Supplemental Information. Antagonistic Activities of Sox2. and Brachyury Control the Fate Choice. of Neuro-Mesodermal Progenitors
|
|
|
- Andrea Jacobs
- 6 years ago
- Views:
Transcription
1 Developmental Cell, Volume 42 Supplemental Information Antagonistic Activities of Sox2 and Brachyury Control the Fate Choice of Neuro-Mesodermal Progenitors Frederic Koch, Manuela Scholze, Lars Wittler, Dennis Schifferl, Smita Sudheer, Phillip Grote, Bernd Timmermann, Karol Macura, and Bernhard G. Herrmann
2 A Koch_Figure S1 1.4% / 0.2% 2.5% B / Group I Wnt3a Fgf8 Rspo3 Group II Group III Group IV Group V + - / Lef1 Foxc1 Foxf1 Osr1 Sp8 Nkx1-2 Sox1 Sox3 C 2 Pseudotime Component Component 1
3 Figure S1. Related to Figure 1. (A) FACS plot of cells from hearts of / embryos, used to set-up the gating in Figure 1B. (B) Genome browser snapshots showing normalized read densities across examples of differentially expressed genes clustered into groups I-V. (C) Same PCA plot showing the cell trajectory as in Figure 1F, but colored according to pseudotime.
4 Koch_Figure S2 A B d2 d3 / / 2% 83% 98% 17% C / JM8 JM8 T Sox2 Tbx6 * H3 * 70kDa 55kDa 55kDa 40kDa 55kDa 40kDa 25kDa 15kDa D Group I Group II Group III Group IV Group V Sox2 T Wnt3a Lef1 Tbx3 Sp8 Sox3 E peaks 228 genes 542 peaks 319 genes Frequency Distance from T to Closest Sox2 Peak Maximum (bp)
5 Figure S2. Related to Figure 2. (A) Fluorescence microscopy of cells containing and reporters, differentiated in vitro from ES cells to NMPs. The majority of reporter-positive cells are /. Scale bars, 50µm. (B) FACS analysis of cells containing and reporters, differentiated in vitro from ES cells to NMPs either at day 2 (d2, left) or day 3 (d3, right) of differentiation. (C) Western blotting confirming that both T and Sox2 protein are co-expressed at day 3 (d3) of differentiation marking the NMP stage, in wild-type JM8 cells () (right) as well as in cells containing and reporters (left). The expression of Tbx6 at d3 and d4 indicates that the NMP stage is transient, followed by differentiation to (presomitic) mesoderm. Controls are undifferentiated ES-cells grown in 2i + LIF medium (d0); histone H3 was used as a loading control; * indicates background bands. (D) Genome browser screenshots showing T and Sox2 -Seq read densities across exemplary genes from groups I-V and their surrounding genomic regions. (E) To investigate whether T and Sox2 co-bind the same potential regulatory elements at the 574 shared putative target genes (see Figure 2C), the distances from the maxima of T peaks (n=1990) to the closest Sox2 peak associated with the same gene were retrieved. In total, 324 and 542 (314 and 524 when correcting for peak associations to two genes due to intergenic binding) are found within 1kb and 5kb respectively, corresponding to 228 and 319 of the 574 putative target genes.
6 Koch_Figure S3 A Brightfield merge B Neg. (head) CE TS % 1.3% 66.0% 31.1% T 2J/2J T 2J/2J 99.2% 0.6% 47.4% 49.4% C n = 4, ,541 2,372 D Sox2 Promoter Intergenic Genic n = 2, e T bits 1 0 motif frequency distance from peak summit (bp) β-catenin Rspo3
7 Figure S3. Related to Figure 3. (A) Fluorescence microscopy pictures of embryos obtained by tetraploid complementation using either wild-type () or mutant T 2J/2J ES cells containing the reporter, verifying the correct reporter expression in the posterior mesoderm. Scale bars, 500µm. (B) FACS profiles of positive cells sorted from caudal ends (CE) of wild-type () or T 2J/2 embryos of stage TS13, used for differential RNA-Seq analysis. Forebrains (heads) were used as a negative control and the relative amounts of cells in each subpopulation in and T 2J/2J mutants are indicated in percentages. (C) Analysis of β-catenin -Seq data showing the distribution of binding sites, the de novo motif analysis and the frequency distribution of that motif around the peak maxima. (D) Exemplary genome browser screenshot showing Sox2, T and β-catenin binding near the Rspo3 locus.
8 A B Koch_Figure S4 Neg. (heart) CE TS12 CE TS13 T 2J/2J T 2J/2J 0% 0.1% / 0% 1.3% / 0% 51.1% 31.1% / 26.0% 9.7% / 26.3% 53.7% / 17.6% 10.7% 26.3% / 16.4% 0.9% 11.3% 14.4% C TS12 TS13 T 2J/2J normalized frequency (x10,000) normalized frequency (x10,000) / / bp 200bp 400bp 600bp 0bp 200bp 400bp 600bp 0bp 200bp 400bp 600bp 0bp 200bp 400bp 600bp 0bp 200bp 400bp 600bp 0bp 200bp 400bp 600bp bp 200bp 400bp 600bp 0bp 200bp 400bp 600bp 0bp 200bp 400bp 600bp 0bp 200bp 400bp 600bp 0bp 200bp 400bp 600bp / / bp 200bp 400bp 600bp D Before Normalization 1600 After Normalization Mass Mass
9 Figure S4. Related to Figure 4. (A) Fluorescence microscopy pictures of embryos obtained by tetraploid complementation using either wild-type () or T 2J/2J ES cells containing the and reporters, verifying the correct expression patterns. Scale bars, 500µm. (B) FACS profiles of and/or reporter-positive cells isolated from caudal ends of wild-type (, upper) or T 2J/2J embryos (lower panel) of stages TS12 (left) and TS13 (right), used for in vivo ATAC-Seq analysis. Samples from hearts (left) were used as negative controls; the relative amounts of cells in each subpopulation are indicated in percentages. (C) Frequency distribution plots of DNA fragment sizes sequenced in ATAC- Seq experiments of sorted cell populations. (D) Box plots of genomic coverage distributions of all samples across different chromosomes shown before (left) and after (right) normalization.
