SUPPLEMENTARY INFORMATION
|
|
|
- Ernest Smith
- 5 years ago
- Views:
Transcription
1 doi: /nature10202 Supplementary Figure 1: Rapid generation of neurons from human ES cells. a, Phase contrast image of feeder-free culture of human ES cells infected with Brn2, Ascl1, Myt1l (BAM) and EGFP 2 days after infection. b-d, BAM-ES cells rapidly acquired neuronal morphologies within 7 days after dox treatment based on EGFP fluorescence microscopy. Inserts show enlarged area of typical morphology of cells at different stages. Scale bars indicate 100 μm. e-g, Scatter data plots of membrane resistance (Rin, e), membrane capacitance (Cm, f) and resting membrane potential (RMP, g) showing the degree of heterogeneity of these parameters. 1
2 Supplementary Figure 2: Quantification and representative images of human ES cells infected with 1-3 BAM factors. a, Quantification of MAP2-positive cells with the indicated 1-3 factor combinations of the genes Brn2 (B), Ascl1 (A) and Myt1l (M) 8 days after infection (average and SD of three randomly taken pictures (AxioCam) with a 20 objective on a Zeiss Axio Observer microscope). The total amount of virus was kept constant between different factor combinations and the same numbers of ES cells were seeded in each condition before infection. b-h, Representative images showing the typical morphology of cells in different conditions. While several conditions produce similar numbers of MAP2-positive cells, the BAM condition appears to generate cells with the most complex neuronal morphologies. Scale bar in b is applicable to all images and indicates 20 μm. 2
3 Supplementary Figure 3: Generation of neurons from human ips cells. a-b, Eight days after induction, BAM-infected ips cells displayed complex neuronal morphologies and expressed the pan-neuronal markers MAP2 (a) and Tuj1 (b). c, Representative traces of action potentials (black traces, 3 traces were overlapped) in response to ramp current injections (lower panel) 15 days after induction. Action potentials could be blocked by application of 10-6 M TTX. Membrane potential was maintained at around 63mV. d, Wholecell currents in an ips-in cell. Insert, fast activating and inactivating sodium currents. Lower panel indicates the protocol applied. e, Whole-cell currents in the presence of 10-6 M TTX. Open arrowhead indicates that prominent voltage-dependent sodium currents were blocked by TTX. Lower panel indicates the protocol applied. f, Membrane potential changes including action potential generations in response to step current injections (lower panel) in an in cell generated from BAMN-infected ips cells. Scale bars: 10μm (a,b). 3
4 Supplementary Figure 4: RT-PCR and immunofluorescence characterization of primary human fibroblast lines a, The human fetal fibroblast lines (HFF-A, B, C) and the human postnatal fibroblast lines (HPF- A, B) were grown for 2 weeks in three different conditions each: (i) in MEF media (DMEM+10% serum), (ii) N3 media a serum-free media supporting the survival of various neural cell types, (iii) N3 media + GFs = N3 media supplemented with EGF and FGF2, a condition known to promote neural progenitor growth. The cells were fixed and stained for antibodies known to recognize the human Brn2, GFAP, Sox2, MAP2, PSA-NCAM, Tuj1, Sox10 4
5 and Pan-Cytokeratin (clone Lu-5). For all antibody stainings except Tuj1 no single positive cell could be detected in 100, ,000 cells screened per condition and cell line. The Tuj1 antibodies weakly labeled the majority of the fibroblasts mostly in a diffuse cytoplasmatic pattern. The staining intensity in fibroblasts was orders of magnitudes lower than that in neurons (compare to positive control staining below). Our RT-PCR analysis on the bulk population (see below) and single cell level (see Fig. 2h) confirmed expression of tubulins on the RNA level. We were not surprised to see low level Tuj1 expression in fibroblasts as we have seen the same phenomena in mouse fibroblasts before 11. Note: For this reason when in cells were quantified in this manuscript we included only bright Tuj1-positive cells with a round soma and processes of at least the length 3 of the diameter of the soma. We conclude that none of the cultures have the potential to spontaneously differentiate into neurons. Shown are representative images of the stained fibroblast lines HFF-A and HPF-B under the indicated culture conditions. Bottom panel: positive control stainings: Brn2 (transfected fibroblasts), GFAP (mouse glia), Sox2 (mouse precursor cells), MAP2 (5 week old HFF-iN cells), PSA-NCAM (5 week old HFF-iN cells), Tuj1 (5 week old HFF-iN cells), Sox10 (human melanoma), Pan-Cytokeratin staining (clone Lu- 5, human keratinocytes). b, RT-PCR analysis of the three fetal and two of the three foreskin fibroblast lines generated. Fibroblasts were grown in different conditions as described above. Human total brain RNA (Invitrogen) was used for positive control reactions except otherwise stated. As expected, nestin, Tuj1, and sox9 mrnas were detected in all samples. Brn1, Gfap, and Pax6 were not detectable. For some lines we detected low level expression of p75 and Sox10. This may suggest the presence of small numbers of neural crest-derived cell types in these cultures. Please note that we tested lines HFF-A and HPF-B also for the presence of Sox10-postive cells by immunofluorescence (see above). While the positive control sample gave a strong and specific staining pattern we could not detect any positive cells in either line (out of about 100,000 cells screened). Thus, either the RNA signal stems from cells with extremely low Sox10 expression levels (that would not be typical for neural crest stem cells) or from very few high Sox10-expressing cells (less than 1:100,000). Ncx1 (also known as Tlx2) was not detected in any sample or media condition. Msx1 and Slug transcripts were detected in all samples, which was not surprising given their broad-spectrum expressions in endoderm and mesoderm originated tissues (see e.g. Cobaleda et al. 2007, Ann. Rev. Genet. Vol. 41, 41-61; Hatano et al., 1997 Anat. Embryol. 195(5):419-25; Davidson 1995, Trends Genet. 11(10):405-11). Human 5
