Supplementary Figures Montero et al._supplementary Figure 1
|
|
|
- Ethelbert McBride
- 6 years ago
- Views:
Transcription
1 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1
2 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres do not show TERRA features. (A) Confocal microscopy images of double RNA-FISH using probes targeting either TAR1, DDX11L or WASH transcripts (red) and TERRA-UUAGGG track (green). At least 50 nuclei were analyzed in all the cases. AS: probe complementary to the transcript arising from the positive DNA strand; S: probe complementary to the transcript from the negative DNA strand. Scale bar: 10 µm. (B) Confocal microscopy images from RNA-FISH with the different RNA-FISH probes in the absence or presence of RNase. Scale bar: 10 µm. (C) Northern blotting using 32 P-dCTP-labelled probes on total RNA from HCT116 cells DKO (for the DNMT1 and 3b genes) or HCT116 wild-type, HeLa and U2OS treated and untreated with 5 Azacytidin (Aza) for 72 hours. The RNA was used to prepare two identical membranes, in one of them the TERRA probe was hybridized first and, upon stripping, the WASH probe. In the other membrane, the DDX probe was hybridized first and then the TAR1 probe. 18S was included as a loading control. The expected size for DDX transcripts is kb and for WASH Kb. For TAR1 region, no annotated transcripts have been described so far. *Unspecific band due to cross-hybridization with rrna 18S and 28S. (D) The RNA extracted in (B) was also used for RNA dot-blot to detect either TERRA- UUAGGG track, DDX11L or WASH transcripts; 18S serves as loading control. All the hybridizations were performed on the same membrane one after the other upon stripping. (Graphs) Dot-blot quantification normalized by 18S (mean values±s.e.m., n=3 biological replicates). The Student s t-test was used for the statistical analysis of the comparison between untreated and Aza-treated cells. One-way Anova with Tukey post test was used for the statistical analysis of the comparison of one cell line with the others (*p < 0.05, **p < 0.01 and ***p < 0.001).
3 Montero et al_suppl. Info 3 Supplementary Figure 2. Different combinations of grnas are able to delete the 20q and Xp loci in different cell lines. IMR90 non-transformed human fibroblasts, HeLa, HCT166 and U2OS cell lines were transfected with different combinations of Cas9-GFP-plasmids containing two different grnas to delete 10Kb of the 20q and XpYp region enriched in TERRA transcripts. Two-days upon transfection total DNA was isolated from the different cells lines and PCR using specifics primers to detect the deletion was performed and visualized in ethidium bromide gels. The individual grnas used in each combination (C1, C2, C3 and C4) (grna mix) are shown below. The primers for the grnas synthesis can be found in the Supplementary Table 5. In both cases control PCRs (Ctrl I and Ctrl II) using DNA for non treated cells are shown. * DNA amplification found in the treated cells PCR but not in the control PCR.
4 Montero et al_suppl. Info 4 Montero et al._supplementary Figure 3
5 Montero et al_suppl. Info 5 Supplementary Figure 3. Regenotyping of false homozygous clones upon expansion. (A) Ethidium bromide gels showing the genotype for the CRISPR and wild-type allele in the different clones of the Hela, HCT116 and U2OS cells obtained upon the deletion of the 20q and Xp loci before and after expansion. The primers to detect the deletion were designed to obtain a PCR product of 219 bp for the 20q deletion and of 407bp for the Xp deletion but other sizes can be expected depending on how the CRISPR cut&repair was performed by the cell. The DNA marker legend is shown at the left bottom. Ethidium bromide gels with different expositions are due to the run of the different samples on different days. The white strips in the gels indicate the removal of irrelevant samples from that gel (B) Ethidium bromide gels showing the two different CRISPR alleles in the 20q-KO clones A2 and B4
6 Montero et al_suppl. Info 6 Supplementary Figure 4. Total TERRA levels upon deletion of the 20q locus. Quantification of TERRA signal obtained in the Northern blot from Fig. 3B normalized by 18S. Fold change is shown
7 Montero et al_suppl. Info 7 Montero et al._supplementary Figure 5
8 Montero et al_suppl. Info 8 Supplementary Figure 5. Off-target mutation analysis in the 20q-TERRA KO cells. (A) TIDE (Tracking of Indels by DEcomposition) analysis to study the four top of targets for the grnas (START2 and END1) used to generate the 20q locus deletion. The analysis was performed with DNA pools from either wild type cells or cells bearing the 20q deletion, named C3 (from which the clones A2, B4 and C4 were obtained from). A DNA that did not undergo CRISPR treatment was used as reference. (B) TIDE analysis for the off-targets 3, grna END1 using DNAs from the 20q-KO clones A2, B4 and C4 and from cells that did not undergo CRSIPR treatment. A DNA from other set of cells that did not undergo CRISPR treatment was used as reference (C) T7 endonuclease assay was performed on DNAs from the regions of interest (off-target2/start2, off-target 3/END1 and off-target 4/END1) obtained from pool of cells bearing the 20q deletion (C3) or from the 20q-KO clones. Two identical DNAs except for a C to G mutation in one of them (named C and G) were used as positive control. The ethidium bromide gels shows the different DNAs upon T7 endonuclease digestion.
9 Montero et al_suppl. Info 9 Montero et al._supplementary Figure 6
10 Montero et al_suppl. Info 10 Supplementary Figure 6 Detection of shelterins in 20q-TERRA KO cells. (A) Quantification of immunoprecipitated telomeric repeats upon TRF2- or TRF1-ChIP. Values were obtained after normalization to telomeric DNA input and corrected by the differences in the telomere length between the clones and wild-type cells (see Figure 4). Error bars correspond to the 2-3 independent experiments (mean values± s.e.m., n=2-3 independent experiments). One-way Anova with Tukey post test was used for statistical analysis (**p < 0.01). (B) Representative images of TRF2 and TRF1 immunoflorescence (green) in wild-type cells and 20q-KO TERRA cells (clone C4). Merge images with DAPI are also shown (Graphs). Quantification of the mean fluorescence intensity of TRF1 and TRF2 is represented (mean values± s.e.m., n=3 independent experiments). The Student s t-test was used for statistical analysis (*p < 0.05). Scale bar: 10 µm.