10 A Koch_Figure S5 T 2J/2J / B T 2J/2J C chr5 chr6 chr10 chr11 T 2J/2J / T 2J/2J chr4 chr3 chr1 chr7 chr9 chr12 chr8 chr13 chr15 chr16 chr14 chr2 chr17 chrx chr18 chr /
11 Figure S5. Related to Figure 4. (A) Genome-browser snapshot of chromosome 2 showing ATAC-Seq data profiles derived from FACS purified cells of wild-type () or T 2J/2J TS12 embryos relative to stage-matched wild-type NMP cells. (B) Genome-wide correlation matrix of all ATAC-Seq datasets derived from indicated cell types at and genotypes at TS12 relative to their respective stage-matched wild-type NMP dataset. (C) Heatmap of the genome-wide Hilbert curve of global ATAC-Seq data (relative to the NMP dataset) derived from FACSpurified, / or cells from TS12 wild-type () or mutant (T 2J/2J ) embryos. The map of the underlying Hilbert curves shows the location of each chromosome.
12 Koch_Figure S6 A Brightfield mcherry B Neg. (head) CE TS14 Tbx6 -/- Tbx6 -/- 99.0% 99.4% Tbx6 mch + Tbx6 mch + 0.9% 39.2% 59.5% Tbx6::mCherry Tbx6::mCherry Tbx6 mch + Tbx6 mch + 0.5% 33.5% 65.8% Tbx6::mCherry Tbx6::mCherry C KO F T D E Tbx6 mch + Tbx6 mch + KO KO KO KO Msgn1 Wnt3a Foxf1 Fgf8 G H Tbx6 T Tbx6 T Fgf8 Msgn1 Tbx6 mch + KO Tbx6 Sox1 Sox3 Sox3
13 Figure S6. Related to Figure 5. (A) Fluorescence microscopy pictures of embryos obtained by tetraploid complementation using wild-type () or Tbx6 -/- ES cells containing the Tbx6::mCherry reporter, verifying the correct expression pattern at TS14. In the mcherry signal is mainly observed in paraxial mesoderm, however the signal persists in several somite pairs due to mcherry stability. For FACS sorting, only caudal end material posterior to presomite -2 in wild-type, representing mostly presomitic mesoderm tissue was isolated by microdissection. (B) FACS profiles of Tbx6::mCherry cells sorted from (upper panel) or Tbx6 -/- embryos (lower panel) of stage TS14 used for transcriptome or ATAC-Seq analysis. Samples from heads were used as negative controls; the relative amounts of cells in each subpopulation are indicated in percentages. (C-E) Exemplary genome-browser views of RNA-seq read densities from dysregulated genes behaving similarly in T 2J/2J and Tbx6 -/- knock-outs (C), genes downregulated in T 2J/2J and upregulated in Tbx6 -/- knock-outs (D), and genes upregulated in T 2J/2J and unchanged in Tbx6 -/- knock-outs (E). (F-H) Exemplary genome-browser views of T and Tbx6 -seq read densities from dysregulated genes behaving similarly in T 2J/2J and Tbx6 -/- knock-outs (F), downregulated in T 2J/2J and upregulated in Tbx6 -/- knock-outs (G), and upregulated in T 2J/2J and unchanged in Tbx6 -/- knock-outs (H).
Antagonistic Activities of Sox2 and Brachyury Control the Fate Choice of Neuro-Mesodermal
 Article Antagonistic Activities of Sox2 and Brachyury Control the Fate Choice of Neuro-Mesodermal Progenitors Highlights d Brachyury is essential for the maintenance of neuromesodermal progenitors d d
Article Antagonistic Activities of Sox2 and Brachyury Control the Fate Choice of Neuro-Mesodermal Progenitors Highlights d Brachyury is essential for the maintenance of neuromesodermal progenitors d d
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
of NOBOX. Supplemental Figure S1F shows the staining profile of LHX8 antibody (Abcam, ab41519), which is a
 SUPPLEMENTAL MATERIALS AND METHODS LHX8 Staining During the course of this work, it came to our attention that the NOBOX antibody used in this study had been discontinued. There are a number of other nuclear
SUPPLEMENTAL MATERIALS AND METHODS LHX8 Staining During the course of this work, it came to our attention that the NOBOX antibody used in this study had been discontinued. There are a number of other nuclear
Supporting Information
 Supporting Information Ho et al. 1.173/pnas.81288816 SI Methods Sequences of shrna hairpins: Brg shrna #1: ccggcggctcaagaaggaagttgaactcgagttcaacttccttcttgacgnttttg (TRCN71383; Open Biosystems). Brg shrna
Supporting Information Ho et al. 1.173/pnas.81288816 SI Methods Sequences of shrna hairpins: Brg shrna #1: ccggcggctcaagaaggaagttgaactcgagttcaacttccttcttgacgnttttg (TRCN71383; Open Biosystems). Brg shrna
Supporting Information
 Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Nature Genetics: doi: /ng Supplementary Figure 1. H3K27ac HiChIP enriches enhancer promoter-associated chromatin contacts.