6 spinal cord total RNA (DV Biologics) was used for positive control reactions for Ncx1, Msx1 and Slug. In summary, the absence of some and presence of other neural crest-associated markers shows that the proportion of neural crest stem cells must be below our detection limit. Importantly, even the potentially few numbers of neural crest cells in the cultures (i) have no endogenous neurogenic potential (ii) could not explain the relatively high efficiency rates to generate neuronal cells. Please also note that we focused our analysis on those lines in which neither Sox10 nor p75 mrna expression was detected (HFF-A and HPF-B). 6
7 Supplementary Figure 5: Morphological and functional properties of IMR-90-derived BAM-iN cells. a, Human IMR-90 fetal lung fibroblasts infected with BAM and EGFP. Twenty days after dox treatment the cells exhibited Tuj1 expression and typical neuronal morphologies. In contrast, cells infected with the EGFP virus alone did not exhibit any neuron-like morphologies up to 3 weeks post infection. b, Membrane properties of IMR-90 derived in cells. Single, action potential-like responses could be recorded following step current injections (n=4). Membrane potential was held around 66mV. Scale bar:100 μm. 7
8 Supplementary Figure 6: Quantification, morphology and functional characterizations of human fetal fibroblasts infected with different combination of BAMN pool viruses. a, The number of Tuj1-positive neuronal cells were determined in the various indicated 2-, 3, and 4- Factor conditions (B=Brn2, A=Ascl1, M=Myt1l, N=Neurod1). Shown are average numbers and standard deviations per 20 visual field (n=8 fields). b, Left panel; Tuj1 staining of human fibroblast under indicated conditions; right panels; Representative membrane potential changes in response to ramp current injections. Cells were maintained at membrane potential as indicated 8
9 in each trace. A 500 pa ramp with 1sec duration protocol was applied. n equals the number of neurons recorded from three independent cultures. Bar=50 m. While the 2-Factor condition BN was surprisingly efficient to generate cells with neuronal morphology, action potentials were observed in a subpopulation of cells. On the other hand, the BAMN condition yielded similar amount of neuronal cells but those were able to generate repetitive action potentials. 9
10 Supplementary Figure 7: Generation of human in cells from multiple HFF lines. HFF-A derived in cells express Tuj1 (green) and Neurofilament (NF, red) (a), Synaptotagmin 1 (Syt1, red) (b). Immunofluorescence analysis of HFF-B derived in cells with Tuj1(c) and PSA-NCAM (d) antibodies. e, Representative traces of membrane potentials in response to step hypopolarization and depolarizations. f-g, Immunofluorescence analysis of HFF-C derived in cells with Tuj1 (f) and PSA-NCAM (g) antibodies. h, Representative traces of membrane potentials in response to step hypoplarization and depolarizations. i, Scatter plots showing the distribution of intrinsic membrane properties between individual cells (Rin = input resistance, Cm = capacitance, RMP = resting membrane potential). Bar=10 m. 10
11 Supplementary Figure 8: Single cell collection guided by fluorescence and light microscope. a, A merged image of fluorescence and DIC images showing the morphology of an in cell generated from HFF cells infected with BAMN and EGFP. A glass pipette was placed near the cell body. b-c, DIC images showing the cell body of the in cell was aspirated into the pipette tip. d, Merged fluorescence and bright-field images shows that a single fluorescent cell was collected. Aspirated cells were collected into individual PCR tubes containing 5 µl 2 cellsdirect reaction buffer (Invitrogen) by positive pressure and processed for single-cell gene profiling using Fluidigm technology. Bar=10 µm. 11
12 Supplementary Figure 9: in cells derived from HFF cells express Tbr1 and Peripherin a-c, Representative images of HFF-iN cells expressing GFP (a), Tbr1 (b), and DAPI (c). d, Merged picture of a-c. The arrowhead indicates a Tbr1-positive in cell; the asterisk indicates a Tbr1-negative in cell. e and f, Representative images of HFF-iN cells expressing peripherin. g, An merged picture of EGFP (green) and peripherin staining (red) showing a peripherin-positive and a peripherin-negative in cell. Scale bars: in (d)=10 µm, applies to a-c; in (e)=50 µm and in (g)=10 µm and applies to (f). 12