11 Montero et al_suppl. Info 11 Supplementary Figure 7. Comparison of 20q-TERRA transcripts with Xp-RNAs. Raw data from RNA-seq samples (IP-EV and IP-T) 1 was downloaded and aligned to the human subtelomeric reference genome. Next, transcripts were modeled by Cufflinks in the chr.20q and chr.xp subtelomeric regions of interest (corresponding to the regions to be deleted with CRISPR/Cas9) and pairwise alignments performed (see Materials and Methods). The snapshot shows (from top to bottom) modeled transcripts in the different IPs, RNA-seq reads (dense format) and the modeled one-exon transcript for each condition
12 Montero et al_suppl. Info 12 Supplementary Figure 8. Expression analysis of the 20q-TERRA transcripts with respect the Xp-RNAs. (A) Raw data from RNA-seq samples (IP-EV and IP-T) (Porro et al., 2014) was downloaded and aligned to the human subtelomeric reference genome. FPKM (fragments per kilobase of transcript per million fragments mapped) in the chr.20q and chr.xp subtelomeric regions of interest (corresponding to the regions to be deleted with CRISPR/Cas9) were calculated for each of the IPs and input. (Graph). Quantification of the FPKM in each IP normalized by their corresponding input. (B) Detection of the 20q-TERRA or Xp transcripts by PCR using different pair of primers in wild-type cells, 20q-KO clones (A2, B4, C4) and Xp-KO clone (D8). The fold RNA enrichment normalized by GAPDH is shown. Primers 20q/qPCR1-3 are designed within the 20q TERRA locus. Primer Xp/qPCR 1 and 2 are designed within the Xp locus but only the Xp/qPCR2 is designed within the Xp deletion performed in clone D8.
13 Montero et al_suppl. Info 13 Supplementary Figure 9. Raw gels and blots from Figure 2E and 3B. (A) Ethidium bromide gels showing the CRISPR allele for the deletion of the 20q and Xp loci detected by PCR in different clones of the U2OS cells. The white strips in the gels indicate the removal of irrelevant samples from that gel. X are irrelevant samples. (B) Northern blotting using 32P-dCTPlabelled probe against TERRA-UUAGGG tract in the U2OS cells WT or KO for the 20q or Xp loci. 18S was included as a loading control. *Unspecific band due to cross-hybridization with rrna 18S and 28S.
14 Montero et al_suppl. Info 14 Supplementary Tables Supplementary Table 1. Chromosome ends amplified by probes against TAR1, DDX and WASH Probe Amplified region TAR1 1q,2q,4q,10q,21q,22q DDX 1p,2,9p,12p,15q,16p,Xq,Yq WASH 1p,9p,12p,Xq,Yq Supplementary Table 2. TAR1 probe specificity recognizing different chromosome ends Max score Total score Query cove E value Ident Probe origin Chr1q % 0 100% 1q Chr2q % 0 100% 2q,4q Chr5q % 0 99% 2q,4q Chr6q % 0 99% 10q Chr9p % 1.00E % 10q Chr10q % 0 100% 10q Chr13q % 0 98% 10q Chr15q % 1.00E % 10q Chr16p % 2.00E % 10q Chr22q % 0 100% 22q ChrXq % 4.00E % 10q Supplementary Table 3. DDX probe specificity recognizing different chromosome ends Max score Total score Query cove E value Ident Probe origin Chr1p % 0 100% 1p Chr9p % 0 100% 9p,Xq,Yq Chr12p % 0 100% 12p Chr15q % 0 100% 15q Chr16p % 0 100% 16p Chr19p % 0 100% 1p ChrXq % 0 100% 9p,Xq,Yq Supplementary Table 4. WASH probe specificity recognizing different chromosome ends Max score Total score Query cove E value Ident Probe origin Chr1p % 0 100% 1p Chr9p % 0 100% 9p
15 Montero et al_suppl. Info 15 Chr12p % 0 100% 12p Chr15q % 0 99% 1p Chr16p % 0 98% 1p Chr19p % 0 99% 1p ChrXq % 0 100% Xq,Yq Supplementary Table 5. Primers for grna cloning Region Primer Sequence (5-3 ) Chr20q grna20q-start1 CACCgTATAGCGGCGGCACGCCGCC R-gRNA20q-start1 AAACGGCGGCGTGCCGCCGCTATAc grna20q-start2 CACCgCAGGCTGGCGCGACGTGCGG R-gRNA20q-start2 AAACCCGCACGTCGCGCCAGCCTGc grna20q-end1 CACCgCCCACCACCTTAGCGGATCA R-gRNA20q-end1 AAACTGATCCGCTAAGGTGGTGGGc grna20q-end2 CACCgAAATTTTACTTCATCGGAGG R-gRNA20q-end2 AAACCCTCCGATGAAGTAAAATTTc ChrXpYp grnaxpyp-start1 CACCgCATGGTGGGGACCCGATGCT R-gRNAXpYp-start1 AAACAGCATCGGGTCCCCACCATGc grnaxpyp-start2 CACCGCCACCATATCATACTCGGA R-gRNAXpYp-start2 AAACTCCGAGTATGATATGGTGGC grnaxpyp-end1 CACCgAACCGGGTGGGTACGTCAAC R-gRNAXpYp-end1 AAACGTTGACGTACCCACCCGGTTc grnaxpyp-end2 CACCGGTCTACACCGGCTTCATCC R-gRNAXpYp-end2 AAACGGATGAAGCCGGTGTAGACC Supplementary Table 6. grna combinations Combination#1: grna-start1+grna-end1 Combination #2: grna-start1+grna-end2 Combination #3: grna-start2+grna-end1 Combination #4: grna-start2+grna-end2 Supplementary Table 7. Primers used to amplify the KO alelle grna combination Primer Sequence (5-3 ) #1,#2,#3,#4 F-20qKOI CCCGGGTCTGTGTTAAGC #1,#3 R-20qKOI GATCAGGGCTGTCTTAATGC #2,#4 R-0qKOII TCATCACACCTGCATATACTGT #1,#2,#3,#4 F-pYpKO GGCAAAGGGAGCAGTCAT #1,#2,#3,#4 R-pYpKO GACGCAACTTCACAGTTACA