 Supplementary Figure 1 H3K27ac HiChIP enriches enhancer promoter-associated chromatin contacts. (a) Schematic of chromatin contacts captured in H3K27ac HiChIP. (b) Loop call overlap for cohesin HiChIP
Supplementary Figure 1 H3K27ac HiChIP enriches enhancer promoter-associated chromatin contacts. (a) Schematic of chromatin contacts captured in H3K27ac HiChIP. (b) Loop call overlap for cohesin HiChIP
Nature Biotechnology: doi: /nbt Supplementary Figure 1. sndrop-seq overview.
 Supplementary Figure 1 sndrop-seq overview. A. sndrop-seq method showing modifications needed to process nuclei, including bovine serum albumin (BSA) coating and droplet heating to ensure complete nuclear
Supplementary Figure 1 sndrop-seq overview. A. sndrop-seq method showing modifications needed to process nuclei, including bovine serum albumin (BSA) coating and droplet heating to ensure complete nuclear
Authors: Takuro Horii, Masamichi Yamamoto, Sumiyo Morita, Mika Kimura, Yasumitsu Nagao, Izuho Hatada *
 SUPPLEMENTARY INFORMATION Title: p53 Suppresses Tetraploid Development in Mice Authors: Takuro Horii, Masamichi Yamamoto, Sumiyo Morita, Mika Kimura, Yasumitsu Nagao, Izuho Hatada * Supplementary Methods
SUPPLEMENTARY INFORMATION Title: p53 Suppresses Tetraploid Development in Mice Authors: Takuro Horii, Masamichi Yamamoto, Sumiyo Morita, Mika Kimura, Yasumitsu Nagao, Izuho Hatada * Supplementary Methods
Spermatazoa. Sertoli cells. Morc1 WT adult. Morc1 KO adult. Morc1 WT P14.5. Morc1 KO P14.5. Germ cells. Germ cells. Spermatids.
 a. Spermatazoa Sertoli cells Morc1 WT adult c. Morc1 KO adult Morc1 WT P1.5 Morc1 KO P1.5 Morc1 WT adult Morc1 KO adult Germ cells Germ cells Spermatids Spermatozoa Supplementary Figure 1. Confirmation
a. Spermatazoa Sertoli cells Morc1 WT adult c. Morc1 KO adult Morc1 WT P1.5 Morc1 KO P1.5 Morc1 WT adult Morc1 KO adult Germ cells Germ cells Spermatids Spermatozoa Supplementary Figure 1. Confirmation
Zhang et al., RepID facilitates replication Initiation. Supplemental Information:
 Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Table 1: Oligo designs. A list of ATAC-seq oligos used for PCR.
 Ad1_noMX: Ad2.1_TAAGGCGA Ad2.2_CGTACTAG Ad2.3_AGGCAGAA Ad2.4_TCCTGAGC Ad2.5_GGACTCCT Ad2.6_TAGGCATG Ad2.7_CTCTCTAC Ad2.8_CAGAGAGG Ad2.9_GCTACGCT Ad2.10_CGAGGCTG Ad2.11_AAGAGGCA Ad2.12_GTAGAGGA Ad2.13_GTCGTGAT
Ad1_noMX: Ad2.1_TAAGGCGA Ad2.2_CGTACTAG Ad2.3_AGGCAGAA Ad2.4_TCCTGAGC Ad2.5_GGACTCCT Ad2.6_TAGGCATG Ad2.7_CTCTCTAC Ad2.8_CAGAGAGG Ad2.9_GCTACGCT Ad2.10_CGAGGCTG Ad2.11_AAGAGGCA Ad2.12_GTAGAGGA Ad2.13_GTCGTGAT
Supplementary Data. Generation and Characterization of Mixl1- Inducible Embryonic Stem Cells Under the Control of Oct3/4 Promoter or CAG Promoter
 Supplementary Data Generation and Characterization of Mixl1- Inducible Embryonic Stem Cells Under the Control of Oct3/4 Promoter or CAG Promoter We first introduced an expression unit composed of a tetresponse
Supplementary Data Generation and Characterization of Mixl1- Inducible Embryonic Stem Cells Under the Control of Oct3/4 Promoter or CAG Promoter We first introduced an expression unit composed of a tetresponse
SUPPLEMENTARY INFORMATION
 18Pura Brain Liver 2 kb 9Mtmr Heart Brain 1.4 kb 15Ppara 1.5 kb IPTpcc 1.3 kb GAPDH 1.5 kb IPGpr1 2 kb GAPDH 1.5 kb 17Tcte 13Elov12 Lung Thymus 6 kb 5 kb 17Tbcc Kidney Embryo 4.5 kb 3.7 kb GAPDH 1.5 kb
18Pura Brain Liver 2 kb 9Mtmr Heart Brain 1.4 kb 15Ppara 1.5 kb IPTpcc 1.3 kb GAPDH 1.5 kb IPGpr1 2 kb GAPDH 1.5 kb 17Tcte 13Elov12 Lung Thymus 6 kb 5 kb 17Tbcc Kidney Embryo 4.5 kb 3.7 kb GAPDH 1.5 kb
Supplemental Figure 1.
 Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Galaxy Platform For NGS Data Analyses
 Galaxy Platform For NGS Data Analyses Weihong Yan wyan@chem.ucla.edu Collaboratory Web Site http://qcb.ucla.edu/collaboratory http://collaboratory.lifesci.ucla.edu Workshop Outline ü Day 1 UCLA galaxy
Galaxy Platform For NGS Data Analyses Weihong Yan wyan@chem.ucla.edu Collaboratory Web Site http://qcb.ucla.edu/collaboratory http://collaboratory.lifesci.ucla.edu Workshop Outline ü Day 1 UCLA galaxy
Abcam.com. hutton.ac.uk. Ipmdss.dk. Bo Gong and Eva Chou
 Abcam.com Bo Gong and Eva Chou Ipmdss.dk hutton.ac.uk What is a homeotic gene? A gene which regulates the developmental fate of anatomical structures in an organism Why study them? Understand the underlying
Abcam.com Bo Gong and Eva Chou Ipmdss.dk hutton.ac.uk What is a homeotic gene? A gene which regulates the developmental fate of anatomical structures in an organism Why study them? Understand the underlying
Nature Genetics: doi: /ng Supplementary Figure 1. ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets.
 Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Information
 Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
Supplemental Materials
 Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Introduction to genome biology
 Introduction to genome biology Lisa Stubbs Deep transcritpomes for traditional model species from ENCODE (and modencode) Deep RNA-seq and chromatin analysis on 147 human cell types, as well as tissues,
Introduction to genome biology Lisa Stubbs Deep transcritpomes for traditional model species from ENCODE (and modencode) Deep RNA-seq and chromatin analysis on 147 human cell types, as well as tissues,
What we ll do today. Types of stem cells. Do engineered ips and ES cells have. What genes are special in stem cells?
 Do engineered ips and ES cells have similar molecular signatures? What we ll do today Research questions in stem cell biology Comparing expression and epigenetics in stem cells asuring gene expression
Do engineered ips and ES cells have similar molecular signatures? What we ll do today Research questions in stem cell biology Comparing expression and epigenetics in stem cells asuring gene expression
Do engineered ips and ES cells have similar molecular signatures?
 Do engineered ips and ES cells have similar molecular signatures? Comparing expression and epigenetics in stem cells George Bell, Ph.D. Bioinformatics and Research Computing 2012 Spring Lecture Series
Do engineered ips and ES cells have similar molecular signatures? Comparing expression and epigenetics in stem cells George Bell, Ph.D. Bioinformatics and Research Computing 2012 Spring Lecture Series
Supplemental Figure 1 A
 Supplemental Figure A prebleach postbleach 2 min 6 min 3 min mh2a.-gfp mh2a.2-gfp mh2a2-gfp GFP-H2A..9 Relative Intensity.8.7.6.5 mh2a. GFP n=8.4 mh2a.2 GFP n=4.3 mh2a2 GFP n=2.2 GFP H2A n=24. GFP n=7.
Supplemental Figure A prebleach postbleach 2 min 6 min 3 min mh2a.-gfp mh2a.2-gfp mh2a2-gfp GFP-H2A..9 Relative Intensity.8.7.6.5 mh2a. GFP n=8.4 mh2a.2 GFP n=4.3 mh2a2 GFP n=2.2 GFP H2A n=24. GFP n=7.
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Characterization of Hi-C/CHi-C dynamics and enhancer identification. (a) Scatterplot of Hi-C read counts supporting contacts between domain boundaries. Contacts enclosing domains
Supplementary Figure 1 Characterization of Hi-C/CHi-C dynamics and enhancer identification. (a) Scatterplot of Hi-C read counts supporting contacts between domain boundaries. Contacts enclosing domains
Supplemental Figure legends Figure S1. (A) (B) (C) (D) Figure S2. Figure S3. (A-E) Figure S4. Figure S5. (A, C, E, G, I) (B, D, F, H, Figure S6.
 Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
a previous report, Lee and colleagues showed that Oct4 and Sox2 bind to DXPas34 and Xite
 SUPPLEMENTARY INFORMATION doi:1.138/nature9496 a 1 Relative RNA levels D12 female ES cells 24h Control sirna 24h sirna Oct4 5 b,4 Oct4 Xist (wt) Xist (mut) Tsix 3' (wt) Tsix 5' (wt) Beads Oct4,2 c,2 Xist
SUPPLEMENTARY INFORMATION doi:1.138/nature9496 a 1 Relative RNA levels D12 female ES cells 24h Control sirna 24h sirna Oct4 5 b,4 Oct4 Xist (wt) Xist (mut) Tsix 3' (wt) Tsix 5' (wt) Beads Oct4,2 c,2 Xist
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
SUPPLEMENTARY INFORMATION
 SUPPLEMENTRY INFORMTION DOI:.38/ncb Kdmb locus kb Long isoform Short isoform Long isoform Jmj XX PHD F-box LRR Short isoform XX PHD F-box LRR Target Vector 3xFlag Left H Neo Right H TK LoxP LoxP Kdmb Locus
SUPPLEMENTRY INFORMTION DOI:.38/ncb Kdmb locus kb Long isoform Short isoform Long isoform Jmj XX PHD F-box LRR Short isoform XX PHD F-box LRR Target Vector 3xFlag Left H Neo Right H TK LoxP LoxP Kdmb Locus
From Variants to Pathways: Agilent GeneSpring GX s Variant Analysis Workflow
 From Variants to Pathways: Agilent GeneSpring GX s Variant Analysis Workflow Technical Overview Import VCF Introduction Next-generation sequencing (NGS) studies have created unanticipated challenges with
From Variants to Pathways: Agilent GeneSpring GX s Variant Analysis Workflow Technical Overview Import VCF Introduction Next-generation sequencing (NGS) studies have created unanticipated challenges with
Development 142: doi: /dev : Supplementary Material
 Development 142: doi:1.42/dev.11687: Supplementary Material Figure S1 - Relative expression of lineage markers in single ES cell and cardiomyocyte by real-time quantitative PCR analysis. Single ES cell
Development 142: doi:1.42/dev.11687: Supplementary Material Figure S1 - Relative expression of lineage markers in single ES cell and cardiomyocyte by real-time quantitative PCR analysis. Single ES cell
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum. Blood-stage Development
 Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
Basics of RNA-Seq. (With a Focus on Application to Single Cell RNA-Seq) Michael Kelly, PhD Team Lead, NCI Single Cell Analysis Facility
 2018 ABRF Meeting Satellite Workshop 4 Bridging the Gap: Isolation to Translation (Single Cell RNA-Seq) Sunday, April 22 Basics of RNA-Seq (With a Focus on Application to Single Cell RNA-Seq) Michael Kelly,
2018 ABRF Meeting Satellite Workshop 4 Bridging the Gap: Isolation to Translation (Single Cell RNA-Seq) Sunday, April 22 Basics of RNA-Seq (With a Focus on Application to Single Cell RNA-Seq) Michael Kelly,
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Suppl. Table S1. Characteristics of DHS regions analyzed by bisulfite sequencing. No. CpGs analyzed in the amplicon. Genomic location specificity
 Suppl. Table S1. Characteristics of DHS regions analyzed by bisulfite sequencing. DHS/GRE Genomic location Tissue specificity DHS type CpG density (per 100 bp) No. CpGs analyzed in the amplicon CpG within
Suppl. Table S1. Characteristics of DHS regions analyzed by bisulfite sequencing. DHS/GRE Genomic location Tissue specificity DHS type CpG density (per 100 bp) No. CpGs analyzed in the amplicon CpG within
C24. ros1-1. ros1-1 rdm18-1. ros1-1 rdm18-2. ros1-1 nrpe1
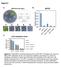 Figure S1 Methylation level 1 0.8 0.6 0.4 0.2 0 p35s methylation levels 24 ros1-1 ros1-1 rdm18-1 ros1-1 rdm18-2 ros1-1 nrpe1 mg mhg mhh Figure S1. RM18/PKL promotes silencing at the p35s-npt II transgene
Figure S1 Methylation level 1 0.8 0.6 0.4 0.2 0 p35s methylation levels 24 ros1-1 ros1-1 rdm18-1 ros1-1 rdm18-2 ros1-1 nrpe1 mg mhg mhh Figure S1. RM18/PKL promotes silencing at the p35s-npt II transgene
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
GENOME 371, Problem Set 6
 GENOME 371, Problem Set 6 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been
GENOME 371, Problem Set 6 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Generation of App knock-in mice reveals deletion mutations protective against Alzheimer s. disease-like pathology. Nagata et al.
 Generation of App knock-in mice reveals deletion mutations protective against Alzheimer s disease-like pathology Nagata et al. Supplementary Fig 1. Previous App knock-in model did not show Aβ accumulation
Generation of App knock-in mice reveals deletion mutations protective against Alzheimer s disease-like pathology Nagata et al. Supplementary Fig 1. Previous App knock-in model did not show Aβ accumulation
HC70AL SUMMER 2014 PROFESSOR BOB GOLDBERG Gene Annotation Worksheet
 HC70AL SUMMER 2014 PROFESSOR BOB GOLDBERG Gene Annotation Worksheet NAME: DATE: QUESTION ONE Using primers given to you by your TA, you carried out sequencing reactions to determine the identity of the
HC70AL SUMMER 2014 PROFESSOR BOB GOLDBERG Gene Annotation Worksheet NAME: DATE: QUESTION ONE Using primers given to you by your TA, you carried out sequencing reactions to determine the identity of the
Supplemental Materials
 Supplemental Materials Supplemental Figure S. Phenotypic assessment of alb4 mutant plants under different stress conditions. (A) High-light stress and drought stress. Wild-type (WT) and alb4 mutant plants
Supplemental Materials Supplemental Figure S. Phenotypic assessment of alb4 mutant plants under different stress conditions. (A) High-light stress and drought stress. Wild-type (WT) and alb4 mutant plants
Nature Genetics: doi: /ng.3556 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE. Supplementary Figure 1
 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
Supplementary Information
 Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Nature Immunology: doi: /ni.3694
 Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplemental Data. Zhou et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. Confirmation of mutant mapping results. (A) Complementation assay with stably transformed genomic fragments (ComN-N) (2 kb upstream of TSS and 1.5 kb downstream of TES) and CaMV
Supplemental Figure 1. Confirmation of mutant mapping results. (A) Complementation assay with stably transformed genomic fragments (ComN-N) (2 kb upstream of TSS and 1.5 kb downstream of TES) and CaMV
Nature Genetics: doi: /ng Supplementary Figure 1. Liz/Zdbf2 dynamics in vivo and in vitro.