13 Supplementary Figure 10: Endogenous and exogenous mrna expression levels of BAMN factors. The transcript levels of the endogenous (human) genes and exogenous (mouse) genes were quantified in human fetal fibroblasts that were (i) uninfected (before), (ii) infected but in absence of dox (no dox), (iii) 48h after infection and dox treatment (dox 48 h), and (iv) cultured for 2 weeks in dox and another 10 days without dox (dox withdrawal, dox WD). Quantitative RT-PCR reactions were performed using SYBR Green (Applied Biosystems, see methods) according to manufacturer s recommendations, relative mrna levels were calculated by the delta-delta-ct method and normalized to mrna levels of each gene at 48 hours after addition of dox. Primers were designed to specifically amplify either the human UTR or the mouse coding sequence of the respective cdnas. Primer sequences are listed in Suppl. Table
14 Supplementary Figure 11: HFF-iN cells do not require continuous expression of exogenous BAMN factors to retain a neuronal phenotype. HFF-iN cells 5 weeks after BAMN infection. Cells were treated with dox for the initial 2 weeks of culture, and cultured in the absence of dox for the remaining 3 weeks. a-c, Immunofluorescence analyses were performed for Tuj1 (a), MAP2 (b) and PSA-NCAM (c) to confirm retention of neuronal characteristics. d, HFF-iN cell generated action potentials upon current injections. Bar=20 m. 14
15 Supplementary Figure 12: Membrane properties of HFF-iN cells a, Spontaneous action potentials recorded from an in cell 25 days after infection (observed in 1/42 cells recorded). b, Representative traces of induced action potentials by a ramp depolarization (500 pa over 1 sec) with or without 1 M TTX. Two traces from each condition were overlayed. TTX effectively blocked the formation of action potentials. c, Representative 15
16 traces of whole-cell currents measured in voltage-clamp mode. Cell was held at -70 mv, voltage was increased stepwise from -90 mv to +60 mv in 10 mv intervals. d, Current-voltage relationship of outward whole-cell currents recorded from HFF-iN cells. Currents were measured at 50 ms before the end of the pulse (arrow indicated in c). Cells were held at -70 mv, 500 ms durations of voltage steps from -90 mv to +80 mv, at 10 mv intervals, were applied (n=3). e. Inward current response following application of 10-4 M GABA from a pipette using a picospritzer (n=4); lower trace shows that application of picrotoxin, a specific antagonist of GABA A receptors, blocks the GABA-induced current response. The internal solution contained ~135 mm Cl - explaining the inward current response. Cells were held at -70 mv. f. Inward current responses following application of 10-3 M L-glutamate from a pipette using a picospritzer (n=4). Lower panel indicates that glutamate-induced current response could be blocked by CNQX, a specific antagonist of AMPA receptors. Cells were held at -70 mv. g, Scatter plots showing the distribution of intrinsic membrane properties between individual cells of HFF-A and HPF-B at indicated time points after infection (Rin = input resistance, Cm = capacitance, RMP = resting membrane potential). Note the observed heterogeneity. 16
17 Supplementary Figure 13: Gene expression analyses of in cells derived from HPFs a-c, Representative images of HPF cultures 6 weeks after infection with BAMN and EGFP viruses (a), Tbr1 (b), and DAPI (c). d, Merged picture of a-c. Almost all postnatal fibroblasts expressed Tbr1 in the nucleus (asterisks) but the signal was much weaker than in in cells. ImageJ (NIH) software was used to quantify the numbers of Tbr1-positive in cells expressing higher levels than the average fibroblast expression level. To achieve this, images of Tbr1 staining were taken at constant exposure time (20 objectives). Background fluorescence was subtracted from the whole picture by threshold level to exclude the signal of the weakly stained fibroblast nuclei. Resulting Tbr1/DAPI-positive structures per in cells was quantified. e, Representative images of HPF-iN cells expressing Peripherin. f, Single cell gene expression profiling using Fluidigm dynamic arrays. Rows represent the evaluated genes and columns represent individual cells. Heatmap (blue to red) represents the threshold Ct values as indicated. Scale bars=10 µm. 17
18 Supplementary Figure 14: in cells derived from skin fibroblasts of an 11-year-old human a, Representative pictures of Tuj1 (left panels) and MAP2 (right panels) staining of in cells generated from a primary culture of dermal fibroblasts 21 days after BAMN transduction. b, Representative traces of membrane potential changes in response to a ramp current injection. c, Representative traces of whole-cell currents in response to a ramp voltage depolarization. Fast activating and inactivating sodium currents were shown. Bar=50 m 18
19 Supplementary Figure 15: in cells derived from HPF cells receive synaptic inputs from mouse cortical neurons. HPFs were infected with BAMN-factor and human synapsin promoter-driving td-tomato. Fourteen days after transduction, in cells expressing td-tomato were insolated by FACS and seeded on mouse cortical neuronal cultures established 5 days before. Fourteen days after replating (28 days after dox), two out of six recorded cells showed unambiguous synaptic responses. The trace shows an example of the spontaneous bursting synaptic activity recorded from one in cell integrated into the mouse cortical neuronal network in vitro. The lower panel shows expanded traces indicating the kinetics of the synaptic responses. 19