16 Montero et al_suppl. Info 16 Supplementary Table 8. Primers to detect transcripts from the 20q and Xp loci Region Primer Sequence (5-3 ) 20q F120q-qpcr CTGGTGCCAGAGTGGATT R120q-qpcr F220q-qpcr R220q-qpcr F320q-qpcr CACCTGTTCTCTTTGTCTGG ACATGGGCGATACTCAGG CCCACTACTGTGCCTCAA GAAGTTGCTGGGTTCTATGG R320q-qpcr ATGGTGCAGACACTGTGG Xp F1xp-qpcr GCAAAGAGTGAAAGAACGAAGCTT R1xp-qpcr CCCTCTGAAAGTGGACCTATCA F2xp-qpcr R2xp-qpcr CTTGAGCTCTCACCACTCAC GCCACGATAGCTTCTTCC Supplementary Table 9. Chromosomal rearrangements found by CGH array Chr Cytoband Start Stop Gain Loss Deletion pval chr3 p p , , , E-97 chr4 q24 - q , , , E-09 chr6 p p , , , ,900e-324 chr6 p p , , , E-80 chr6 p p , , , E-07 chr6 p p , , , E-42 chr6 q , , , E-43 chr7 p p , , , E-92 chr7 p , , , E-08 chr9 p , , , E-42 chr9 p p , , , ,900e-324 chr9 p p , , , E-11 chr9 p p ,181,035 0, , E-62 chr9 p p , , , E-29 chr10 p p , , ,917, E-27 chr10 q q , , , ,900e-324 chr10 q q , , , E-11 chr12 p p , , , E-61 chr12 p , , ,189, E-23 chr14 q q , , , ,900e-324 chr14 q q , , , E-17 chr15 q12 - q , , , E-09 chr15 q14 - q , , , ,900e-324 chr15 q14 - q , , , E-65 chr15 q23 - q , , , E-74 chr15 q23 - q , , , E-12 chr18 p p , , , ,900e-324
17 Montero et al_suppl. Info 17 chr18 p , , ,636, E-13 chr18 p , , , E-11 chr18 p , , , E-10 chr19 p , , , E-32 chr19 q11 - q , , , E-109 chr21 p , , , E-36 chr21 q q , , , ,900e-324 chr21 q , , , E-19 Supplementary Table 10. Percentage of identity between the different modeled transcripts in the 20q and Xp loci. IP-EV: IP-sh scramble; IP-T: IP-shTRF2 1 Chr.Xp transcripts Chr.20q transcripts IP-T Chr.Xp transcripts Chr.20q transcripts IP-EV Supplementary Table 11. Percentage of identity, size (bp) and presence of DNA repeats in the 20q-modeled transcripts with more than 50% homology with the Xp-modeled transcripts. IP-EV: IP-sh scramble; IP-T: IP-shTRF2 1 20q RNAs %Homology with Xp Transcript size Repeats IP-T % 72 bp LTR % 546 bp SINE/LINE % 167 bp -
18 Montero et al_suppl. Info 18 IP-EV % 90 bp SINE/LINE % 185 bp LTR % 219 bp LINE/LTR Supplementary Table 12. Percentage of Identity between the one-exon modeled transcript from the 20q and Xp loci. IP-EV: IP-sh scramble; IP-T: IP-shTRF2 1 20q-RNAs vs Xp-RNAs IP-EV 37.70% IP-T 39.20% Supplementary Table 13. Primers for RNA-FISH and Northern blot probes Region Primer Sequence (5-3 ) TAR1 F-ProbeTAR1 GCAGAGTTCTTCTCAGGTCA R-ProbeTAR1 TGTCATGTGTGCATTAGGAA F-T7ProbeTAR1 ccaagcttctaatacgactcactatagggagagcagagttcttctcaggtca R-T7ProbeTAR1 ccaagcttctaatacgactcactatagggagatgtcatgtgtgcattaggaa DDX11L F-ProbeDDX11L TGTGTGGAAGTTCACTCCTG R-ProbeDDX11L CAGGATGGAAGACAGATTGG F-T7ProbeDDX11L ccaagcttctaatacgactcactatagggagatgtgtggaagttcactcctg R-T7ProbeDDX11L ccaagcttctaatacgactcactatagggagacaggatggaagacagattgg WASH F-ProbeWASH GCCTCTTAAGAACACAGTGG R-ProbeWASH ATCTCTGGGAAAGGACCTG F-T7ProbeWASH ccaagcttctaatacgactcactatagggagagcctcttaagaacacagtgg R-T7ProbeWASH ccaagcttctaatacgactcactatagggagaatctctgggaaaggacctg Chr20q F-ProbeChr20q CATTGCTGGTGGAAGACAG R-ProbeChr20q GGACCCTCTGTAACAAATGAC F-T7ProbeChr20q ccaagcttctaatacgactcactatagggagacattgctggtggaagacag R-T7ProbeChr20q ccaagcttctaatacgactcactatagggagaggaccctctgtaacaaatgac ChrXpYp F-ProbeChrXpYp AACCTGCCGTATCTCACC R-ProbeChrXpYp ACACTCCTGTAGTCCCACCT F-T7ProbeChrXpYp ccaagcttctaatacgactcactatagggagaaacctgccgtatctcacc R-T7ProbeChrXpYp ccaagcttctaatacgactcactatagggagaacactcctgtagtcccacct Supplementary Table 14. Primers for promoter region cloning Region Primer Sequence (5-3 ) 20qPr1 F-0qPr1 CGCGGTACCAAGCCTAAGTGTGGCAAGTC R-0qPr1 CCCGCTCGAGCAAGGTGTGTGCTGAGAAAG 20qPr2 F-0qPr2 CGCGGTACCGGAATCATGAAGTATGTGACC
