated with or without BMP4+BMP8A and cell lysates were collected for Western analysis
kda 65 45 45 35 Supplementary Figure 1. Western analysis of p-smad1/5/8 of differentiated hescs. H9 hescs were differentiated with or without BMP4+BMP8A and cell lysates were collected for Western analysis