20 Supplementary Table 1: Electrophysiological characterizations of human ES-iN and HFFiN cells. Electrophysiological Parameters in human in cells Average ± S.E. N p Value hes-in cells Membrane Input Resistance (GΩ) Day ± Day ± p<0.001 Day ± p<0.001 Membrane Capacitance (pf) Day ± Day ± p<0.001 Day ± p<0.001 RMP (mv) Day ± Day ± p<0.001 Day ± p<0.001 After-hyperpolarization potential (mv) Day 0 n.a. n.a. 12 Day ± Day ± AP (Spontaneous and Induced) Spontaneous Induced Total neurons recorded Day 0 n.d. n.d. 12 Day Day Human Fetal Fibroblast (HFF) - in cells Membrane Input Resistance (GΩ) HFF-A (Day 14-25) 1.77 ± HFF-A (Day 34-35) 1.11 ± p<0.05 HFF-B (Day 24) 1.24 ± HFF-C (Day 24) 1.35 ± Membrane Capacitance (pf) HFF-A (Day 14-25) ± HFF-A (Day 34-35) ± p<0.001 HFF-B (Day 24) ±
21 HFF-C (Day 24) 17.4 ± RMP (mv) HFF-A (Day 14-25) ± HFF-A (Day 34-35) ± n.s. HFF-B (Day 24) ± HFF-C (Day 24) ± AP (Spontaneous and Induced) Spontaneous Induced Total neurons recorded 28 (9 with AP like HFF-A (Day 14-25) 1 responses) 42 HFF-A (Day 34-35) HFF-B (Day 24) HFF-C (Day 24) Synaptic activities (co-cultured with mouse cortical neurons) HFF-A Spontaneous only Evoked only Spontaneous & Evoked Total neurons recorded Day Day Human Postnatal Fibroblast (HPF) - in cells (Day 28 to 35 after infection) Membrane Input Resistance (GΩ) HPF-A 1.56 ± HPF-B 1.58 ± HPF-C 1.34 ± Membrane Capacitance (pf) HPF-A 23.2 ± HPF-B ± HPF-C 19.8 ± RMP (mv) HPF-A -39 ± HPF-B ± HPF-C -46 N.A 2 AP (Spontaneous and Induced) Spontaneous Induced Total neurons recorded HPF-A HPF-B HPF-C
22 Synaptic activities (co-cultured with mouse cortical neurons) HPF Spontaneous only Evoked only Spontaneous & Evoked Total neurons recorded Day Note: hes-in cells were induced using BAM pool and HFF-iN cells and HPF-iN cells were induced using BAMN pool of transcription factors. AP: Action Potential; HFF: Human fetal fibroblast (lines A, -B, and C); HPF: Human postnatal fibroblast (lines A, -B and C); RMP: Resting Membrane Potential. Student t-tests were used for statistical analysis. 22
23 Supplementary Table 2 Primers used in this study for Fluidigm dynamic array RT-PCR, RT-PCR, and quantitative RT-PCR. Gene Symbol NCBI acc. No. primer sequences sense/antisense method ACTB NM_ ccaaccgcgagaagatga sense F ccagaggcgtacagggatag antisense F CAMKIIB NM_ gagccatcctcaccaccat sense F tattcgtctggggcttgact antisense F DCX NM_ catccccaacacctcagaag sense F ggaggttccgtttgctga antisense F GAD65 M ctcattgccttcacgtctga sense F gctgtctgttccaatccctaa antisense F GAD67 NM_ atggtgatgggatattttctcc sense F gccatgccctttgtcttaac antisense F GFAP NM_ ccaacctgcagattcgaga sense F tcttgaggtggccttctgac antisense F GAPDH NM_ agccacatcgctcagacac sense F gcccaatacgaccaaatcc antisense F MAP2 NM_ cctgtgttaagcggaaaacc sense F agagactttgtcctttgcctgt antisense F NCAM NM_ taccgcggcaagaacatc sense F ccacctgcagagaaactgc antisense F OLIGO2 NM_ agctcctcaaatcgcatcc sense F atagtcgtcgcagctttcg antisense F PAVLB NM_ caaaattggggttgacgaa sense F ggggcagtcagtgcttctta antisense F SYN1 NM_ gacggaagggatcacatcat sense F ctggtggtcaccaatgagc antisense F TUBB3 NM_ gcaactacgtgggcgact sense F cgaggcacgtacttgtgaga antisense F TH NM_ gccaaggacaagctcagg sense F agcgtgtacgggtcgaact antisense F VGAT AK ccacgacaagcccaaaat sense F taggcccagcacgaacat antisense F VGAT NM_ cacgacaagcccaaaatcac sense F NM_ agatgatgagaaacaaccccag antisense F vglut1 ENST cacgtggtggtgcagaaa sense F cgtgtatgaggccgacagt antisense F VGluT1 NM_ tcaataacagcacgacccac sense NM_ tcctggaatctgagtgacaatg antisense F F 23
24 Gene Symbol NCBI acc. No. primer sequences sense/antisense method VGLUT2 ENST aatcactcggccagatctaca sense F cgtcagctcgattgtctcc antisense F VGLUT3 AJ tggccacaacctctttttg sense F ccatccaatgtactgcacca antisense F NESTIN NM_ agacttccctcagctttcaggaccc sense R ctctggtccaggcaggacgct antisense R BRN1 NM_ tctattgtgcactcggacgcgg sense R ggtggctgctgggggctg antisense R GFAP NM_ gcacgaacgagtccctggagag sense R aatgacctctccatcccgcatctc antisense R PAX6 NM_ gccagcaacacacctagtca sense R ggggaaatgagtcctgttga antisense R p75 NM_ ctgcaagcagaacaagcaag sense R ggcctcatgggtaaaggagt antisense R SOX9 NM_ ggcgagcactcggggcaatc sense R gcctgctgcttggacatccacac antisense R SOX10 NM_ gaaagaccacccggactaca sense R tccaactcagccacatcaaa antisense R TUJ1 NM_ gagcggatcagcgtctactacaa sense R gatactcctcacgcaccttgct antisense R mascl1 NM_ agggatcctacgaccctctta sense Q accagttggtaaagtccagcag antisense Q mbrn2 NM_ gacacgccgacctcagac sense Q gatccgcctctgcttgaat antisense Q mmyt1l NM_ cctatgaggaccagtctcca sense Q tcttttctgttcgagggatctt antisense Q mneurod1 NM_ gtcccagcccactaccaat sense Q caagaaagtccgagggttga antisense Q hgapdh NM_ agccacatcgctcagacac sense Q gcccaatacgaccaaatcc antisense Q hascl1 NM_ caagagagcgcagccttag sense Q gcaaaagtcagtgctgaacg antisense Q hbrn2 NM_ aataaggcaaaaggaaagcaact sense Q caaaacacatcattacacctgct antisense Q hmyt1l NM_ caatggaaagggattttaagca sense Q tttgagattatgtaccaacgttagatg antisense Q hneurod1 NM_ gttattgtgttgccttagcacttc sense Q agtgaaatgaattgctcaaattgt antisense Q Note: F: primers for Fluidigm assays; R: primers for RT-PCR; Q: primers for qrt-pcr. 24
Supplementary Data. Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells
 Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and
 Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
NEURONAL CELL CULTURE MATRIX FOR BETTER MAINTENANCE AND SURVIVAL OF NEURONAL CELL CULTURES IN TISSUE CULTURE.
 NEURONAL CELL CULTURE MATRIX FOR BETTER MAINTENANCE AND SURVIVAL OF NEURONAL CELL CULTURES IN TISSUE CULTURE. D. R. Aguirre, N. DiMassa, Chrystal Johnson, H. Eran, R. Perez, C.V.R. Sharma, M.V.R. Sharma,
NEURONAL CELL CULTURE MATRIX FOR BETTER MAINTENANCE AND SURVIVAL OF NEURONAL CELL CULTURES IN TISSUE CULTURE. D. R. Aguirre, N. DiMassa, Chrystal Johnson, H. Eran, R. Perez, C.V.R. Sharma, M.V.R. Sharma,
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Generation of NSCs from hpscs in SDC medium.