19 Montero et al_suppl. Info 19 R-0qPr2 CCCGCTCGAGGGCCTAGTATCCGGAATATACA 20qPr3 F-0qPr3 CGCGGTACCTGTGCGTGTTTCTGTGTC R-0qPr3 CCCGCTCGAGGTTCAAGGCCCTCCACAG 20qPr4 F-0qPr4 CGCGGTACCGTGGAGTCAGACCACACG R-0qPr4 CCCGCTCGAGTACCAGAAGCTGGAGAAAGG Supplementary Table 15. Primers for the amplification of DNA regions with predicted off-target mutations grna Primer Sequence (5-3 ) grna Start2 grna End1 F-gS2Off1 TACAAGGTCCGGAGAAGC R-gS2Off1 GAGCGGAGGAGGGAGGAC F-gS2Off2 TGAGTACCAAGGGTCACCTC R-gS2Off2 GGACGGCTGGTTCTCAGG F-gS2Off3 GGATGATCTGGGAGAAGC R-gS2Off3 GGGAATCTGCAGGACAAAC F-gS2Off4 GGGCAGGTAAGGACGTTT R-gS2Off4 GCTCCTCACTCCCAAATC F-gE1Off1 CTCTAGTTCATGCAGGATGC R-gE1Off1 GAGTGAGAAGAGAGCCTTGG F-gE1Off2 AATCCAGGAGGCAGAGGT R-gE1Off2 CAGGTCAGGGAGTCAACAAC F-gE1Off3 TAGGTCACAGAGCAGTAGGG R-gE1Off3 GTCCCTAAGGCATTTAGCA F-gE1Off4 TTTCCCAACTATTTGCTGAT R-gE1Off4 GTTCTGCACCTTATAAACCAG Supplementary References 1. Porro, A. et al. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun 5, 5379 (2014).
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
hnrnp D/AUF1 Rabbit IgG hnrnp M
 Mouse IgG Goat IgG Rabbit IgG Mouse IgG hnrnp F Goat IgG Mouse IgG Kb 6 4 3 2 15 5 Supplementary Figure S1. In vivo binding of TERRA-bound RBPs to target RNAs. Immunoprecipitation (IP) assay using 3 mg
Mouse IgG Goat IgG Rabbit IgG Mouse IgG hnrnp F Goat IgG Mouse IgG Kb 6 4 3 2 15 5 Supplementary Figure S1. In vivo binding of TERRA-bound RBPs to target RNAs. Immunoprecipitation (IP) assay using 3 mg
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplemental Figure 1 A
 Supplemental Figure A prebleach postbleach 2 min 6 min 3 min mh2a.-gfp mh2a.2-gfp mh2a2-gfp GFP-H2A..9 Relative Intensity.8.7.6.5 mh2a. GFP n=8.4 mh2a.2 GFP n=4.3 mh2a2 GFP n=2.2 GFP H2A n=24. GFP n=7.
Supplemental Figure A prebleach postbleach 2 min 6 min 3 min mh2a.-gfp mh2a.2-gfp mh2a2-gfp GFP-H2A..9 Relative Intensity.8.7.6.5 mh2a. GFP n=8.4 mh2a.2 GFP n=4.3 mh2a2 GFP n=2.2 GFP H2A n=24. GFP n=7.
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Supplementary Information
 Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
CRISPR RNA-guided activation of endogenous human genes
 CRISPR RNA-guided activation of endogenous human genes Morgan L Maeder, Samantha J Linder, Vincent M Cascio, Yanfang Fu, Quan H Ho, J Keith Joung Supplementary Figure 1 Comparison of VEGF activation induced
CRISPR RNA-guided activation of endogenous human genes Morgan L Maeder, Samantha J Linder, Vincent M Cascio, Yanfang Fu, Quan H Ho, J Keith Joung Supplementary Figure 1 Comparison of VEGF activation induced
Isolation of single-base genome-edited human ips cells without
 Nature Methods Isolation of single-base genome-edited human ips cells without antibiotic selection Yuichiro Miyaoka, Amanda H. Chan, Luke M. Judge, Jennie Yoo, Miller Huang, Trieu D. Nguyen, Paweena P.
Nature Methods Isolation of single-base genome-edited human ips cells without antibiotic selection Yuichiro Miyaoka, Amanda H. Chan, Luke M. Judge, Jennie Yoo, Miller Huang, Trieu D. Nguyen, Paweena P.
Nature Biotechnology: doi: /nbt Supplementary Figure 1. In vitro validation of OTC sgrnas and donor template.
 Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Allele-specific locus binding and genome editing by CRISPR at the
 Supplementary Information Allele-specific locus binding and genome editing by CRISPR at the p6ink4a locus Toshitsugu Fujita, Miyuki Yuno, and Hodaka Fujii Supplementary Figure Legends Supplementary Figure
Supplementary Information Allele-specific locus binding and genome editing by CRISPR at the p6ink4a locus Toshitsugu Fujita, Miyuki Yuno, and Hodaka Fujii Supplementary Figure Legends Supplementary Figure
CFTR-null wt CFTR-null 1.0. Probe: Neo R. Figure S1
 A. B. 4.0 3.0 2.0 1.0 4.0 3.0 2.0 1.0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Probe: Neo R CFTR-null wt CFTR-null Figure S1 A. 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 10kb 8kb CFTR-null wt B. Probe: CFTR
A. B. 4.0 3.0 2.0 1.0 4.0 3.0 2.0 1.0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Probe: Neo R CFTR-null wt CFTR-null Figure S1 A. 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 10kb 8kb CFTR-null wt B. Probe: CFTR
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1. Validation of CDK9-inhibitor treatment.
 Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Nature Genetics: doi: /ng Supplementary Figure 1. ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets.
 Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplemental Data. Zhou et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. Confirmation of mutant mapping results. (A) Complementation assay with stably transformed genomic fragments (ComN-N) (2 kb upstream of TSS and 1.5 kb downstream of TES) and CaMV
Supplemental Figure 1. Confirmation of mutant mapping results. (A) Complementation assay with stably transformed genomic fragments (ComN-N) (2 kb upstream of TSS and 1.5 kb downstream of TES) and CaMV
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
BIO 202 Midterm Exam Winter 2007
 BIO 202 Midterm Exam Winter 2007 Mario Chevrette Lectures 10-14 : Question 1 (1 point) Which of the following statements is incorrect. a) In contrast to prokaryotic DNA, eukaryotic DNA contains many repetitive
BIO 202 Midterm Exam Winter 2007 Mario Chevrette Lectures 10-14 : Question 1 (1 point) Which of the following statements is incorrect. a) In contrast to prokaryotic DNA, eukaryotic DNA contains many repetitive
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Zhang et al., RepID facilitates replication Initiation. Supplemental Information:
 Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nucleaseinduced
 Supplementary Data Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nucleaseinduced mutations Suresh Ramakrishna 1, Seung Woo Cho 2, Sojung Kim 2, Myungjae Song 1, Ramu Gopalappa
Supplementary Data Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nucleaseinduced mutations Suresh Ramakrishna 1, Seung Woo Cho 2, Sojung Kim 2, Myungjae Song 1, Ramu Gopalappa
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Protocols for cell lines using CRISPR/CAS
 Protocols for cell lines using CRISPR/CAS Procedure overview Map Preparation of CRISPR/CAS plasmids Expression vectors for guide RNA (U6-gRNA) and Cas9 gene (CMV-p-Cas9) are ampicillin-resist ant and stable
Protocols for cell lines using CRISPR/CAS Procedure overview Map Preparation of CRISPR/CAS plasmids Expression vectors for guide RNA (U6-gRNA) and Cas9 gene (CMV-p-Cas9) are ampicillin-resist ant and stable
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
SUPPLEMENT MATERIALS FOR CURTIN,
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 SUPPLEMENT MATERIALS FOR CURTIN, et al. Validating genome-wide association candidates: Selecting, testing, and characterizing
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 SUPPLEMENT MATERIALS FOR CURTIN, et al. Validating genome-wide association candidates: Selecting, testing, and characterizing
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Supplemental Figure legends Figure S1. (A) (B) (C) (D) Figure S2. Figure S3. (A-E) Figure S4. Figure S5. (A, C, E, G, I) (B, D, F, H, Figure S6.
 Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter
 CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter SUPPLEMENTARY DATA See Supplementary Sequence File: 1 Supplementary Figure S1: The total protein was extracted from
CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter SUPPLEMENTARY DATA See Supplementary Sequence File: 1 Supplementary Figure S1: The total protein was extracted from
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for
 Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
BIOLOGY - CLUTCH CH.20 - BIOTECHNOLOGY.
 !! www.clutchprep.com CONCEPT: DNA CLONING DNA cloning is a technique that inserts a foreign gene into a living host to replicate the gene and produce gene products. Transformation the process by which
!! www.clutchprep.com CONCEPT: DNA CLONING DNA cloning is a technique that inserts a foreign gene into a living host to replicate the gene and produce gene products. Transformation the process by which
GENETICS EXAM 3 FALL a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size.
 Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Bootcamp: Molecular Biology Techniques and Interpretation
 Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09861 & &' -(' ()*+ ')(+,,(','-*+,&,,+ ',+' ' 23,45/0*6787*9:./09 ;78?4?@*+A786?B- &' )*+*(,-* -(' ()*+ ')(+,,(','-*+,&,,+ ',+'./)*+*(,-*..)*+*(,-*./)*+*(,-*.0)*+*(,-*..)*+*(,-*
doi:10.1038/nature09861 & &' -(' ()*+ ')(+,,(','-*+,&,,+ ',+' ' 23,45/0*6787*9:./09 ;78?4?@*+A786?B- &' )*+*(,-* -(' ()*+ ')(+,,(','-*+,&,,+ ',+'./)*+*(,-*..)*+*(,-*./)*+*(,-*.0)*+*(,-*..)*+*(,-*
GENETICS - CLUTCH CH.15 GENOMES AND GENOMICS.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF GENOMICS Genomics is the study of genomes in their entirety Bioinformatics is the analysis of the information content of genomes - Genes, regulatory sequences,
!! www.clutchprep.com CONCEPT: OVERVIEW OF GENOMICS Genomics is the study of genomes in their entirety Bioinformatics is the analysis of the information content of genomes - Genes, regulatory sequences,
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
 Xi et al. Genome Biology (2015) 16:231 DOI 10.1186/s13059-015-0791-1 RESEARCH A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
Xi et al. Genome Biology (2015) 16:231 DOI 10.1186/s13059-015-0791-1 RESEARCH A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Negative selection analysis of sgrnas targeting all Brd4 exons comparing day 2 to day 10 time points. Systematic evaluation of 64 Brd4 sgrnas in negative selection experiments, targeting
Supplementary Figure 1 Negative selection analysis of sgrnas targeting all Brd4 exons comparing day 2 to day 10 time points. Systematic evaluation of 64 Brd4 sgrnas in negative selection experiments, targeting
of NOBOX. Supplemental Figure S1F shows the staining profile of LHX8 antibody (Abcam, ab41519), which is a