 Supplementary igure 1 Liz/Zdbf2 dynamics in vivo and in vitro. (a) Schematic of Liz/Zdbf2 regulation during early development. Summary from Duffié et al. 18. Liz transcript initiates from the unmethylated
Supplementary igure 1 Liz/Zdbf2 dynamics in vivo and in vitro. (a) Schematic of Liz/Zdbf2 regulation during early development. Summary from Duffié et al. 18. Liz transcript initiates from the unmethylated
Supplemental Data. Hachez et al. Plant Cell (2014) /tpc Suppl. Figure 1A
 Suppl. Figure 1A Suppl. Figure 1B Supplemental Figure 1: Results of the commercial screening of interactants using split ubiquitin technique. (A) Isolated preys (192) using the bait construct pbt3-n- as
Suppl. Figure 1A Suppl. Figure 1B Supplemental Figure 1: Results of the commercial screening of interactants using split ubiquitin technique. (A) Isolated preys (192) using the bait construct pbt3-n- as
To assess the localization of Citrine fusion proteins, we performed antibody staining to
 Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
Browser Exercises - I. Alignments and Comparative genomics
 Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
Browser Exercises - I Alignments and Comparative genomics 1. Navigating to the Genome Browser (GBrowse) Note: For this exercise use http://www.tritrypdb.org a. Navigate to the Genome Browser (GBrowse)
ChIP-seq data analysis with Chipster. Eija Korpelainen CSC IT Center for Science, Finland
 ChIP-seq data analysis with Chipster Eija Korpelainen CSC IT Center for Science, Finland chipster@csc.fi What will I learn? Short introduction to ChIP-seq Analyzing ChIP-seq data Central concepts Analysis
ChIP-seq data analysis with Chipster Eija Korpelainen CSC IT Center for Science, Finland chipster@csc.fi What will I learn? Short introduction to ChIP-seq Analyzing ChIP-seq data Central concepts Analysis
Supplemental Figure 1. Sanger sequence analysis of 27 FA families confirms
 SUPPLEMENTARY FIGURE LEGENDS Supplemental Figure 1. Sanger sequence analysis of 27 FA families confirms mutations. The family ID, gene mutated and the Sanger sequence traces showing each mutation are presented.
SUPPLEMENTARY FIGURE LEGENDS Supplemental Figure 1. Sanger sequence analysis of 27 FA families confirms mutations. The family ID, gene mutated and the Sanger sequence traces showing each mutation are presented.
Summary MicroRNAs (mirnas) are genomically encoded small RNAs used by organisms to regulate the expression of proteins generated from messenger RNA
 Summary MicroRNAs (mirnas) are genomically encoded small RNAs used by organisms to regulate the expression of proteins generated from messenger RNA transcripts. The in vivo requirement of specific mirnas
Summary MicroRNAs (mirnas) are genomically encoded small RNAs used by organisms to regulate the expression of proteins generated from messenger RNA transcripts. The in vivo requirement of specific mirnas
A Repressor Complex Governs the Integration of
 Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and
 Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and Tin TFBS motifs and located in the flanking or intronic
Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and Tin TFBS motifs and located in the flanking or intronic
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Nature Methods: doi: /nmeth.4396
 Supplementary Figure 1 Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC. (a) Heatmap-based representation of ATAC-seq quality control
Supplementary Figure 1 Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC. (a) Heatmap-based representation of ATAC-seq quality control
Gene expression analysis. Biosciences 741: Genomics Fall, 2013 Week 5. Gene expression analysis
 Gene expression analysis Biosciences 741: Genomics Fall, 2013 Week 5 Gene expression analysis From EST clusters to spotted cdna microarrays Long vs. short oligonucleotide microarrays vs. RT-PCR Methods
Gene expression analysis Biosciences 741: Genomics Fall, 2013 Week 5 Gene expression analysis From EST clusters to spotted cdna microarrays Long vs. short oligonucleotide microarrays vs. RT-PCR Methods
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
Sundari Chetty, Felicia Walton Pagliuca, Christian Honore, Anastasie Kweudjeu, Alireza Rezania, and Douglas A. Melton
 A simple tool to improve pluripotent stem cell differentiation Sundari Chetty, Felicia Walton Pagliuca, Christian Honore, Anastasie Kweudjeu, Alireza Rezania, and Douglas A. Melton Supplementary Information
A simple tool to improve pluripotent stem cell differentiation Sundari Chetty, Felicia Walton Pagliuca, Christian Honore, Anastasie Kweudjeu, Alireza Rezania, and Douglas A. Melton Supplementary Information
Supporting Information
 Supporting Information Kilian et al. 10.1073/pnas.1105861108 SI Materials and Methods Determination of the Electric Field Strength Required for Successful Electroporation. The transformation construct
Supporting Information Kilian et al. 10.1073/pnas.1105861108 SI Materials and Methods Determination of the Electric Field Strength Required for Successful Electroporation. The transformation construct
Understanding embryonic head development. ANAT2341 Tennille Sibbritt Embryology Unit Children s Medical Research Institute
 Understanding embryonic head development ANAT2341 Tennille Sibbritt Embryology Unit Children s Medical Research Institute Head malformations among the most common category of congenital malformations in
