Spermatazoa. Sertoli cells. Morc1 WT adult. Morc1 KO adult. Morc1 WT P14.5. Morc1 KO P14.5. Germ cells. Germ cells. Spermatids.
|
|
|
- Nickolas Cooper
- 5 years ago
- Views:
Transcription
1 a. Spermatazoa Sertoli cells Morc1 WT adult c. Morc1 KO adult Morc1 WT P1.5 Morc1 KO P1.5 Morc1 WT adult Morc1 KO adult Germ cells Germ cells Spermatids Spermatozoa Supplementary Figure 1. Confirmation that Morc1tg/tg males are infertile. a, Histology of Morc1+/+ and Morc1tg/tg adults. b-c, Detection of germ cells in Morc1+/+ and Morc1tg/tg adults by immunofluorescence (IF). is a marker of germ cell identity. Scale bars indicate 10μm.
2 a. Ladder WT ES cells xflag-morc1 Morc1 expression (relative to Rrm) kda 10 kda 110 kda 80 kda * * xflag Morc1 E1.5 E1.5 E1.5 E18.5 c. Morc1 het E1.5 Morc1 het E18.5 LINE1-ORF Composite LINE1-ORF Composite Morc1 KO E1.5 Morc1 KO E18.5 LINE1-ORF1p Composite LINE1-ORF1p Composite Supplementary Figure. Transposon derepression in Morc1 tg/tg arises concurrently with its expession in the germline. a, Relative Morc1 mrna expression in whole testis as measured by RT-PCR. Expression is shown relative to the housekeeping gene Rrm. Six technical replicates per timepoint were analyzed, mean and standard error are plotted. b, Western blot confirming that anti-morc1 coiled coil antibody recognize an overexpressed Flag-MORC1 in mouse embryonic stem cells. c, Immunoflourescent detection of LINE1-ORF1p protein in late embryogenesis (E1.5, E18.5). is a marker ofgerm cell identity. Scale bars indicate 0μm.
3 a. E1.5 E18.5 P.5 P5.5 GFP GFP GFP GFP SSC-A Lo, EPCAM Hi, MHCI - + P10.5 SSC-A Lo EPCAM Hi MHCI - SSC-A EpCAM FSC-A Somatic cells Somatic cells + FSC-A MHC I Supplementary Figure. Strategies for sorting pure germ cells. a, Flow cytometry plots showing a distinct egfp + germ population in Oct-IRES-eGfp. egfp + cells could be used to sort embryonic and early postnatal germ cells effectively but was not useful after P.5. b, Sorting a SSC-A Lo, EPCAM Hi, MHCI - population of cells from P10.5 whole testis, yields a 9.8% pure population of Hi (germ) cells. The EpCAM Lo somatic cell population is shown as a control. is an intracellular protein and thus is not an optimal marker for sorting cells for RNA-Seq applications. Therefore, the SSC-A Lo, EPCAM Hi, MHCI - population was used to sort P10.5 germ cells for whole genome-bisulfite sequencing and RNA-seq experiments.
4 a. P10.5 RT-PCR (whole testis) P10.5 RNA-seq (whole testis) P10.5 RNA-seq (sorted) Relative transposon expression (Morc1 KO/Morc1 het) 10 8 IAP-Ez GLN Etn 10 8 IAP-Ey IAP-Ez IAP-LTR1 IAP-LTR GLN Etn LINE1 A LINE1 F LINE1 Gf LINE1 T IAP-Ey IAP-Ez IAP-LTR1 IAP-LTR GLN Etn LINE1 A LINE1 F LINE1 Gf LINE1 T.0 P10.5 RNA-seq (whole testis) Relative transposon expression (Morc1 het/morc1 WT) IAP-Ey IAP-Ez IAP-LTR1 IAP-LTR GLN Etn LINE1 A LINE1 F LINE1 Gf LINE1 T Supplementary Figure. Validation of sequencing data and strategy. a, RT-qPCR for select transposon classes at P10.5 in whole testis yields a similar pattern of upregulation in Morc1 tg/tg as observed by RNA-Seq of whole testis and sorted germ cells, confirming the validity of each approach. The degree of upregulation is somewhat stronger in the sorted cells, probably because transcripts from the unaffected somatic cells blunt the increase in whole testis. b, There is no substantial increase in transposon expression in Morc1 tg/+ mice relative to Morc1 +/+, allowing us to use Morc1 tg/+ littermates of Morc1 tg/tg mice as controls in subsequent experiments. - replicates per genotype were analyzed, mean and standard error are indicated.
5 a. P DNA transposon putative pirna/mirna sense/antisense primary/secondary Het KO Fraction of pirnas by origin SINE LTR LINE Non-transposon Het KO c. d. 5 pirna abundance (% mirna level) 1 Sense/antisense Het KO All LTR Het KO IAP subfamily Het KO Het KO Het KO Het KO All pirna LTR IAP Het KO e. f. Primary/secondary 10 8 Het KO Het KO Het KO All pirna LTR IAP Het KO Normalized pirna (nt-0nt) abundance over upregulated repeats Morc1 het Black: All Blue: Sense Red: Antisense Morc1 KO TSS TES TSS TES Supplementary Figure 5. Increased pirna reads deriving from transposon elements upregulated at P10.5 a, Relative pirna abundance and characteristics in P10.5 Morc1 tg/tg and Morc1 tg/+ testes. b, pirnas from P10.5 testes are plotted by transposon class. Consistent with overexpression of various LTR retrotransposons at this timepoint, a higher fraction of pirna are derived from LTR retrotransposons. c, A large increase of pirna derived from IAP and in Morc1 tg/tg is observed. d, The ratio of primary/secondary and e, sense/antisense pirna of IAP derived sequence is plotted for Morc1 tg/+ and Morc1 tg/tg. An increase in primary, sense transcript is observed relative to other species, consistent with pirnas being processed directly from transposons. f, Average distribution of sense and antisense pirna over transposons upregulated in Morc1 tg/tg. Middle region represents the body of the transposon, and the flanking regions represent the 5 (left) and (right) regions outside of the transposon. Both flanking regions were scaled to the same lengths as for the middle regions.
6 a IAP-Ez-int 9 IAPLTR1 1 IAPLTR 99 9 IAP-d-int IAP-Ey-int 11 IAPEY 0 Control Region HypoDMR 111 (Morc1 KO/Morc1 het) IAP Ez IAPLTR1 IAPLTR IAPEy-int IAPEY_LTR IAPEY_LTR IAPEy-int IAPEY IAP-d-int IAP-Ey-int IAPEY IAPEY 7 0 c. E1.5 E18.5 P.5 P10.5 Timepoint (Sorted germ cells) Percentage methylation Morc1 het Morc1 KO IAPLTR1 IAPLTR IAPLTR IAPLTR IAPLTREY IAPLTREY Supplementary Figure. Hypomethylation of IAP elements in Morc1 tg/tg. a-b, IAP elements show hypomethylation in the Morc1 tg/tg, however there is weak correspondence between the extent of upregulation (compare (a) to (b)) and frequency of hypomethylated DMRs. c, Overall, there is modest global hypomethylation of all IAP LTRs in Morc1 tg/tg. - replicates per genotype and timepoint were analyzed, mean and standard error are indicated.
7 Nebulin (Neb) TSS locus DMR E1.5 Het RNA-seq E1.5 KO RNA-seq P10.5 Het RNA-seq P10.5 KO RNA-seq E1.5 het 5mC E1.5 KO 5mC P.5 het 5mC P.5 KO 5mC P10.5 het 5mC P10.5 KO 5mC Neb TSS RLTR7 RLTR10A Supplementary Figure 7. Coordinated transposon and gene silencing at the Nebulin transcription start site. Pooled, uniquely mapping RNA-seq reads and BS-seq data is shown relative to the Nebulin locus. The TSS of the Nebulin gene is hypomethylated in Morc1 tg/tg, probably because of adjacent RLTR10A and RLTR7 LTRs. The Nebulin gene is correspondingly upregulated at P10.5. Each CG is represented as by a bar, with the height of the bar indicating the frequency with which the CG is methylated. A dot at a position indicates no methylation. At least one read must map to the CG for a bar to appear.
8 Rasgrf1 locus Paternally imprinted region Paternally imprinted region (inset right) Rasgrf1 TSS DMR E1.5 Het RNA-seq E1.5 Het RNA-seq E1.5 KO RNA-seq E1.5 KO RNA-seq P10.5 Het RNA-seq P10.5 Het RNA-seq P10.5 KO RNA-seq P10.5 KO RNA-seq E1.5 het 5mC E1.5 het 5mC E1.5 KO 5mC E1.5 KO 5mC P.5 het 5mC P.5 het 5mC P.5 KO 5mC P.5 KO 5mC P10.5 het 5mC P10.5 het 5mC P10.5 KO 5mC P10.5 KO 5mC Supplementary Figure 8. Hypomethylation of the imprinting control region of the Rasgrf1 locus. (Left) The paternally imprinted region of Rasgrf1 is indicated relative to the transcription start site of the gene (Right), transcription and methylation over the paternally imprinted region is shown. Part of the paternally imprinted region is overexpressed and hypomethylated in the Morc1 tg/tg mutant relative to the control.
9 Cdkl locus a. Hypermethylated DMRs Hypermethylated DMRs Hypomethylated DMRs E1.5 Het RNA-seq E1.5 KO RNA-seq P.5 het 5mC P.5 KO 5mC CDKL TSS RLTR10A Unannotated transcript locus Hypermethylated DMRs Hypomethylated DMR E1.5 Het RNA-seq E1.5 KO RNA-seq P.5 het 5mC P.5 KO 5mC IAPLTR1 Supplementary Figure 9. Hypermethylation of select regions is linked to transcriptional readthrough. The Cdkl (a) promoter and the probable promoter of an unannotated transcript (b) are hypomethylated in the Morc1 tg/tg. There is a corresponding increase in reads deriving from these transcripts and regions of hypermethylation develop in the transcript body.
10 a. Early 5mC Late 5mC No 5mC Early 5mC Late 5mC Early 5mC DMR DMR DMR E1.5 het 5mC E1.5 het 5mC E1.5 KO 5mC E1.5 KO 5mC P.5 het 5mC P.5 het 5mC P.5 KO 5mC P.5 KO 5mC P10.5 het 5mC P10.5 het 5mC P10.5 KO 5mC P10.5 KO 5mC Supplementary Figure 10. Hypomethylated regions in Morc1 tg/tg are methylated more slowly than other loci. Two regions (a) and (b) that show both early and late methylation are shown. Hypomethylated DMRs are late-methylating.
11 a. Log (HKme/input) in E1.5 WT germ cells Hypo DMR Control Log (ChIP/input) in E1.5 WT germ cells HB0ac Log (ChIP/input) in E1.5 WT germ cells HK7ac Log (ChIP/input) in E1.5 WT germ cells HK7me HypoDMR Control DMR HypoDMR Control DMR HypoDMR Control DMR Supplementary Figure 11. Regions hypomethylated in Morc1 tg/tg have elevated HKme at E1.5. a-b, Abundance of ChIP-seq reads over control and hypomethylated DMRs are represented by boxplots. Each DMR corresponds to one point on the boxplot. (a) Morc1 tg/tg DMRs have elevated HKme at E1.5, indicating that these regions are sites of transcriptional initiation at the onset of de novo methylation. (b) HB0ac, KK7ac and HK7me abundances are similar over hypomethylated and control DMRs.
12 a. E1.5 ATAC-seq germ cell libraries Size distribution of fragments in representative ATAC-seq library 500bp 00bp 00bp nuc nuc 1nuc bp Short insert count bp 1 nuc nuc nuc size c. E1.5 germ cells (10k cells) Relative ATAC-seq read abundance 10 5 group RPKM > < RPKM < 100 < RPKM < 0 1 < RPKM < 0. < RPKM < 1 RPKM < 0. 0 TSS TES Supplementary Figure 1. Technical validation of the ATAC-seq method. a) ATAC-seq libraries using material from 10k embryonic germ cells produced an appropriate nucleosome-banding pattern, consistent with the tendency of primer insertion events to occur in nucleosome linker regions. b) The distribution of fragment sizes in the ATAC-seq library shows periodicity consistent with the 10.5bp pitch of DNA and the 180bp periodicity of nucleosome size. c) Metaplot of ATAC-seq reads relative to TSS in embryonic germ cells. A higher density of ATAC-seq reads is observed proximal to the TSS of the gene, with higher expressed genes showing greater ATAC-seq peaks. This confirms the potential of ATAC-seq as a method to find transcriptional start sites in embryonic germ cells.
13 a. c. Increased transposon expression Maintained transposon expression E1.5 to P10.5 E1.5 to P10.5 DMR DMR DMR Reduced transposon expression E1.5 to P10.5 E1.5 Het ATAC-seq E1.5 KO ATAC-seq P10.5 het ATAC-seq P10.5 KO ATAC-seq E1.5 Het RNA-seq E1.5 KO RNA-seq P10.5 het RNA-seq P10.5 KO RNA-seq P.5 het 5mC P.5 KO 5mC MERVK10C IAPLTR RLTR10B MMERGLN Supplementary Figure 1. ATAC-seq peaks follow the pattern of expression in the embryonic and postnatal germline. a-c, ATAC-seq, expression, and methylation data are shown at three loci. In all three loci, transposons are expressed at E1.5 and an ATAC-seq peak is present. Where transposon expression is sustained at P10.5 in Morc1 tg/tg (a), (b), an peak of ATAC-seq reads is also present. Where transposon expression is silenced however, the ATAC-seq peak is lost (c).
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
of NOBOX. Supplemental Figure S1F shows the staining profile of LHX8 antibody (Abcam, ab41519), which is a
 SUPPLEMENTAL MATERIALS AND METHODS LHX8 Staining During the course of this work, it came to our attention that the NOBOX antibody used in this study had been discontinued. There are a number of other nuclear
SUPPLEMENTAL MATERIALS AND METHODS LHX8 Staining During the course of this work, it came to our attention that the NOBOX antibody used in this study had been discontinued. There are a number of other nuclear
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
6/256 1/256 0/256 1/256 2/256 7/256 10/256. At3g06290 (SAC3B)
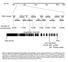 Chr.III M 5M 1M 15M 2M 23M BAC clones F22F7 F1A16 F24F17 F24P17 T8E24 F17A9 F21O3 F17A17 F18C1 F2O1 F28L1 F5E6 F3E22 T1B9 MLP3 Number of recombinants 6/256 1/256 /256 1/256 2/256 7/256 1/256 At3g629 (SAC3B)
Chr.III M 5M 1M 15M 2M 23M BAC clones F22F7 F1A16 F24F17 F24P17 T8E24 F17A9 F21O3 F17A17 F18C1 F2O1 F28L1 F5E6 F3E22 T1B9 MLP3 Number of recombinants 6/256 1/256 /256 1/256 2/256 7/256 1/256 At3g629 (SAC3B)
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplemental Figure 1.
 Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplementary Materials
 Molaro et al., 1 Supplementary Materials Two waves of de novo methylation during mouse germ cell development Antoine Molaro, Ilaria Falciatori, Emily Hodges, Alexei A. Aravin, Krista Marran, Shahin Rafii,
Molaro et al., 1 Supplementary Materials Two waves of de novo methylation during mouse germ cell development Antoine Molaro, Ilaria Falciatori, Emily Hodges, Alexei A. Aravin, Krista Marran, Shahin Rafii,
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb
 Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
gypsy fold change in steady state RNA levels RnpS1 Acn mago tsu GFP armi sirna mediated depletion against indicated genes in OSCs
 Hayashi_FigS1 fold change in steady state RNA levels 1000 100 10 1 gypsy GFP armi mago tsu Acn RnpS1 btz sirna mediated depletion against indicated genes in OSCs Supplementary Figure S1. The EJC is required
Hayashi_FigS1 fold change in steady state RNA levels 1000 100 10 1 gypsy GFP armi mago tsu Acn RnpS1 btz sirna mediated depletion against indicated genes in OSCs Supplementary Figure S1. The EJC is required
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for
 Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Characterization of Hi-C/CHi-C dynamics and enhancer identification. (a) Scatterplot of Hi-C read counts supporting contacts between domain boundaries. Contacts enclosing domains
Supplementary Figure 1 Characterization of Hi-C/CHi-C dynamics and enhancer identification. (a) Scatterplot of Hi-C read counts supporting contacts between domain boundaries. Contacts enclosing domains
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Nature Genetics: doi: /ng Supplementary Figure 1. ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets.
 Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Nature Genetics: doi: /ng.3556 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE. Supplementary Figure 1
 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
Supplementary Table S1
 Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Simultaneous profiling of transcriptome and DNA methylome from a single cell
 Additional file 1: Supplementary materials Simultaneous profiling of transcriptome and DNA methylome from a single cell Youjin Hu 1, 2, Kevin Huang 1, 3, Qin An 1, Guizhen Du 1, Ganlu Hu 2, Jinfeng Xue
Additional file 1: Supplementary materials Simultaneous profiling of transcriptome and DNA methylome from a single cell Youjin Hu 1, 2, Kevin Huang 1, 3, Qin An 1, Guizhen Du 1, Ganlu Hu 2, Jinfeng Xue
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Nature Immunology: doi: /ni.3694
 Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplemental Figure legends Figure S1. (A) (B) (C) (D) Figure S2. Figure S3. (A-E) Figure S4. Figure S5. (A, C, E, G, I) (B, D, F, H, Figure S6.
 Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Watson BM Gene Capitolo 11
 Watson BM Gene Capitolo 11 Le proteine della trasposizione ? target-site primed reverse transcription Watson et al., BIOLOGIA MOLECOLARE DEL GENE, Zanichelli editore S.p.A. Trimeric structure for an
Watson BM Gene Capitolo 11 Le proteine della trasposizione ? target-site primed reverse transcription Watson et al., BIOLOGIA MOLECOLARE DEL GENE, Zanichelli editore S.p.A. Trimeric structure for an
C24. ros1-1. ros1-1 rdm18-1. ros1-1 rdm18-2. ros1-1 nrpe1
 Figure S1 Methylation level 1 0.8 0.6 0.4 0.2 0 p35s methylation levels 24 ros1-1 ros1-1 rdm18-1 ros1-1 rdm18-2 ros1-1 nrpe1 mg mhg mhh Figure S1. RM18/PKL promotes silencing at the p35s-npt II transgene
Figure S1 Methylation level 1 0.8 0.6 0.4 0.2 0 p35s methylation levels 24 ros1-1 ros1-1 rdm18-1 ros1-1 rdm18-2 ros1-1 nrpe1 mg mhg mhh Figure S1. RM18/PKL promotes silencing at the p35s-npt II transgene
ChampionChIP Quick, High Throughput Chromatin Immunoprecipitation Assay System
 ChampionChIP Quick, High Throughput Chromatin Immunoprecipitation Assay System Liyan Pang, Ph.D. Application Scientist 1 Topics to be Covered Introduction What is ChIP-qPCR? Challenges Facing Biological
ChampionChIP Quick, High Throughput Chromatin Immunoprecipitation Assay System Liyan Pang, Ph.D. Application Scientist 1 Topics to be Covered Introduction What is ChIP-qPCR? Challenges Facing Biological
Nature Methods: doi: /nmeth.4396
 Supplementary Figure 1 Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC. (a) Heatmap-based representation of ATAC-seq quality control
Supplementary Figure 1 Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC. (a) Heatmap-based representation of ATAC-seq quality control
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplemental Figure 1 A
 Supplemental Figure A prebleach postbleach 2 min 6 min 3 min mh2a.-gfp mh2a.2-gfp mh2a2-gfp GFP-H2A..9 Relative Intensity.8.7.6.5 mh2a. GFP n=8.4 mh2a.2 GFP n=4.3 mh2a2 GFP n=2.2 GFP H2A n=24. GFP n=7.
Supplemental Figure A prebleach postbleach 2 min 6 min 3 min mh2a.-gfp mh2a.2-gfp mh2a2-gfp GFP-H2A..9 Relative Intensity.8.7.6.5 mh2a. GFP n=8.4 mh2a.2 GFP n=4.3 mh2a2 GFP n=2.2 GFP H2A n=24. GFP n=7.
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1. Validation of CDK9-inhibitor treatment.
 Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Figure S1. nuclear extracts. HeLa cell nuclear extract. Input IgG IP:ORC2 ORC2 ORC2. MCM4 origin. ORC2 occupancy
 A nuclear extracts B HeLa cell nuclear extract Figure S1 ORC2 (in kda) 21 132 7 ORC2 Input IgG IP:ORC2 32 ORC C D PRKDC ORC2 occupancy Directed against ORC2 C-terminus (sc-272) MCM origin 2 2 1-1 -1kb
A nuclear extracts B HeLa cell nuclear extract Figure S1 ORC2 (in kda) 21 132 7 ORC2 Input IgG IP:ORC2 32 ORC C D PRKDC ORC2 occupancy Directed against ORC2 C-terminus (sc-272) MCM origin 2 2 1-1 -1kb
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Distribution of mirnas between lncrna and protein-coding genes. Pie chart showing distribution of human mirna between protein coding and lncrna genes. To the right, lncrna mirna
Supplementary Figure 1 Distribution of mirnas between lncrna and protein-coding genes. Pie chart showing distribution of human mirna between protein coding and lncrna genes. To the right, lncrna mirna
Supplementary Tables. Primers and probes. Target Primer / probe sequences Chemistry Human HBB F: AACTGTGTTCACTAGCAACCTCAAA
 Supplementary Tables Primers and probes Target Primer / probe sequences Chemistry H F: AACTGTGTTCACTAGCAACCTCAAA promoter R: ACAGGGCAGTAACGGCAGACT H Downstream Actb alpha (162) alpha (162.) (Pro) (16)
Supplementary Tables Primers and probes Target Primer / probe sequences Chemistry H F: AACTGTGTTCACTAGCAACCTCAAA promoter R: ACAGGGCAGTAACGGCAGACT H Downstream Actb alpha (162) alpha (162.) (Pro) (16)
Embryonic germ cell formation. Maintainance of pluripotency
 hvasa 2.5 kb promoter egfp 3 UTR(1kb) 1. Stable integration of germ cell reporter into hescs 2. Adherent differentiation and FACS isolation for VASA:GFP+ cells 4. Silencing or overexpression of the human
hvasa 2.5 kb promoter egfp 3 UTR(1kb) 1. Stable integration of germ cell reporter into hescs 2. Adherent differentiation and FACS isolation for VASA:GFP+ cells 4. Silencing or overexpression of the human
SUPPLEMENTARY INFORMATION
 Supplementary Discussion Interestingly, a recent study demonstrated that knockdown of Tet1 alone would lead to dysregulation of differentiation genes, but only minor defects in ES maintenance and no change
Supplementary Discussion Interestingly, a recent study demonstrated that knockdown of Tet1 alone would lead to dysregulation of differentiation genes, but only minor defects in ES maintenance and no change
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09861 & &' -(' ()*+ ')(+,,(','-*+,&,,+ ',+' ' 23,45/0*6787*9:./09 ;78?4?@*+A786?B- &' )*+*(,-* -(' ()*+ ')(+,,(','-*+,&,,+ ',+'./)*+*(,-*..)*+*(,-*./)*+*(,-*.0)*+*(,-*..)*+*(,-*
doi:10.1038/nature09861 & &' -(' ()*+ ')(+,,(','-*+,&,,+ ',+' ' 23,45/0*6787*9:./09 ;78?4?@*+A786?B- &' )*+*(,-* -(' ()*+ ')(+,,(','-*+,&,,+ ',+'./)*+*(,-*..)*+*(,-*./)*+*(,-*.0)*+*(,-*..)*+*(,-*
Epigenetic control of tomato fruit development. Key Laboratories of Molecular Epigenetics of MOE, Northeast Normal University, Changchun, China
 Epigenetic control of tomato fruit development Silin Zhong 1,2*,, Zhangjun Fei 1,3,*,, Yun-Ru Chen 1, Yi Zheng 1, Mingyun Huang 1, Julia Vrebalov 1, Ryan McQuinn 1, Nigel Gapper 1, Bao Liu 2, Jenny Xiang
Epigenetic control of tomato fruit development Silin Zhong 1,2*,, Zhangjun Fei 1,3,*,, Yun-Ru Chen 1, Yi Zheng 1, Mingyun Huang 1, Julia Vrebalov 1, Ryan McQuinn 1, Nigel Gapper 1, Bao Liu 2, Jenny Xiang
Fig. S1. Phenotypic analysis and assessment of meiotic silencing in Dgcr8 and Dicer cko males.
 Fig. S1. Phenotypic analysis and assessment of meiotic silencing in Dgcr8 and Dicer cko males. A-B Photographic representation of whole testes removed from Dgcr8 (A) and Dicer (B) litters (wild type (WT):
Fig. S1. Phenotypic analysis and assessment of meiotic silencing in Dgcr8 and Dicer cko males. A-B Photographic representation of whole testes removed from Dgcr8 (A) and Dicer (B) litters (wild type (WT):
Zhang et al., RepID facilitates replication Initiation. Supplemental Information:
 Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Integrated NGS Sample Preparation Solutions for Limiting Amounts of RNA and DNA. March 2, Steven R. Kain, Ph.D. ABRF 2013
 Integrated NGS Sample Preparation Solutions for Limiting Amounts of RNA and DNA March 2, 2013 Steven R. Kain, Ph.D. ABRF 2013 NuGEN s Core Technologies Selective Sequence Priming Nucleic Acid Amplification
Integrated NGS Sample Preparation Solutions for Limiting Amounts of RNA and DNA March 2, 2013 Steven R. Kain, Ph.D. ABRF 2013 NuGEN s Core Technologies Selective Sequence Priming Nucleic Acid Amplification
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Design and sequence of system 1 for targeted demethylation.
 Supplementary Figure 1 Design and sequence of system 1 for targeted demethylation. (a) Design of system 1 for targeted demethylation. TET1CD was fused to a catalytic inactive Cas9 nuclease (dcas9) and
Supplementary Figure 1 Design and sequence of system 1 for targeted demethylation. (a) Design of system 1 for targeted demethylation. TET1CD was fused to a catalytic inactive Cas9 nuclease (dcas9) and
Supplementary Data. Supplementary Table S1.
 Supplementary Data Supplementary Table S1. Primer Sequences for Real-Time Reverse Transcription-Polymerase Chain Reaction Gene Forward 5?3 Reverse 3?5 Housekeeping gene 36B4 TCCAGGCTTTGGGCATCA CTTTATCAGCTGCACATCACTCAGA
Supplementary Data Supplementary Table S1. Primer Sequences for Real-Time Reverse Transcription-Polymerase Chain Reaction Gene Forward 5?3 Reverse 3?5 Housekeeping gene 36B4 TCCAGGCTTTGGGCATCA CTTTATCAGCTGCACATCACTCAGA
a previous report, Lee and colleagues showed that Oct4 and Sox2 bind to DXPas34 and Xite
 SUPPLEMENTARY INFORMATION doi:1.138/nature9496 a 1 Relative RNA levels D12 female ES cells 24h Control sirna 24h sirna Oct4 5 b,4 Oct4 Xist (wt) Xist (mut) Tsix 3' (wt) Tsix 5' (wt) Beads Oct4,2 c,2 Xist
SUPPLEMENTARY INFORMATION doi:1.138/nature9496 a 1 Relative RNA levels D12 female ES cells 24h Control sirna 24h sirna Oct4 5 b,4 Oct4 Xist (wt) Xist (mut) Tsix 3' (wt) Tsix 5' (wt) Beads Oct4,2 c,2 Xist
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna
 Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Table S1. Primers used in the study
 Table S1. Primers used in the study Primer name Application Sequence I1F16 Genotyping GGCAAGTGAGTGAGTGCCTA I1R11 Genotyping CCCACTCGTATTGACGCTCT V19 Genotyping GGGTCTCAAAGTCAGGGTCA D18Mit184-F Genotyping
Table S1. Primers used in the study Primer name Application Sequence I1F16 Genotyping GGCAAGTGAGTGAGTGCCTA I1R11 Genotyping CCCACTCGTATTGACGCTCT V19 Genotyping GGGTCTCAAAGTCAGGGTCA D18Mit184-F Genotyping
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Nature Biotechnology: doi: /nbt Supplementary Figure 1. In vitro validation of OTC sgrnas and donor template.
 Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
SUPPLEMENTARY INFORMATION
 Supplemental Figure 1. Expression levels of cell type specific marker genes in each cell type. (a) Root meristem of ProCYCD5:GFP. GFP is specifically expressed in lower columella. (b) (e) Expression levels
Supplemental Figure 1. Expression levels of cell type specific marker genes in each cell type. (a) Root meristem of ProCYCD5:GFP. GFP is specifically expressed in lower columella. (b) (e) Expression levels
SUPPLEMENTARY INFORMATION
 SUPPLEMENTRY INFORMTION DOI:.38/ncb Kdmb locus kb Long isoform Short isoform Long isoform Jmj XX PHD F-box LRR Short isoform XX PHD F-box LRR Target Vector 3xFlag Left H Neo Right H TK LoxP LoxP Kdmb Locus
SUPPLEMENTRY INFORMTION DOI:.38/ncb Kdmb locus kb Long isoform Short isoform Long isoform Jmj XX PHD F-box LRR Short isoform XX PHD F-box LRR Target Vector 3xFlag Left H Neo Right H TK LoxP LoxP Kdmb Locus
Retrotransposons as a major source of epigenetic variations in the mammalian genome
 Epigenetics ISSN: 1559-2294 (Print) 1559-2308 (Online) Journal homepage: https://www.tandfonline.com/loi/kepi20 Retrotransposons as a major source of epigenetic variations in the mammalian genome Muhammad
Epigenetics ISSN: 1559-2294 (Print) 1559-2308 (Online) Journal homepage: https://www.tandfonline.com/loi/kepi20 Retrotransposons as a major source of epigenetic variations in the mammalian genome Muhammad
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State
 Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature875 a promoter firefly luciferase CNS b Supplementary Figure 1. Dual luciferase assays on enhancer activity of CNS1, 2, and 3. a. promoter sequence was inserted upstream of firefly luciferase
doi: 1.138/nature875 a promoter firefly luciferase CNS b Supplementary Figure 1. Dual luciferase assays on enhancer activity of CNS1, 2, and 3. a. promoter sequence was inserted upstream of firefly luciferase
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Candidate region (0.74 Mb) ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC ATC TCT GGG ACT CAT TAG CAG GAG GCT AGC
 A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure S1: (A) Schematic representation of the Jarid2 Flox/Flox
 Supplementary Figure S1: (A) Schematic representation of the Flox/Flox locus and the strategy used to visualize the wt and the Flox/Flox mrna by PCR from cdna. An example of the genotyping strategy is
Supplementary Figure S1: (A) Schematic representation of the Flox/Flox locus and the strategy used to visualize the wt and the Flox/Flox mrna by PCR from cdna. An example of the genotyping strategy is
Supplementary Fig. 1.
 Supplementary Fig. 1. RNAi-induced reduction in E(z) or Esc or mutations in Polycomb Group genes or corto partially rescues oogenesis of piwi mutants. (a) In vitro reconstituted PRC2 complexes (represented
Supplementary Fig. 1. RNAi-induced reduction in E(z) or Esc or mutations in Polycomb Group genes or corto partially rescues oogenesis of piwi mutants. (a) In vitro reconstituted PRC2 complexes (represented
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
Experimental genetics - 2 Partha Roy
 Partha Roy Experimental genetics - 2 Making genetically altered animal 1) Gene knock-out k from: a) the entire animal b) selected cell-type/ tissue c) selected cell-type/tissue at certain time 2) Transgenic
Partha Roy Experimental genetics - 2 Making genetically altered animal 1) Gene knock-out k from: a) the entire animal b) selected cell-type/ tissue c) selected cell-type/tissue at certain time 2) Transgenic
Supplementary. germ division-specific
 Figure Legends Supplementary Figure 1 DNA microarray data of representative genes for germ cells and/or two-cell embryos. PGC specification genes such as Blimp1 are shown separately from other germ cell
Figure Legends Supplementary Figure 1 DNA microarray data of representative genes for germ cells and/or two-cell embryos. PGC specification genes such as Blimp1 are shown separately from other germ cell
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
Rassf1a -/- Sav1 +/- mice. Supplementary Figures:
 1 Supplementary Figures: Figure S1: Genotyping of Rassf1a and Sav1 mutant mice. A. Rassf1a knockout mice lack exon 1 of the Rassf1 gene. A diagram and typical PCR genotyping of wildtype and Rassf1a -/-
1 Supplementary Figures: Figure S1: Genotyping of Rassf1a and Sav1 mutant mice. A. Rassf1a knockout mice lack exon 1 of the Rassf1 gene. A diagram and typical PCR genotyping of wildtype and Rassf1a -/-
Topics in Molecular Biology ( ) 3 rd Lecture Hall, Room 504 Thursday 1pm-2:50pm
 Topics in Molecular Biology (01139620) 3 rd Lecture Hall, Room 504 Thursday 1pm-2:50pm Instructor: Ming Tian Biology Building 633 E-mail: mingtian@pku.edu.cn Phone: 6275-6459 Office hours: Monday 1pm-4pm
Topics in Molecular Biology (01139620) 3 rd Lecture Hall, Room 504 Thursday 1pm-2:50pm Instructor: Ming Tian Biology Building 633 E-mail: mingtian@pku.edu.cn Phone: 6275-6459 Office hours: Monday 1pm-4pm
Chemical and/or enzymatic deamination across various sequencing methods to localize modified cytosines.
 Supplementary Figure 1 Chemical and/or enzymatic deamination across various sequencing methods to localize modified cytosines. (a) Upon treatment with bisulfite under acidic conditions and at elevated
Supplementary Figure 1 Chemical and/or enzymatic deamination across various sequencing methods to localize modified cytosines. (a) Upon treatment with bisulfite under acidic conditions and at elevated
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
SUPPLEMENTAL FIGURE LEGENDS Suppl. Fig. 1. Notch and myostatin expression is unaffected in the absence of p65 during postnatal development. A & B. Myostatin and Notch-1 expression levels were determined
Parts of a standard FastQC report
 FastQC FastQC, written by Simon Andrews of Babraham Bioinformatics, is a very popular tool used to provide an overview of basic quality control metrics for raw next generation sequencing data. There are
FastQC FastQC, written by Simon Andrews of Babraham Bioinformatics, is a very popular tool used to provide an overview of basic quality control metrics for raw next generation sequencing data. There are
Supplementary Information
 Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
SUPPLEMENTARY INFORMATION
 doi:1.138/nature11233 Supplementary Figure S1 Sample Flowchart. The ENCODE transcriptome data are obtained from several cell lines which have been cultured in replicates. They were either left intact (whole
doi:1.138/nature11233 Supplementary Figure S1 Sample Flowchart. The ENCODE transcriptome data are obtained from several cell lines which have been cultured in replicates. They were either left intact (whole
administration of tamoxifen. Bars show mean ± s.e.m (n=10-11). P-value was determined by
 Supplementary Figure 1. Chimerism of CD45.2 + GFP + cells at 1 month post transplantation No significant changes were detected in chimerism of CD45.2 + GFP + cells between recipient mice repopulated with
Supplementary Figure 1. Chimerism of CD45.2 + GFP + cells at 1 month post transplantation No significant changes were detected in chimerism of CD45.2 + GFP + cells between recipient mice repopulated with
AD BD TOC1. Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between
 AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
AD X BD TOC1 AD BD X PIFΔAD PIF TOC1 TOC1 PIFΔAD PIF N TOC1 TOC1 C1 PIFΔAD PIF C1 TOC1 TOC1 C PIFΔAD PIF C TOC1 Supplementary Figure 1: Yeast two-hybrid assays showing the interaction between PIF and TOC1
Allele-specific locus binding and genome editing by CRISPR at the
 Supplementary Information Allele-specific locus binding and genome editing by CRISPR at the p6ink4a locus Toshitsugu Fujita, Miyuki Yuno, and Hodaka Fujii Supplementary Figure Legends Supplementary Figure
Supplementary Information Allele-specific locus binding and genome editing by CRISPR at the p6ink4a locus Toshitsugu Fujita, Miyuki Yuno, and Hodaka Fujii Supplementary Figure Legends Supplementary Figure
Ramp1 EPD0843_4_B11. EUCOMM/KOMP-CSD Knockout-First Genotyping
 EUCOMM/KOMP-CSD Knockout-First Genotyping Introduction The majority of animals produced from the EUCOMM/KOMP-CSD ES cell resource contain the Knockout-First-Reporter Tagged Insertion allele. As well as
EUCOMM/KOMP-CSD Knockout-First Genotyping Introduction The majority of animals produced from the EUCOMM/KOMP-CSD ES cell resource contain the Knockout-First-Reporter Tagged Insertion allele. As well as
Usp14 EPD0582_2_G09. EUCOMM/KOMP-CSD Knockout-First Genotyping
 EUCOMM/KOMP-CSD Knockout-First Genotyping Introduction The majority of animals produced from the EUCOMM/KOMP-CSD ES cell resource contain the Knockout-First-Reporter Tagged Insertion allele. As well as
EUCOMM/KOMP-CSD Knockout-First Genotyping Introduction The majority of animals produced from the EUCOMM/KOMP-CSD ES cell resource contain the Knockout-First-Reporter Tagged Insertion allele. As well as
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplemental Figure 1 A
 Supplemental Figure 1 A Supplemental Data. Han et al. (2016). Plant Cell 10.1105/tpc.15.00997 BamHI -1616 bp SINE repeats SalI ATG SacI +1653 bp HindIII LB HYG p35s pfwa LUC Tnos RB pfwa-bamhi-f: CGGGATCCCGCCTTTCTCTTCCTCATCTGC
Supplemental Figure 1 A Supplemental Data. Han et al. (2016). Plant Cell 10.1105/tpc.15.00997 BamHI -1616 bp SINE repeats SalI ATG SacI +1653 bp HindIII LB HYG p35s pfwa LUC Tnos RB pfwa-bamhi-f: CGGGATCCCGCCTTTCTCTTCCTCATCTGC
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in
 Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Non-coding Function & Variation, MPRAs II. Mike White Bio /5/18
 Non-coding Function & Variation, MPRAs II Mike White Bio 5488 3/5/18 MPRA Review Problem 1: Where does your CRE DNA come from? DNA synthesis Genomic fragments Targeted regulome capture Problem 2: How do
Non-coding Function & Variation, MPRAs II Mike White Bio 5488 3/5/18 MPRA Review Problem 1: Where does your CRE DNA come from? DNA synthesis Genomic fragments Targeted regulome capture Problem 2: How do
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1
 Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Supplemental Table/Figure Legends
 MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
Infiltrating immune cells do not differ between T2E and non-t2e samples and represent a small fraction of total cellularity.
 Supplementary Figure 1 Infiltrating immune cells do not differ between T2E and non-t2e samples and represent a small fraction of total cellularity. (a) ESTIMATE immune score from mrna expression array
Supplementary Figure 1 Infiltrating immune cells do not differ between T2E and non-t2e samples and represent a small fraction of total cellularity. (a) ESTIMATE immune score from mrna expression array
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Chromatin immunoprecipitation (ChIP) ES and FACS-sorted GFP+/Flk1+ cells were fixed in 1% formaldehyde and sonicated until fragments of an average
 Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted
Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted
Supplemental Information. Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis
 Supplemental Information Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis Yunpeng Bai ;, Lujuan Zhang #, Hongming Zhou #, Yuanshu Dong #, Qi Zeng, Weinian Shou, and Zhong-Yin Zhang
Supplemental Information Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis Yunpeng Bai ;, Lujuan Zhang #, Hongming Zhou #, Yuanshu Dong #, Qi Zeng, Weinian Shou, and Zhong-Yin Zhang
RNA-Sequencing analysis
 RNA-Sequencing analysis Markus Kreuz 25. 04. 2012 Institut für Medizinische Informatik, Statistik und Epidemiologie Content: Biological background Overview transcriptomics RNA-Seq RNA-Seq technology Challenges
RNA-Sequencing analysis Markus Kreuz 25. 04. 2012 Institut für Medizinische Informatik, Statistik und Epidemiologie Content: Biological background Overview transcriptomics RNA-Seq RNA-Seq technology Challenges
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter
 CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter SUPPLEMENTARY DATA See Supplementary Sequence File: 1 Supplementary Figure S1: The total protein was extracted from
CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter SUPPLEMENTARY DATA See Supplementary Sequence File: 1 Supplementary Figure S1: The total protein was extracted from
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
Figure S1. Replication initiation sites at efficient ORIs do not coincide with nucleosomedepleted
 Figure S1. Replication initiation sites at efficient ORIs do not coincide with nucleosomedepleted regions in the mouse genome. Typical examples of SNS profiles (Cayrou et al., 11) and nucleosome occupancies
Figure S1. Replication initiation sites at efficient ORIs do not coincide with nucleosomedepleted regions in the mouse genome. Typical examples of SNS profiles (Cayrou et al., 11) and nucleosome occupancies
Mammalian non-cg methylations are conserved and cell-type specific and may have been involved in the evolution of transposon elements
 Mammalian non-cg methylations are conserved and cell-type specific and may have been involved in the evolution of transposon elements Weilong Guo, Michael Zhang, Hong Wu Supplementary Figures Fig. S1-S16
Mammalian non-cg methylations are conserved and cell-type specific and may have been involved in the evolution of transposon elements Weilong Guo, Michael Zhang, Hong Wu Supplementary Figures Fig. S1-S16
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
Supplemental Information. Antagonistic Activities of Sox2. and Brachyury Control the Fate Choice. of Neuro-Mesodermal Progenitors
 Developmental Cell, Volume 42 Supplemental Information Antagonistic Activities of Sox2 and Brachyury Control the Fate Choice of Neuro-Mesodermal Progenitors Frederic Koch, Manuela Scholze, Lars Wittler,
Developmental Cell, Volume 42 Supplemental Information Antagonistic Activities of Sox2 and Brachyury Control the Fate Choice of Neuro-Mesodermal Progenitors Frederic Koch, Manuela Scholze, Lars Wittler,
H3K36me3 polyclonal antibody
 H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
* * IgL locus (V - J ) λ λ
 Supplementary Figures and Figure Legend Supplementary Figure S1 A C F Vav-P-BCL2 doner mice 8 6 4 2 Lymphoma free (%)1 Tumor Pathology Score Retroviral transduction Vav-P-BCL2 HPCs HPC + shrna VavP-Bcl2
Supplementary Figures and Figure Legend Supplementary Figure S1 A C F Vav-P-BCL2 doner mice 8 6 4 2 Lymphoma free (%)1 Tumor Pathology Score Retroviral transduction Vav-P-BCL2 HPCs HPC + shrna VavP-Bcl2
Analyzing ChIP-seq data. R. Gentleman, D. Sarkar, S. Tapscott, Y. Cao, Z. Yao, M. Lawrence, P. Aboyoun, M. Morgan, L. Ruzzo, J. Davison, H.
 Analyzing ChIP-seq data R. Gentleman, D. Sarkar, S. Tapscott, Y. Cao, Z. Yao, M. Lawrence, P. Aboyoun, M. Morgan, L. Ruzzo, J. Davison, H. Pages Biological Motivation Chromatin-immunopreciptation followed
Analyzing ChIP-seq data R. Gentleman, D. Sarkar, S. Tapscott, Y. Cao, Z. Yao, M. Lawrence, P. Aboyoun, M. Morgan, L. Ruzzo, J. Davison, H. Pages Biological Motivation Chromatin-immunopreciptation followed