 SUPPLEMENTAL MATERIALS AND METHODS LHX8 Staining During the course of this work, it came to our attention that the NOBOX antibody used in this study had been discontinued. There are a number of other nuclear
SUPPLEMENTAL MATERIALS AND METHODS LHX8 Staining During the course of this work, it came to our attention that the NOBOX antibody used in this study had been discontinued. There are a number of other nuclear
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
SUPPLEMENTARY INFORMATION doi:10.1038/nature09937 a Name Position Primersets 1a 1b 2 3 4 b2 Phenotype Genotype b Primerset 1a D T C R I E 10000 8000 6000 5000 4000 3000 2500 2000 1500 1000 800 Donor (D)
6/256 1/256 0/256 1/256 2/256 7/256 10/256. At3g06290 (SAC3B)
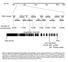 Chr.III M 5M 1M 15M 2M 23M BAC clones F22F7 F1A16 F24F17 F24P17 T8E24 F17A9 F21O3 F17A17 F18C1 F2O1 F28L1 F5E6 F3E22 T1B9 MLP3 Number of recombinants 6/256 1/256 /256 1/256 2/256 7/256 1/256 At3g629 (SAC3B)
Chr.III M 5M 1M 15M 2M 23M BAC clones F22F7 F1A16 F24F17 F24P17 T8E24 F17A9 F21O3 F17A17 F18C1 F2O1 F28L1 F5E6 F3E22 T1B9 MLP3 Number of recombinants 6/256 1/256 /256 1/256 2/256 7/256 1/256 At3g629 (SAC3B)
Fatchiyah
 Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
SUPPLEMENTARY INFORMATION
 Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers Wataru Sakai, Elizabeth M. Swisher, Beth Y. Karlan, Mukesh K. Agarwal, Jake Higgins, Cynthia Friedman, Emily Villegas,
Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers Wataru Sakai, Elizabeth M. Swisher, Beth Y. Karlan, Mukesh K. Agarwal, Jake Higgins, Cynthia Friedman, Emily Villegas,
Chapter 6 - Molecular Genetic Techniques
 Chapter 6 - Molecular Genetic Techniques Two objects of molecular & genetic technologies For analysis For generation Molecular genetic technologies! For analysis DNA gel electrophoresis Southern blotting
Chapter 6 - Molecular Genetic Techniques Two objects of molecular & genetic technologies For analysis For generation Molecular genetic technologies! For analysis DNA gel electrophoresis Southern blotting
Before starting, write your name on the top of each page Make sure you have all pages
 Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Protocol for Genome Editing via the RNA-guided Cas9 Nuclease in. Zebrafish Embryos 1
 Protocol for Genome Editing via the RNA-guided Cas9 Nuclease in Zebrafish Embryos 1 1. In vitro synthesis of capped Cas9 mrna The full length of humanized Cas9 cdnas with double NLS were cloned into pxt7
Protocol for Genome Editing via the RNA-guided Cas9 Nuclease in Zebrafish Embryos 1 1. In vitro synthesis of capped Cas9 mrna The full length of humanized Cas9 cdnas with double NLS were cloned into pxt7
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM Part I: Definitions. Homology: Reverse transcriptase. Allostery: cdna library
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
Supplementary Information for: CRISPR/Cas9-based target validation. for p53-reactivating model compounds
 for: CRISPR/Cas9-based target validation for p53-reactivating model compounds Michael Wanzel 1,5, Jonas B. Vischedyk 1, Miriam P. Gittler 1, Niklas Gremke 1, Julia R. Seiz 1, Mirjam Hefter 1, Magdalena
for: CRISPR/Cas9-based target validation for p53-reactivating model compounds Michael Wanzel 1,5, Jonas B. Vischedyk 1, Miriam P. Gittler 1, Niklas Gremke 1, Julia R. Seiz 1, Mirjam Hefter 1, Magdalena
Multiple choice questions (numbers in brackets indicate the number of correct answers)
 1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
- 1 - Supplemental Data
 - 1-1 Supplemental Data 2 3 4 5 6 7 8 9 Supplemental Figure S1. Differential expression of AtPIP Genes in DC3000-inoculated plants. Gene expression in leaves was analyzed by real-time RT-PCR and expression
- 1-1 Supplemental Data 2 3 4 5 6 7 8 9 Supplemental Figure S1. Differential expression of AtPIP Genes in DC3000-inoculated plants. Gene expression in leaves was analyzed by real-time RT-PCR and expression
Nature Immunology: doi: /ni Supplementary Figure 1. Control experiments for Figure 1.
 Supplementary Figure 1 Control experiments for Figure 1. (a) Localization of SETX in untreated or PR8ΔNS1 virus infected A549 cells (4hours). Nuclear (DAPI) and Tubulin staining are shown. SETX antibody
Supplementary Figure 1 Control experiments for Figure 1. (a) Localization of SETX in untreated or PR8ΔNS1 virus infected A549 cells (4hours). Nuclear (DAPI) and Tubulin staining are shown. SETX antibody
Phenotype analysis: biological-biochemical analysis. Genotype analysis: molecular and physical analysis
 1 Genetic Analysis Phenotype analysis: biological-biochemical analysis Behaviour under specific environmental conditions Behaviour of specific genetic configurations Behaviour of progeny in crosses - Genotype
1 Genetic Analysis Phenotype analysis: biological-biochemical analysis Behaviour under specific environmental conditions Behaviour of specific genetic configurations Behaviour of progeny in crosses - Genotype
Genomic DNA fragments identified by ChIP-chip assay for MR [28] corresponding to two
![Genomic DNA fragments identified by ChIP-chip assay for MR [28] corresponding to two Genomic DNA fragments identified by ChIP-chip assay for MR [28] corresponding to two](/thumbs/89/98549522.jpg) Supplemental Materials and Methods Genomic DNA fragments identified by ChIP-chip assay for MR [28] corresponding to two upstream regions of the human Klf9 gene (-5139 to -5771 bp and -3875 to -4211 bp)
Supplemental Materials and Methods Genomic DNA fragments identified by ChIP-chip assay for MR [28] corresponding to two upstream regions of the human Klf9 gene (-5139 to -5771 bp and -3875 to -4211 bp)
Supplementary Figure 1. CRISPR/Cas9-induced targeted mutations in TaGASR7, TaDEP1, TaNAC2, TaPIN1, TaLOX2 and TaGW2 genes in wheat protoplasts.
 Supplementary Figure 1. CRISPR/Cas9-induced targeted mutations in TaGASR7, TaDEP1, TaNAC2, TaPIN1, TaLOX2 and TaGW2 genes in wheat protoplasts. Lanes 1 and 2: digested CRISPR/Cas9-transformed protoplasts;
Supplementary Figure 1. CRISPR/Cas9-induced targeted mutations in TaGASR7, TaDEP1, TaNAC2, TaPIN1, TaLOX2 and TaGW2 genes in wheat protoplasts. Lanes 1 and 2: digested CRISPR/Cas9-transformed protoplasts;
Supplementary Figures and Figure legends
 Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Analyses of ECTRs by C-circle and T-circle assays.
 Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
The Biotechnology Toolbox
 Chapter 15 The Biotechnology Toolbox Cutting and Pasting DNA Cutting DNA Restriction endonuclease or restriction enzymes Cellular protection mechanism for infected foreign DNA Recognition and cutting specific
Chapter 15 The Biotechnology Toolbox Cutting and Pasting DNA Cutting DNA Restriction endonuclease or restriction enzymes Cellular protection mechanism for infected foreign DNA Recognition and cutting specific
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Genome-wide genetic screening with chemically-mutagenized haploid embryonic stem cells
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 Supplementary Information Genome-wide genetic screening with chemically-mutagenized haploid embryonic stem cells Josep V. Forment 1,2, Mareike Herzog
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 Supplementary Information Genome-wide genetic screening with chemically-mutagenized haploid embryonic stem cells Josep V. Forment 1,2, Mareike Herzog
Inhibition of HSV-1 Replication by Gene Editing Strategy. Running Title: Targeting HSV-1 Infection by CRISPR/Cas9
 SUPPLEMENTARY MATERIALS 2/4/16 Inhibition of HSV-1 Replication by Gene Editing Strategy Running Title: Targeting HSV-1 Infection by CRISPR/Cas9 Pamela C. Roehm 1,2,3*, Masoud Shekarabi 1, Hassen Wollebo
SUPPLEMENTARY MATERIALS 2/4/16 Inhibition of HSV-1 Replication by Gene Editing Strategy Running Title: Targeting HSV-1 Infection by CRISPR/Cas9 Pamela C. Roehm 1,2,3*, Masoud Shekarabi 1, Hassen Wollebo
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Design and sequence of system 1 for targeted demethylation.
 Supplementary Figure 1 Design and sequence of system 1 for targeted demethylation. (a) Design of system 1 for targeted demethylation. TET1CD was fused to a catalytic inactive Cas9 nuclease (dcas9) and
Supplementary Figure 1 Design and sequence of system 1 for targeted demethylation. (a) Design of system 1 for targeted demethylation. TET1CD was fused to a catalytic inactive Cas9 nuclease (dcas9) and
Marker types. Potato Association of America Frederiction August 9, Allen Van Deynze
 Marker types Potato Association of America Frederiction August 9, 2009 Allen Van Deynze Use of DNA Markers in Breeding Germplasm Analysis Fingerprinting of germplasm Arrangement of diversity (clustering,
Marker types Potato Association of America Frederiction August 9, 2009 Allen Van Deynze Use of DNA Markers in Breeding Germplasm Analysis Fingerprinting of germplasm Arrangement of diversity (clustering,
Protocol CRISPR Genome Editing In Cell Lines
 Protocol CRISPR Genome Editing In Cell Lines Protocol 2: HDR donor plasmid applications (gene knockout, gene mutagenesis, gene tagging, Safe Harbor ORF knock-in) Notes: 1. sgrna validation: GeneCopoeia
Protocol CRISPR Genome Editing In Cell Lines Protocol 2: HDR donor plasmid applications (gene knockout, gene mutagenesis, gene tagging, Safe Harbor ORF knock-in) Notes: 1. sgrna validation: GeneCopoeia
CELL BIOLOGY - CLUTCH CH TECHNIQUES IN CELL BIOLOGY.
 !! www.clutchprep.com CONCEPT: LIGHT MICROSCOPE A light microscope uses light waves and magnification to view specimens Can be used to visualize transparent, and some cellular components - Can visualize
!! www.clutchprep.com CONCEPT: LIGHT MICROSCOPE A light microscope uses light waves and magnification to view specimens Can be used to visualize transparent, and some cellular components - Can visualize
Puro. Knockout Detection (KOD) Kit
 Puro Knockout Detection (KOD) Kit Cat. No. CC-03 18 Oct. 2016 Contents I. Kit Contents and Storage II. Product Overview III. Methods Experimental Outline Genomic DNA Preparation Obtain Hybrid DNA Digest
Puro Knockout Detection (KOD) Kit Cat. No. CC-03 18 Oct. 2016 Contents I. Kit Contents and Storage II. Product Overview III. Methods Experimental Outline Genomic DNA Preparation Obtain Hybrid DNA Digest
Bi 8 Lecture 4. Ellen Rothenberg 14 January Reading: from Alberts Ch. 8
 Bi 8 Lecture 4 DNA approaches: How we know what we know Ellen Rothenberg 14 January 2016 Reading: from Alberts Ch. 8 Central concept: DNA or RNA polymer length as an identifying feature RNA has intrinsically
Bi 8 Lecture 4 DNA approaches: How we know what we know Ellen Rothenberg 14 January 2016 Reading: from Alberts Ch. 8 Central concept: DNA or RNA polymer length as an identifying feature RNA has intrinsically
Supplementary Materials
 Supplementary Materials Supplementary Methods Supplementary Discussion Supplementary Figure 1 Calculated frequencies of embryo cells bearing bi-allelic alterations. Targeted indel mutations induced by
Supplementary Materials Supplementary Methods Supplementary Discussion Supplementary Figure 1 Calculated frequencies of embryo cells bearing bi-allelic alterations. Targeted indel mutations induced by
Cell line CDH1 mrna AS-paRNA S-paRNA
 a b Cell line mrna AS-paRNA S-paRNA HCT116 P53 +/+ 39.47 + + HCT116 P53 -/- 35.36 + + LNCAP 1562.4 + - MCF7 C1 1864.76 + - MCF7 C2 1812.81 + - Supplementary Figure 1. Promoter-associated transcripts at