Understanding embryonic head development ANAT2341 Tennille Sibbritt Embryology Unit Children s Medical Research Institute Head malformations among the most common category of congenital malformations in
Supplementary Information. c d e
 Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
Introduction to genome biology
 Introduction to genome biology Lisa Stubbs We ve found most genes; but what about the rest of the genome? Genome size* 12 Mb 95 Mb 170 Mb 1500 Mb 2700 Mb 3200 Mb #coding genes ~7000 ~20000 ~14000 ~26000
Introduction to genome biology Lisa Stubbs We ve found most genes; but what about the rest of the genome? Genome size* 12 Mb 95 Mb 170 Mb 1500 Mb 2700 Mb 3200 Mb #coding genes ~7000 ~20000 ~14000 ~26000
Development of genetically modified live attenuated parasites as potential vaccines against visceral leishmaniasis
 Development of genetically modified live attenuated parasites as potential vaccines against visceral leishmaniasis Poonam Salotra National Institute of Pathology (ICMR) New Delhi Impact of Visceral Leishmaniasis
Development of genetically modified live attenuated parasites as potential vaccines against visceral leishmaniasis Poonam Salotra National Institute of Pathology (ICMR) New Delhi Impact of Visceral Leishmaniasis
Supplementary Information. Isl2b regulates anterior second heart field development in zebrafish
 Supplementary Information Isl2b regulates anterior second heart field development in zebrafish Hagen R. Witzel 1, Sirisha Cheedipudi 1, Rui Gao 1, Didier Y.R. Stainier 2 and Gergana D. Dobreva 1,3* 1 Origin
Supplementary Information Isl2b regulates anterior second heart field development in zebrafish Hagen R. Witzel 1, Sirisha Cheedipudi 1, Rui Gao 1, Didier Y.R. Stainier 2 and Gergana D. Dobreva 1,3* 1 Origin
RNAseq Applications in Genome Studies. Alexander Kanapin, PhD Wellcome Trust Centre for Human Genetics, University of Oxford
 RNAseq Applications in Genome Studies Alexander Kanapin, PhD Wellcome Trust Centre for Human Genetics, University of Oxford RNAseq Protocols Next generation sequencing protocol cdna, not RNA sequencing
RNAseq Applications in Genome Studies Alexander Kanapin, PhD Wellcome Trust Centre for Human Genetics, University of Oxford RNAseq Protocols Next generation sequencing protocol cdna, not RNA sequencing
GENOTYPING BY PCR PROTOCOL FORM MUTANT MOUSE REGIONAL RESOURCE CENTER North America, International
 Please provide the following information required for genetic analysis of your mutant mice. Please fill in form electronically by tabbing through the text fields. The first 2 pages are protected with gray
Please provide the following information required for genetic analysis of your mutant mice. Please fill in form electronically by tabbing through the text fields. The first 2 pages are protected with gray
SUPPLEMENTARY INFORMATION. Tolerance of a knotted near infrared fluorescent protein to random circular permutation
 SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Bio5488 Practice Midterm (2018) 1. Next-gen sequencing
 1. Next-gen sequencing 1. You have found a new strain of yeast that makes fantastic wine. You d like to sequence this strain to ascertain the differences from S. cerevisiae. To accurately call a base pair,
1. Next-gen sequencing 1. You have found a new strain of yeast that makes fantastic wine. You d like to sequence this strain to ascertain the differences from S. cerevisiae. To accurately call a base pair,
Simultaneous profiling of transcriptome and DNA methylome from a single cell
 Additional file 1: Supplementary materials Simultaneous profiling of transcriptome and DNA methylome from a single cell Youjin Hu 1, 2, Kevin Huang 1, 3, Qin An 1, Guizhen Du 1, Ganlu Hu 2, Jinfeng Xue
Additional file 1: Supplementary materials Simultaneous profiling of transcriptome and DNA methylome from a single cell Youjin Hu 1, 2, Kevin Huang 1, 3, Qin An 1, Guizhen Du 1, Ganlu Hu 2, Jinfeng Xue
(i) A trp1 mutant cell took up a plasmid containing the wild type TRP1 gene, which allowed that cell to multiply and form a colony
 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been isolated that cannot grow
1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been isolated that cannot grow
Supporting information
 Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna
 Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
GATCGTGCACGATCTCGGCAATTCGGGATGCCGGCTCGTCACCGGTCGCT
 Problem. (pts) A. (5pts) Your colleague professor Eugene Mathew Lateed generated a genome-wide DNA methylation map for normal colon cells using MRE-seq and MeDIP-seq. In an intergenic region, he found
Problem. (pts) A. (5pts) Your colleague professor Eugene Mathew Lateed generated a genome-wide DNA methylation map for normal colon cells using MRE-seq and MeDIP-seq. In an intergenic region, he found
Introduction to Microarray Analysis
 Introduction to Microarray Analysis Methods Course: Gene Expression Data Analysis -Day One Rainer Spang Microarrays Highly parallel measurement devices for gene expression levels 1. How does the microarray
Introduction to Microarray Analysis Methods Course: Gene Expression Data Analysis -Day One Rainer Spang Microarrays Highly parallel measurement devices for gene expression levels 1. How does the microarray
Nature Methods: doi: /nmeth Supplementary Figure 1. DMS-MaPseq data are highly reproducible at elevated DMS concentrations.