 Supplementary Figure 1 Generation of NSCs from hpscs in SDC medium. (a) Q-PCR of pluripotent markers (OCT4, NANOG), neural markers (SOX1, PAX6, N-Cadherin), markers for the other germ layers (T, EOMES,
Supplementary Figure 1 Generation of NSCs from hpscs in SDC medium. (a) Q-PCR of pluripotent markers (OCT4, NANOG), neural markers (SOX1, PAX6, N-Cadherin), markers for the other germ layers (T, EOMES,
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplemental Information. Pioneer Factor NeuroD1 Rearranges. Transcriptional and Epigenetic Profiles. to Execute Microglia-Neuron Conversion
 Neuron, Volume 101 Supplemental Information Pioneer Factor NeuroD1 Rearranges Transcriptional and Epigenetic Profiles to Execute Microglia-Neuron Conversion Taito Matsuda, Takashi Irie, Shutaro Katsurabayashi,
Neuron, Volume 101 Supplemental Information Pioneer Factor NeuroD1 Rearranges Transcriptional and Epigenetic Profiles to Execute Microglia-Neuron Conversion Taito Matsuda, Takashi Irie, Shutaro Katsurabayashi,
Supplementary Information
 Supplementary Information Supplementary Figures Supplementary Figure 1. qrt-pcr analysis of the expression of candidate factors including SOX2, MITF, PAX3, DLX5, FOXD3, LEF1, MSX1, PAX6, SOX2 and SOX9
Supplementary Information Supplementary Figures Supplementary Figure 1. qrt-pcr analysis of the expression of candidate factors including SOX2, MITF, PAX3, DLX5, FOXD3, LEF1, MSX1, PAX6, SOX2 and SOX9
Supplementary Data. Supplementary Materials and Methods Quantification of delivered sirnas. Fluorescence microscopy analysis
 Supplementary Data Supplementary Materials and Methods Quantification of delivered sirnas After transfection, cells were washed in three times with phosphate buffered saline and total RNA was extracted
Supplementary Data Supplementary Materials and Methods Quantification of delivered sirnas After transfection, cells were washed in three times with phosphate buffered saline and total RNA was extracted
Supplementary Figure 1 Characterization of OKSM-iNSCs
 Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
SUPPLEMENTARY INFORMATION
 a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2239 Hepatocytes (Endoderm) Foreskin fibroblasts (Mesoderm) Melanocytes (Ectoderm) +Dox rtta TetO CMV O,S,M,K & N H1 hes H7 hes H9 hes H1 hes-derived EBs H7 hes-derived EBs H9 hes-derived
DOI: 10.1038/ncb2239 Hepatocytes (Endoderm) Foreskin fibroblasts (Mesoderm) Melanocytes (Ectoderm) +Dox rtta TetO CMV O,S,M,K & N H1 hes H7 hes H9 hes H1 hes-derived EBs H7 hes-derived EBs H9 hes-derived
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
SUPPLEMENTARY INFORMATION
 doi:.38/nature08267 Figure S: Karyotype analysis of ips cell line One hundred individual chromosomal spreads were counted for each of the ips samples. Shown here is an spread at passage 5, predominantly
doi:.38/nature08267 Figure S: Karyotype analysis of ips cell line One hundred individual chromosomal spreads were counted for each of the ips samples. Shown here is an spread at passage 5, predominantly
Supplemental Information. Direct Reprogramming of Fibroblasts. via a Chemically Induced XEN-like State
 Cell Stem Cell, Volume 21 Supplemental Information Direct Reprogramming of Fibroblasts via a Chemically Induced XEN-like State Xiang Li, Defang Liu, Yantao Ma, Xiaomin Du, Junzhan Jing, Lipeng Wang, Bingqing
Cell Stem Cell, Volume 21 Supplemental Information Direct Reprogramming of Fibroblasts via a Chemically Induced XEN-like State Xiang Li, Defang Liu, Yantao Ma, Xiaomin Du, Junzhan Jing, Lipeng Wang, Bingqing
A Novel Platform to Enable the High-Throughput Derivation and Characterization of Feeder-Free Human ipscs
 Supplementary Information A Novel Platform to Enable the High-Throughput Derivation and Characterization of Feeder-Free Human ipscs Bahram Valamehr, Ramzey Abujarour, Megan Robinson, Thuy Le, David Robbins,
Supplementary Information A Novel Platform to Enable the High-Throughput Derivation and Characterization of Feeder-Free Human ipscs Bahram Valamehr, Ramzey Abujarour, Megan Robinson, Thuy Le, David Robbins,
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Rapid differentiation of human pluripotent stem cells into functional motor neurons by
 Supplementary Information Rapid differentiation of human pluripotent stem cells into functional motor neurons by mrnas encoding transcription factors Sravan Kumar Goparaju, Kazuhisa Kohda, Keiji Ibata,
Supplementary Information Rapid differentiation of human pluripotent stem cells into functional motor neurons by mrnas encoding transcription factors Sravan Kumar Goparaju, Kazuhisa Kohda, Keiji Ibata,
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1. Serial deletion mutants of BLITz.
 Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Biology Open (2015): doi: /bio : Supplementary information
 Fig. S1. Verification of optimal conditions for the generation of LIF-dependent inscs Representative images (from five repeat experiments) of human primary fibroblasts undergoing reprogramming for 21 days
Fig. S1. Verification of optimal conditions for the generation of LIF-dependent inscs Representative images (from five repeat experiments) of human primary fibroblasts undergoing reprogramming for 21 days
Nature Methods: doi: /nmeth Supplementary Figure 1. Identification of candidate factors for the generation of inhibitory neurons.
 Supplementary Figure 1 Identification of candidate factors for the generation of inhibitory neurons. (a) Tuj1 + cells derived from human ES cells using different transcription factors 3 days after induction.
Supplementary Figure 1 Identification of candidate factors for the generation of inhibitory neurons. (a) Tuj1 + cells derived from human ES cells using different transcription factors 3 days after induction.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature22047 Supplementary discussion Overall developmental timeline and brain regionalization in human whole-brain organoids We first defined the timeline of generation of broadly-defined cell
doi:10.1038/nature22047 Supplementary discussion Overall developmental timeline and brain regionalization in human whole-brain organoids We first defined the timeline of generation of broadly-defined cell
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
SUPPLEMENTARY INFORMATION DOI: 1.138/NMAT3777 Biophysical regulation of epigenetic state and cell reprogramming Authors: Timothy L. Downing 1,2, Jennifer Soto 1,2, Constant Morez 2,3,, Timothee Houssin
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
MeCP2. MeCP2/α-tubulin. GFP mir1-1 mir132
 Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Supplementary Material for Yang et al., Generation of oligodendroglial cells by direct lineage conversion
 Supplementary Material for Yang et al., Generation of oligodendroglial cells by direct lineage conversion Supplementary Figure 1. The presence of Plp::EGFP + /O4 + cells 21 days after transgene induction.
Supplementary Material for Yang et al., Generation of oligodendroglial cells by direct lineage conversion Supplementary Figure 1. The presence of Plp::EGFP + /O4 + cells 21 days after transgene induction.
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Effect of timing of DE dissociation and RA concentration on lung field specification in hpscs. (a) Effect of duration of endoderm induction on expression of
Supplementary Figures Supplementary Figure 1. Effect of timing of DE dissociation and RA concentration on lung field specification in hpscs. (a) Effect of duration of endoderm induction on expression of
Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity
 Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity Supplemental figure 1. The quantification method to determine the
Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity Supplemental figure 1. The quantification method to determine the
Supplemental Table/Figure Legends
 MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
Protocol Using the Reprogramming Ecotropic Retrovirus Set: Mouse OSKM to Reprogram MEFs into ips Cells
 STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblast (MEF) cells into induced pluripotent stem (ips) cells in a 6-well format. Transduction
STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblast (MEF) cells into induced pluripotent stem (ips) cells in a 6-well format. Transduction
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat
 Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Protocol Reprogramming MEFs using the Dox Inducible Reprogramming Lentivirus Set: Mouse OKSM
 STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblasts (MEFs) into induced pluripotent stem (ips) cells in a 6-well format. Transduction
STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblasts (MEFs) into induced pluripotent stem (ips) cells in a 6-well format. Transduction
Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of
 Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Xeno-Free Systems for hesc & hipsc. Facilitating the shift from Stem Cell Research to Clinical Applications
 Xeno-Free Systems for hesc & hipsc Facilitating the shift from Stem Cell Research to Clinical Applications NutriStem Defined, xeno-free (XF), serum-free media (SFM) specially formulated for growth and
Xeno-Free Systems for hesc & hipsc Facilitating the shift from Stem Cell Research to Clinical Applications NutriStem Defined, xeno-free (XF), serum-free media (SFM) specially formulated for growth and
Protocol Using a Dox-Inducible Polycistronic m4f2a Lentivirus to Reprogram MEFs into ips Cells
 STEMGENT Page 1 OVERVIEW The following protocol describes the transduction and reprogramming of one well of Oct4-GFP mouse embryonic fibroblasts (MEF) using the Dox Inducible Reprogramming Polycistronic
STEMGENT Page 1 OVERVIEW The following protocol describes the transduction and reprogramming of one well of Oct4-GFP mouse embryonic fibroblasts (MEF) using the Dox Inducible Reprogramming Polycistronic
Quantitative real-time RT-PCR analysis of the expression levels of E-cadherin
 Supplementary Information 1 Quantitative real-time RT-PCR analysis of the expression levels of E-cadherin and ribosomal protein L19 (RPL19) mrna in cleft and bud epithelial cells of embryonic salivary
Supplementary Information 1 Quantitative real-time RT-PCR analysis of the expression levels of E-cadherin and ribosomal protein L19 (RPL19) mrna in cleft and bud epithelial cells of embryonic salivary
Chemically defined conditions for human ipsc derivation and culture
 Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Supplemental Figure 1 (Figure S1), related to Figure 1 Figure S1 provides evidence to demonstrate Nfatc1Cre is a mouse line that directed gene
 Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Effects of neuroactive agents on axonal growth and pathfinding of. retinal ganglion cells generated from human stem cells
 Effects of neuroactive agents on axonal growth and pathfinding of retinal ganglion cells generated from human stem cells Tadashi Yokoi 1, M.D., Ph.D.; Taku Tanaka 1, Ph.D.; Emiko Matsuzaka 1, Ph.D.; Fuminobu
Effects of neuroactive agents on axonal growth and pathfinding of retinal ganglion cells generated from human stem cells Tadashi Yokoi 1, M.D., Ph.D.; Taku Tanaka 1, Ph.D.; Emiko Matsuzaka 1, Ph.D.; Fuminobu
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Supplemental Table 1: Sequences of real time PCR primers. Primers were intronspanning
 Symbol Accession Number Sense-primer (5-3 ) Antisense-primer (5-3 ) T a C ACTB NM_001101.3 CCAGAGGCGTACAGGGATAG CCAACCGCGAGAAGATGA 57 HSD3B2 NM_000198.3 CTTGGACAAGGCCTTCAGAC TCAAGTACAGTCAGCTTGGTCCT 60
Symbol Accession Number Sense-primer (5-3 ) Antisense-primer (5-3 ) T a C ACTB NM_001101.3 CCAGAGGCGTACAGGGATAG CCAACCGCGAGAAGATGA 57 HSD3B2 NM_000198.3 CTTGGACAAGGCCTTCAGAC TCAAGTACAGTCAGCTTGGTCCT 60
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Information
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Seidel et al. 2016 Page 1 of 13 In vitro field potential monitoring on multi-micro electrode array
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Seidel et al. 2016 Page 1 of 13 In vitro field potential monitoring on multi-micro electrode array
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
SUPPLEMENTARY INFORMATION
 Supplementary Figure S1 Colony-forming efficiencies using transposition of inducible mouse factors. (a) Morphologically distinct growth foci appeared from rtta-mefs 6-8 days following PB-TET-mFx cocktail
Supplementary Figure S1 Colony-forming efficiencies using transposition of inducible mouse factors. (a) Morphologically distinct growth foci appeared from rtta-mefs 6-8 days following PB-TET-mFx cocktail
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking
 Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
PRIMARY NEURONAL CULTURE
 PRIMARY NEURONAL CULTURE Products for Your Research Scientists Helping Scientists WWW.STEMCELL.COM TABLE OF CONTENTS 3 Primary Neuronal Culture NeuroCult SM1 and BrainPhys Neuronal Medium 4 Increased Neuronal
PRIMARY NEURONAL CULTURE Products for Your Research Scientists Helping Scientists WWW.STEMCELL.COM TABLE OF CONTENTS 3 Primary Neuronal Culture NeuroCult SM1 and BrainPhys Neuronal Medium 4 Increased Neuronal
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
SUPPLEMENTARY INFORMATION
 a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well
 Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Protocol Reprogramming Human Fibriblasts using the Dox Inducible Reprogramming Polycistronic Lentivirus Set: Human 4F2A LoxP
 STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of BJ Human Fibroblasts (BJ cells) into induced pluripotent stem (ips) cells in a 6-well format. Transduction efficiency
STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of BJ Human Fibroblasts (BJ cells) into induced pluripotent stem (ips) cells in a 6-well format. Transduction efficiency
Supplementary Figure 1. Generation of B2M -/- ESCs. Nature Biotechnology: doi: /nbt.3860
 Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and
 Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
CHO Na V 1.4 Cell Line
 B SYS GmbH CHO Na V 1.4 Cell Line Specification Sheet B SYS GmbH B SYS GmbH CHO Na V 1.4 Cells Page 2 TABLE OF CONTENTS 1. B'SYS Na V 1.4 CELL LINE...2 1.1 THE VOLTAGE GATED SODIUM CHANNELS NA V 1.4...2
B SYS GmbH CHO Na V 1.4 Cell Line Specification Sheet B SYS GmbH B SYS GmbH CHO Na V 1.4 Cells Page 2 TABLE OF CONTENTS 1. B'SYS Na V 1.4 CELL LINE...2 1.1 THE VOLTAGE GATED SODIUM CHANNELS NA V 1.4...2
Changing fibroblasts into neurons: Direct lineage conversion. Journal Club Uli Herrmann
 Changing fibroblasts into neurons: Direct lineage conversion Journal Club Uli Herrmann 25.6.13 Outline Epigenetic landscape model Reprogramming cells Induced pluripotent stem cells Three papers: Direct
Changing fibroblasts into neurons: Direct lineage conversion Journal Club Uli Herrmann 25.6.13 Outline Epigenetic landscape model Reprogramming cells Induced pluripotent stem cells Three papers: Direct
Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.
 LEGENDS TO SUPPLEMENTARY FIGURES Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.2) were stained with the antibodies oct3/4
LEGENDS TO SUPPLEMENTARY FIGURES Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.2) were stained with the antibodies oct3/4
TITLE: Induced Pluripotent Stem Cells as Potential Therapeutic Agents in NF1
 AD Award Number: W81XWH-10-1-0181 TITLE: Induced Pluripotent Stem Cells as Potential Therapeutic Agents in NF1 PRINCIPAL INVESTIGATOR: Jonathan Chernoff, M.D., Ph.D. CONTRACTING ORGANIZATION: Institute
AD Award Number: W81XWH-10-1-0181 TITLE: Induced Pluripotent Stem Cells as Potential Therapeutic Agents in NF1 PRINCIPAL INVESTIGATOR: Jonathan Chernoff, M.D., Ph.D. CONTRACTING ORGANIZATION: Institute
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplementary information
 Supplementary information Supplementary Figure 1 (a) EM image of the pyramidal layer of wt mice. CA3 pyramidal neurons were selected according to their typical alignment, size and shape for subsequent
Supplementary information Supplementary Figure 1 (a) EM image of the pyramidal layer of wt mice. CA3 pyramidal neurons were selected according to their typical alignment, size and shape for subsequent
Defense Technical Information Center Compilation Part Notice
 UNCLASSIFIED Defense Technical Information Center Compilation Part Notice ADP019695 TITLE: Fabricating Neural and Cardiomyogenic Stem Cell Structures by a Novel Rapid Prototyping--the Inkjet Printing Method
UNCLASSIFIED Defense Technical Information Center Compilation Part Notice ADP019695 TITLE: Fabricating Neural and Cardiomyogenic Stem Cell Structures by a Novel Rapid Prototyping--the Inkjet Printing Method
Supplementary Information
 Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
15 June 2011 Supplementary material Bagriantsev et al.
 Supplementary Figure S1 Characterization of K 2P 2.1 (TREK-1) GOF mutants A, Distribution of the positions of mutated nucleotides, represented by a red x, from a pool of 18 unselected K 2P 2.1 (KCNK2)
Supplementary Figure S1 Characterization of K 2P 2.1 (TREK-1) GOF mutants A, Distribution of the positions of mutated nucleotides, represented by a red x, from a pool of 18 unselected K 2P 2.1 (KCNK2)
Methanol fixation allows better visualization of Kal7. To compare methods for
 Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
gacgacgaggagaccaccgctttg aggcacattgaaggtctcaaacatg
 Supplementary information Supplementary table 1: primers for cloning and sequencing cloning for E- Ras ggg aat tcc ctt gag ctg ctg ggg aat ggc ttt gcc ggt cta gag tat aaa gga agc ttt gaa tcc Tpbp Oct3/4
Supplementary information Supplementary table 1: primers for cloning and sequencing cloning for E- Ras ggg aat tcc ctt gag ctg ctg ggg aat ggc ttt gcc ggt cta gag tat aaa gga agc ttt gaa tcc Tpbp Oct3/4
-RT RT RT -RT XBMPRII
 Base pairs 2000 1650 1000 850 600 500 400 Marker Primer set 1 Primer set 2 -RT RT RT -RT XBMPRII kda 100 75 50 37 25 20 XAC p-xac A 300 B Supplemental figure 1. PCR analysis of BMPRII expression and western
Base pairs 2000 1650 1000 850 600 500 400 Marker Primer set 1 Primer set 2 -RT RT RT -RT XBMPRII kda 100 75 50 37 25 20 XAC p-xac A 300 B Supplemental figure 1. PCR analysis of BMPRII expression and western
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells
 Supplementary Information The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells Bing Zhao 1,3, Zhen Qi 1,3, Yehua Li 1,3, Chongkai Wang 2,
Supplementary Information The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells Bing Zhao 1,3, Zhen Qi 1,3, Yehua Li 1,3, Chongkai Wang 2,
Supplementary Figure 1, Wiel et al
 Supplementary Figure 1, Wiel et al Supplementary Figure 1 ITPR2 increases in benign tumors and decreases in aggressive ones (a-b) According to the Oncomine database, expression of ITPR2 increases in renal
Supplementary Figure 1, Wiel et al Supplementary Figure 1 ITPR2 increases in benign tumors and decreases in aggressive ones (a-b) According to the Oncomine database, expression of ITPR2 increases in renal
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State
 Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
CHO-Tet herg Cell Line
 F B SYS GmbH CHO-Tet herg Cell Line Specification Sheet B SYS GmbH B SYS GmbH CHO-Tet herg Cells Page 2 TABLE OF CONTENTS 1 BACKGROUND...3 1.1 Drug-induced QT Prolongation...3 1.2 Regulatory Issues...3
F B SYS GmbH CHO-Tet herg Cell Line Specification Sheet B SYS GmbH B SYS GmbH CHO-Tet herg Cells Page 2 TABLE OF CONTENTS 1 BACKGROUND...3 1.1 Drug-induced QT Prolongation...3 1.2 Regulatory Issues...3
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
T H E J O U R N A L O F C E L L B I O L O G Y
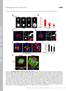 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
Sundari Chetty, Felicia Walton Pagliuca, Christian Honore, Anastasie Kweudjeu, Alireza Rezania, and Douglas A. Melton
 A simple tool to improve pluripotent stem cell differentiation Sundari Chetty, Felicia Walton Pagliuca, Christian Honore, Anastasie Kweudjeu, Alireza Rezania, and Douglas A. Melton Supplementary Information
A simple tool to improve pluripotent stem cell differentiation Sundari Chetty, Felicia Walton Pagliuca, Christian Honore, Anastasie Kweudjeu, Alireza Rezania, and Douglas A. Melton Supplementary Information
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Paul et al.,
 Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in
 Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors
 Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors A Vector GR1 SacI BamHI CTCTGGCTAACTAGGC Insert 5/7nt - G TCGAGAGACCGATTGATCCG Insert 5/7nt - CCTAG 1 G CAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAAC
Figure S1: Insert sequences for GR1, GR2, Qc3c_AgeI and GR3 vectors A Vector GR1 SacI BamHI CTCTGGCTAACTAGGC Insert 5/7nt - G TCGAGAGACCGATTGATCCG Insert 5/7nt - CCTAG 1 G CAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAACAAC
bronchial epithelial cells (I). Bronchi are outlined with dashed line. Scale bars = 25 µm, if not
 Supplemental Figure S1: ronchial epithelial cell polarity and integrity is maintained in bronchi. (A-E) Staining for selected markers of bronchial cell differentiation and intracellular compartments is
Supplemental Figure S1: ronchial epithelial cell polarity and integrity is maintained in bronchi. (A-E) Staining for selected markers of bronchial cell differentiation and intracellular compartments is
Authors: Takuro Horii, Masamichi Yamamoto, Sumiyo Morita, Mika Kimura, Yasumitsu Nagao, Izuho Hatada *
 SUPPLEMENTARY INFORMATION Title: p53 Suppresses Tetraploid Development in Mice Authors: Takuro Horii, Masamichi Yamamoto, Sumiyo Morita, Mika Kimura, Yasumitsu Nagao, Izuho Hatada * Supplementary Methods
SUPPLEMENTARY INFORMATION Title: p53 Suppresses Tetraploid Development in Mice Authors: Takuro Horii, Masamichi Yamamoto, Sumiyo Morita, Mika Kimura, Yasumitsu Nagao, Izuho Hatada * Supplementary Methods
Neural Stem Cell Characterization Kit
 Neural Stem Cell Characterization Kit Catalog No. SCR019 FOR RESEARCH USE ONLY Not for use in diagnostic procedures USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209 Europe +44 (0) 23 8026 2233
Neural Stem Cell Characterization Kit Catalog No. SCR019 FOR RESEARCH USE ONLY Not for use in diagnostic procedures USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209 Europe +44 (0) 23 8026 2233
Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis
 Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis seedlings expressing ERF109pro-GUS. The GUS staining
Supplementary Fig 1. The responses of ERF109 to different hormones and stresses. (a to k) The induced expression of ERF109 in 7-day-old Arabidopsis seedlings expressing ERF109pro-GUS. The GUS staining
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
INNOVATIVE PRODUCTS FOR NEUROSCIENCE RESEARCH
 INNOVATIVE PRODUCTS FOR NEUROSCIENCE RESEARCH NEURAL CELLS & MEDIA STEM CELLS & CULTURE REAGENTS TRANSFECTION CUSTOM SERVICES NEUROSCIENCE PRODUCTS CATALOG TABLE OF CONTENTS Neuroscience research demands
INNOVATIVE PRODUCTS FOR NEUROSCIENCE RESEARCH NEURAL CELLS & MEDIA STEM CELLS & CULTURE REAGENTS TRANSFECTION CUSTOM SERVICES NEUROSCIENCE PRODUCTS CATALOG TABLE OF CONTENTS Neuroscience research demands
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like
 Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like morphology in co-culture with mouse embryonic feeder fibroblasts
Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like morphology in co-culture with mouse embryonic feeder fibroblasts
Development 142: doi: /dev : Supplementary Material
 Development 142: doi:1.42/dev.11687: Supplementary Material Figure S1 - Relative expression of lineage markers in single ES cell and cardiomyocyte by real-time quantitative PCR analysis. Single ES cell
Development 142: doi:1.42/dev.11687: Supplementary Material Figure S1 - Relative expression of lineage markers in single ES cell and cardiomyocyte by real-time quantitative PCR analysis. Single ES cell
A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
Supplementary Fig. 5
 Supplementary Fig. 5 Supplemental Figures legends Supplementary Figure 1 (A) Additional dot plots from CyTOF analysis from untreated group. (B) Gating strategy for assessment of CD11c + NK cells frequency
Supplementary Fig. 5 Supplemental Figures legends Supplementary Figure 1 (A) Additional dot plots from CyTOF analysis from untreated group. (B) Gating strategy for assessment of CD11c + NK cells frequency
SUPPLEMENTARY NOTE 3. Supplememtary Note 3, Wehr et al., Monitoring Regulated Protein-Protein Interactions Using Split-TEV 1
 SUPPLEMENTARY NOTE 3 Fluorescent Proteolysis-only TEV-Reporters We generated TEV reporter that allow visualizing TEV activity at the membrane and in the cytosol of living cells not relying on the addition
SUPPLEMENTARY NOTE 3 Fluorescent Proteolysis-only TEV-Reporters We generated TEV reporter that allow visualizing TEV activity at the membrane and in the cytosol of living cells not relying on the addition
Post-expansion antibody delivery, after epitope-preserving homogenization.
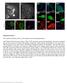 Supplementary Figure 1 Post-expansion antibody delivery, after epitope-preserving homogenization. (a, b) Wide-field fluorescence images of Thy1-YFP-expressing mouse brain hemisphere slice before expansion
Supplementary Figure 1 Post-expansion antibody delivery, after epitope-preserving homogenization. (a, b) Wide-field fluorescence images of Thy1-YFP-expressing mouse brain hemisphere slice before expansion
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Design and sequence of system 1 for targeted demethylation.
 Supplementary Figure 1 Design and sequence of system 1 for targeted demethylation. (a) Design of system 1 for targeted demethylation. TET1CD was fused to a catalytic inactive Cas9 nuclease (dcas9) and
Supplementary Figure 1 Design and sequence of system 1 for targeted demethylation. (a) Design of system 1 for targeted demethylation. TET1CD was fused to a catalytic inactive Cas9 nuclease (dcas9) and
Supplementary Figure 1.
 Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