a b Cell line mrna AS-paRNA S-paRNA HCT116 P53 +/+ 39.47 + + HCT116 P53 -/- 35.36 + + LNCAP 1562.4 + - MCF7 C1 1864.76 + - MCF7 C2 1812.81 + - Supplementary Figure 1. Promoter-associated transcripts at
Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm
 Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm Anja Smith Director R&D Dharmacon, part of GE Healthcare Imagination at work crrna:tracrrna program Cas9 nuclease Active crrna is
Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm Anja Smith Director R&D Dharmacon, part of GE Healthcare Imagination at work crrna:tracrrna program Cas9 nuclease Active crrna is
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Restriction Enzymes (endonucleases)
 In order to understand and eventually manipulate DNA (human or otherwise) an array of DNA technologies have been developed. Here are some of the tools: Restriction Enzymes (endonucleases) In order to manipulate
In order to understand and eventually manipulate DNA (human or otherwise) an array of DNA technologies have been developed. Here are some of the tools: Restriction Enzymes (endonucleases) In order to manipulate
Simple protocol for gene editing using GenCrisprTM Cas9 nuclease
 Simple protocol for gene editing using GenCrisprTM Cas9 nuclease Contents Protocol Step 1: Choose the target DNA sequence Step 2: Design sgrna Step 3: Preparation for sgrna 3.1 In vitro transcription of
Simple protocol for gene editing using GenCrisprTM Cas9 nuclease Contents Protocol Step 1: Choose the target DNA sequence Step 2: Design sgrna Step 3: Preparation for sgrna 3.1 In vitro transcription of
Supplementary Information. Optimization of the production of knock-in alleles by CRISPR/Cas9 microinjection into the mouse zygote
 Supplementary Information Supplementary Figures S1 to S5 and Tables S1 to S3. Optimization of the production of knock-in alleles by CRISPR/Cas9 microinjection into the mouse zygote Aurélien Raveux, Sandrine
Supplementary Information Supplementary Figures S1 to S5 and Tables S1 to S3. Optimization of the production of knock-in alleles by CRISPR/Cas9 microinjection into the mouse zygote Aurélien Raveux, Sandrine
1. Introduction Gene regulation Genomics and genome analyses
 1. Introduction Gene regulation Genomics and genome analyses 2. Gene regulation tools and methods Regulatory sequences and motif discovery TF binding sites Databases 3. Technologies Microarrays Deep sequencing
1. Introduction Gene regulation Genomics and genome analyses 2. Gene regulation tools and methods Regulatory sequences and motif discovery TF binding sites Databases 3. Technologies Microarrays Deep sequencing
Applications and Uses. (adapted from Roche RealTime PCR Application Manual)
 What Can You Do With qpcr? Applications and Uses (adapted from Roche RealTime PCR Application Manual) What is qpcr? Real time PCR also known as quantitative PCR (qpcr) measures PCR amplification as it
What Can You Do With qpcr? Applications and Uses (adapted from Roche RealTime PCR Application Manual) What is qpcr? Real time PCR also known as quantitative PCR (qpcr) measures PCR amplification as it
GT-rich promoters can drive RNA pol II transcription and deposition of H2A.Z in African trypanosomes
 GT-rich promoters can drive RNA pol II transcription deposition of H2A.Z in African trypanosomes Carolin Wedel, Konrad U. Förstner, Ramona Derr T. Nicolai Siegel Appendix Table of Contents Appendix Materials
GT-rich promoters can drive RNA pol II transcription deposition of H2A.Z in African trypanosomes Carolin Wedel, Konrad U. Förstner, Ramona Derr T. Nicolai Siegel Appendix Table of Contents Appendix Materials
Chapter 5. Structural Genomics
 Chapter 5. Structural Genomics Contents 5. Structural Genomics 5.1. DNA Sequencing Strategies 5.1.1. Map-based Strategies 5.1.2. Whole Genome Shotgun Sequencing 5.2. Genome Annotation 5.2.1. Using Bioinformatic
Chapter 5. Structural Genomics Contents 5. Structural Genomics 5.1. DNA Sequencing Strategies 5.1.1. Map-based Strategies 5.1.2. Whole Genome Shotgun Sequencing 5.2. Genome Annotation 5.2.1. Using Bioinformatic
Using mutants to clone genes
 Using mutants to clone genes Objectives: 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype
Using mutants to clone genes Objectives: 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype
Motivation From Protein to Gene
 MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
02 Agenda Item 03 Agenda Item
 01 Agenda Item 02 Agenda Item 03 Agenda Item SOLiD 3 System: Applications Overview April 12th, 2010 Jennifer Stover Field Application Specialist - SOLiD Applications Workflow for SOLiD Application Application
01 Agenda Item 02 Agenda Item 03 Agenda Item SOLiD 3 System: Applications Overview April 12th, 2010 Jennifer Stover Field Application Specialist - SOLiD Applications Workflow for SOLiD Application Application
CRISPR/Cas9 Gene Editing Tools
 CRISPR/Cas9 Gene Editing Tools - Guide-it Products for Successful CRISPR/Cas9 Gene Editing - Why choose Guide-it products? Optimized methods designed for speed and ease of use Complete kits that don t
CRISPR/Cas9 Gene Editing Tools - Guide-it Products for Successful CRISPR/Cas9 Gene Editing - Why choose Guide-it products? Optimized methods designed for speed and ease of use Complete kits that don t
Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric
 Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric common sequences. (a) Schematic illustration of the telomere-proximal site of subtelomeres. The restriction map is based
Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric common sequences. (a) Schematic illustration of the telomere-proximal site of subtelomeres. The restriction map is based