 Supplementary Figure 1 DMS-MaPseq data are highly reproducible at elevated DMS concentrations. a, Correlation of Gini index for 202 yeast mrna regions with 15x coverage at 2.5% or 5% v/v DMS concentrations
Supplementary Figure 1 DMS-MaPseq data are highly reproducible at elevated DMS concentrations. a, Correlation of Gini index for 202 yeast mrna regions with 15x coverage at 2.5% or 5% v/v DMS concentrations
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination.
 Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
% Viability. isw2 ino isw2 ino isw2 ino isw2 ino mM HU 4-NQO CPT
 a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino8 9 2.5.1.15 % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy)
a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino8 9 2.5.1.15 % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy)
Tet proteins influence the balance between neuroectodermal and mesodermal fate choice by inhibiting Wnt signaling
 Tet proteins influence the balance between neuroectodermal and mesodermal fate choice by inhibiting Wnt signaling Xiang Li a,b, Xiaojing Yue a, William A. Pastor a,1, Lizhu Lin c, Romain Georges a, Lukas
Tet proteins influence the balance between neuroectodermal and mesodermal fate choice by inhibiting Wnt signaling Xiang Li a,b, Xiaojing Yue a, William A. Pastor a,1, Lizhu Lin c, Romain Georges a, Lukas
V10: Core Pluripotency Network
 V10: Core Pluripotency Network Mouse ES cells were isolated for the first time in 1981 from mouse blastocysts. Maintenance of the self-renewing state of mouse ES cells requires the cytokine leukemia inhibitory
V10: Core Pluripotency Network Mouse ES cells were isolated for the first time in 1981 from mouse blastocysts. Maintenance of the self-renewing state of mouse ES cells requires the cytokine leukemia inhibitory
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Name Genotype Reference
 Supplemental Data Supplemental Table 1 S. cerevisiae strains. Name Genotype Reference RMY200 UKY403 W303-1a MATa ade2-101 (och) his3 200 lys2-801 (amp) trp1 901 ura3-52 hht1,hhf1::leu2 hht2,hhf2::his3
Supplemental Data Supplemental Table 1 S. cerevisiae strains. Name Genotype Reference RMY200 UKY403 W303-1a MATa ade2-101 (och) his3 200 lys2-801 (amp) trp1 901 ura3-52 hht1,hhf1::leu2 hht2,hhf2::his3
Introduction to Next Generation Sequencing
 The Sequencing Revolution Introduction to Next Generation Sequencing Dena Leshkowitz,WIS 1 st BIOmics Workshop High throughput Short Read Sequencing Technologies Highly parallel reactions (millions to
The Sequencing Revolution Introduction to Next Generation Sequencing Dena Leshkowitz,WIS 1 st BIOmics Workshop High throughput Short Read Sequencing Technologies Highly parallel reactions (millions to
Supplemental Data. Sethi et al. (2014). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
MODULE TSS1: TRANSCRIPTION START SITES INTRODUCTION (BASIC)
 MODULE TSS1: TRANSCRIPTION START SITES INTRODUCTION (BASIC) Lesson Plan: Title JAMIE SIDERS, MEG LAAKSO & WILSON LEUNG Identifying transcription start sites for Peaked promoters using chromatin landscape,
MODULE TSS1: TRANSCRIPTION START SITES INTRODUCTION (BASIC) Lesson Plan: Title JAMIE SIDERS, MEG LAAKSO & WILSON LEUNG Identifying transcription start sites for Peaked promoters using chromatin landscape,
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
Genomes: What we know and what we don t know
 Genomes: What we know and what we don t know Complete draft sequence 2001 October 15, 2007 Dr. Stefan Maas, BioS Lehigh U. What we know Raw genome data The range of genome sizes in the animal & plant kingdoms!
Genomes: What we know and what we don t know Complete draft sequence 2001 October 15, 2007 Dr. Stefan Maas, BioS Lehigh U. What we know Raw genome data The range of genome sizes in the animal & plant kingdoms!
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Figure S1. Unrearranged locus. Rearranged locus. Concordant read pairs. Region1. Region2. Cluster of discordant read pairs, bundle
 Figure S1 a Unrearranged locus Rearranged locus Concordant read pairs Region1 Concordant read pairs Cluster of discordant read pairs, bundle Region2 Concordant read pairs b Physical coverage 5 4 3 2 1
Figure S1 a Unrearranged locus Rearranged locus Concordant read pairs Region1 Concordant read pairs Cluster of discordant read pairs, bundle Region2 Concordant read pairs b Physical coverage 5 4 3 2 1
Nature Genetics: doi: /ng Supplementary Figure 1. High-confidence PRC2 targets and candidate PREs.
 Supplementary Figure 1 High-confidence PRC2 targets and candidate PREs. (a) Flowchart for identification of candidate Arabidopsis PREs. We identified 1504 genomic regions marked by at least 3 of the following:
Supplementary Figure 1 High-confidence PRC2 targets and candidate PREs. (a) Flowchart for identification of candidate Arabidopsis PREs. We identified 1504 genomic regions marked by at least 3 of the following:
