Mock. Control. sirna1
|
|
|
- William Pierce
- 5 years ago
- Views:
Transcription
1 siha CaSki Mock Control sirna1 Supplementary Data Figure 1: Photomicrographs of siha and CaSki cells taken 48 hours after mock transfection, transfection of control sirna or sirna1. (100X magnification). Reduced cell numbers and apoptotic p morphology seen after sirna1 transfection in siha and CaSki cells (HPV-16 positive).
2 Control sirna treated cells sirna1 treated cells sub bg1: % sub G1: 22.8 % Supplementary Data Figure 2: Propidium iodide staining and flow cytometry of control sirna treated (left) and sirna1 treated (right) cells,48 hours after sirna1 transfection. There is an almost two fold increase in the percentage of sub-g1 cells with a corresponding decrease in the percentage of cells in the G2-M phase.
3 Control sirna sirna1 Fold change p p21 GADD45α PUMA β-actin Supplementary Data Figure 3: Western blot of p53 and other p53 responsive proteins in siha cells, 72 hours after sirna1 transfection. There was a significant increase in the levels of all the p53 responsive proteins. (p<0.001 in all cases). Beta-actin was used as a loading control.
4 Supplementary Data Figure 4: Methylation specific-high resolution melting (MS-HRM) of bisulfite treated siha DNA 72 hours after transfection with control sirna or sirna1. Untreated siha DNA (green line) having 1-2 copies of HPV-16 with the enhancer fully unmethylated (0/2 copies methylation) is taken as 0 % methylated control. Untreated CaSki DNA (pink line) having 599/600 copies of the enhancer methylated (99.8 %) is taken as the 100 % methylated control. The melting profiles of control sirna treated (blue line) and sirna1 treated (red line) enhancer amplicons were very similar to that of untreated cells (0% methylated). This proves that there was no DNA methylation in the targeted region after transfection.
5 a) b) Restriction enzyme accessibility assay ratio Unig gested/digested Control sirna sirna1 En nrichment at enh hancer Dimethyl H3K9 ChIP assay Control sirna sirna1 Supplementary Data Figure 5: sirna1 causes a closed chromatin state. a) sirna1 transfection in siha causes a significant 4 fold decrease in BsrGI restriction enzyme accessibility (p < 0.001) at the HPV-16 enhancer. b) sirna1 induces methylation of histone tails as seen by a 3.5 fold enrichment (p < 0.001)at the targeted enhancer using the dimethylated H3K9 ChIP assay.
6 Supplementary Table 1: List of sirnas used in the study sirna ID sirna1 S sirna1 AS sirna(+1) S sirna (+1) AS sirna(-1) S sirna (-1) AS sirna(-2) S sirna (-2) AS sirna (-5) S sirna (-5) AS T7 promoter Sequence (5' to 3') UGCGCCAACGCCUUACAUACCUU GGUAUGUAAGGCGUUGGCGCAUU GCGCCAACGCCUUACAUACCGUU CGGUAUGUAAGGCGUUGGCGCUU AUGCGCCAACGCCUUACAUACUU GUAUGUAAGGCGUUGGCGCAUUU UAUGCGCCAACGCCUUACAUAUU UAUGUAAGGCGUUGGCGCAUAUU CACUAUGCGCCAACGCCUUACUU GUAAGGCGUUGGCGCAUAGUGUU TAATACGACTCACTATAG
7 Supplementary Table 2: List of primers used Primers used for Real Time RT-PCR PrimerID Sequence(5'to3') 18S Forward GTAACCCGTTGAACCCCATT 18S Reverse CCATCCAATCGGTAGTAGCG Actin Forward TCATGAAGTGTGACGTTGACATCCGT Actin Reverse CCTAGAAGCATTTGCGGTGCACGATG POLR2A Forward CATCAAGAGAGTCCAGTTCGG POLR2A Reverse CCCTCAGTCGTCTCTGGGTA PPIA Forward CACCGTGTTCTTCGACATTG PPIA Reverse TTCTGCTGTCTTTGGGACCT B2M Forward TAGCTGTGCTCGCGCTACT B2M Reverse TCTCTGCTGGATGACGTGAG Tubulin Forward TAACCATGAGGGAAATCGTG Tubulin Reverse TCGATGCCATGTTCATCACT GAPDH Forward CCAAGGTCATCCATGACAACTTTGGT GAPDH Reverse TGTTGAAGTCAGAGGAGACCACCTG HPV-16 E6 Forward AGCGACCCAGAAAGTTACCA HPV-16 E6 Reverse GCATAAATCCCGAAAAGCAA HPV-16 E7 Forward ACAAGCAGAACCGGACAGAG HPV-16 E7 Reverse GCCCATTAACAGGTCTTCCA p21 Forward GCATGACAGATTTCTACCACTCC p21 Reverse GCCAGGGTATGTACATGAGGA NOXA Forward GAGATGCCTGGGAAGAAGG NOXA Reverse TTTCTGCCGGAAGTTCAGTT PUMA Forward GACCTCAACGCACAGTACGA PUMA Reverse CACCTAATTGGGCTCCATCT CyclinA2Forward ACCTGGACCCAGAAAACCAT CyclinA2Reverse CACTCACTGGCTTTTCATCTTC Cyclin E Forward AGA AAT GGC CAA AAT CGA CA Cyclin E Reverse CCCGGTCATCATCTTCTTTG
8 Supplementary Table 2 (Contd.) : List of primers used Primers used for Real Time RT-PCR(contd.) PrimerID Fas Forward Fas Reverse OAS1 Forward OAS1 Reverse HPV-18 E6 Forward HPV-18 E6 Reverse HPV-18 E7 Forward HPV-18 E7 Reverse Sequence(5'to3') GCATCTGGACCCTCCTACCT TCCTCAATTCCAATCCCTTG TTCTCCACCTGCTTCACAGA GAGCTCCAGGGCATACTGAG CATGCTGCATGCCATAAATG TGTGTTTCTCTGCGTCGTTG ACCTAAGGCAACATTGCAAG ACAAAGGACAGGGTGTTCAG Primers used for Bisulfite sequencing Bisulfite Outer Forward Bisulfite Outer Reverse Bisulfite Inner Forward(M13 tagged) Bisulfite Inner Reverse(M13 tagged) Primers used for ChIP Chr16ChIP Forward Chr16ChIP Reverse HPV-16 LCR Forward HPV-16 LCR Reverse TAGTTTTATGTTAGTAATTATGGTT ACAACTCTATACATAACTATAATA AGCGGATAACAATTTCACACAGGATGTTTTTTTGATTTGTATTGTTTG CGCCAGGGTTTTCCCAGTCACGACCCTTAAAAATTTAAACCTTATACC GTCTCTTTCTTGTTTTTAAGCTGGG TGAGCTCATTGAGACATTTGG CCAAATCCCTGTTTTCCTGA CGTTGGCGCATAGTGATTTA The HPV-16 LCR Forward and Reverse primers were used for ChIP, Restriction Enzyme Accessibility Assay and quantitation of enhancer associated transcripts
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas.
 Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Primers used for PCR of conductin, SGK1 and GAPDH have been described in (Dehner et al,
 Supplementary METHODS Flow Cytometry (FACS) For FACS analysis, trypsinized cells were fixed in ethanol, rehydrated in PBS and treated with 40μg/ml propidium iodide and 10μ/ml RNase for 30 min at room temperature.
Supplementary METHODS Flow Cytometry (FACS) For FACS analysis, trypsinized cells were fixed in ethanol, rehydrated in PBS and treated with 40μg/ml propidium iodide and 10μ/ml RNase for 30 min at room temperature.
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
SUPPLEMENTAL MATERIAL. Carvajal et al. Supplemental Table 1. List of quantitative PCR primers used for cdna analyses and
 UPPLEMEAL MATERIAL Carvajal et al. upplemental Table 1. List of quantitative PCR primers used for cdna analyses and chromatin immunoprecipitation assays. Figure 1. DNA damage-induced transcriptional repression
UPPLEMEAL MATERIAL Carvajal et al. upplemental Table 1. List of quantitative PCR primers used for cdna analyses and chromatin immunoprecipitation assays. Figure 1. DNA damage-induced transcriptional repression
Supplementary Table S1
 Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Suppl. Table S1. Characteristics of DHS regions analyzed by bisulfite sequencing. No. CpGs analyzed in the amplicon. Genomic location specificity
 Suppl. Table S1. Characteristics of DHS regions analyzed by bisulfite sequencing. DHS/GRE Genomic location Tissue specificity DHS type CpG density (per 100 bp) No. CpGs analyzed in the amplicon CpG within
Suppl. Table S1. Characteristics of DHS regions analyzed by bisulfite sequencing. DHS/GRE Genomic location Tissue specificity DHS type CpG density (per 100 bp) No. CpGs analyzed in the amplicon CpG within
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector.
 Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Supplementary Figure 1. GST pull-down analysis of the interaction of GST-cIAP1 (A, B), GSTcIAP1
 Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot
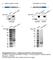 Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot (vector: ~25 kda). (b) Silver staining was used to assess
Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot (vector: ~25 kda). (b) Silver staining was used to assess
Supplemental Figure 1.
 Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. EndoC-βH1 cells were either mock-tranfected (control) or transfected with PolyI:C and analyzed 24 hours later. (A, B) Heatmap from global transcriptomic analysis
SUPPLEMENTARY FIGURES Supplementary Figure 1. EndoC-βH1 cells were either mock-tranfected (control) or transfected with PolyI:C and analyzed 24 hours later. (A, B) Heatmap from global transcriptomic analysis
Supplementary Figure S1
 Supplementary Figure S1 Supplementary Figure S1. CD11b expression on different B cell subsets in anti-snrnp Ig Tg mice. (a) Highly purified follicular (FO) and marginal zone (MZ) B cells were sorted from
Supplementary Figure S1 Supplementary Figure S1. CD11b expression on different B cell subsets in anti-snrnp Ig Tg mice. (a) Highly purified follicular (FO) and marginal zone (MZ) B cells were sorted from
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Supplementary Information
 Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in
 Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or
Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or
Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
Add 5µl of 3N NaOH to DNA sample (final concentration 0.3N NaOH).
 Bisulfite Treatment of DNA Dilute DNA sample to 2µg DNA in 50µl ddh 2 O. Add 5µl of 3N NaOH to DNA sample (final concentration 0.3N NaOH). Incubate in a 37ºC water bath for 30 minutes. To 55µl samples
Bisulfite Treatment of DNA Dilute DNA sample to 2µg DNA in 50µl ddh 2 O. Add 5µl of 3N NaOH to DNA sample (final concentration 0.3N NaOH). Incubate in a 37ºC water bath for 30 minutes. To 55µl samples
b alternative classical none
 Supplementary Figure. 1: Related to Figure.1 a d e b alternative classical none NIK P-IkBa Total IkBa Tubulin P52 (Lighter) P52 (Darker) RelB (Lighter) RelB (Darker) HDAC1 Control-Sh RelB-Sh NF-kB2-Sh
Supplementary Figure. 1: Related to Figure.1 a d e b alternative classical none NIK P-IkBa Total IkBa Tubulin P52 (Lighter) P52 (Darker) RelB (Lighter) RelB (Darker) HDAC1 Control-Sh RelB-Sh NF-kB2-Sh
Supplementary Figure 1A A404 Cells +/- Retinoic Acid
 Supplementary Figure 1A A44 Cells +/- Retinoic Acid 1 1 H3 Lys4 di-methylation SM-actin VEC cfos (-) RA (+) RA 14 1 1 8 6 4 H3 Lys79 di-methylation SM-actin VEC cfos (-) RA (+) RA Supplementary Figure
Supplementary Figure 1A A44 Cells +/- Retinoic Acid 1 1 H3 Lys4 di-methylation SM-actin VEC cfos (-) RA (+) RA 14 1 1 8 6 4 H3 Lys79 di-methylation SM-actin VEC cfos (-) RA (+) RA Supplementary Figure
CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter
 CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter SUPPLEMENTARY DATA See Supplementary Sequence File: 1 Supplementary Figure S1: The total protein was extracted from
CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter SUPPLEMENTARY DATA See Supplementary Sequence File: 1 Supplementary Figure S1: The total protein was extracted from
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Post-translational modification
 Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
Protein expression Western blotting, is a widely used and accepted technique to detect levels of protein expression in a cell or tissue extract. This technique measures protein levels in a biological sample
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1. Validation of CDK9-inhibitor treatment.
 Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Correction: The Leukemia-Associated Mllt10/ Af10-Dot1l Are Tcf4/β-Catenin Coactivators Essential for Intestinal Homeostasis
 CORRECTION Correction: The Leukemia-Associated Mllt10/ Af10-Dot1l Are Tcf4/β-Catenin Coactivators Essential for Intestinal Homeostasis Tokameh Mahmoudi, Sylvia F. Boj, Pantelis Hatzis, Vivian S. W. Li,
CORRECTION Correction: The Leukemia-Associated Mllt10/ Af10-Dot1l Are Tcf4/β-Catenin Coactivators Essential for Intestinal Homeostasis Tokameh Mahmoudi, Sylvia F. Boj, Pantelis Hatzis, Vivian S. W. Li,
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation
 1 2 3 4 5 SUPPLEMENTAL DATA Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation Magalí Nazar, Juan Pablo Nicola, María Laura
1 2 3 4 5 SUPPLEMENTAL DATA Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation Magalí Nazar, Juan Pablo Nicola, María Laura
Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera
 SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplementary Figure 1
 Supplementary Figure 1 Virus infection induces RNF128 expression. (a,b) RT-PCR analysis of Rnf128 (RNF128) mrna expression in mouse peritoneal macrophages (a) and THP-1 cells (b) upon stimulation with
Supplementary Figure 1 Virus infection induces RNF128 expression. (a,b) RT-PCR analysis of Rnf128 (RNF128) mrna expression in mouse peritoneal macrophages (a) and THP-1 cells (b) upon stimulation with
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
Supplementary Figure 1 A
 Supplementary Figure A B M. mullata p53, 3 UTR Luciferase activity (%) mir-5b 8 Le et al. Supplementary Information NC-DP - + - - - - - NC-DP - - + - - - - NC-DP3 - - - + - - - 5b-DP - - - - + + + NC-AS
Supplementary Figure A B M. mullata p53, 3 UTR Luciferase activity (%) mir-5b 8 Le et al. Supplementary Information NC-DP - + - - - - - NC-DP - - + - - - - NC-DP3 - - - + - - - 5b-DP - - - - + + + NC-AS
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends. Figure-S1 Molecular basis of ASS1 deficiency in myxofibrosarcoma: (A)
 Supplementary Figure Legends Figure-S1 Molecular basis of ASS1 deficiency in myxofibrosarcoma: (A) High-resolution oligonucleotide-based array comparative genomic hybridization shows normal DNA copy number
Supplementary Figure Legends Figure-S1 Molecular basis of ASS1 deficiency in myxofibrosarcoma: (A) High-resolution oligonucleotide-based array comparative genomic hybridization shows normal DNA copy number
SUPPLEMENTARY INFORMATION
 Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1
 Supplementary Information p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1 Bei Wang 1, Dandan Niu 1, Liyun Lai 1, & Ee Chee Ren 1,2 1 Singapore Immunology
Supplementary Information p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1 Bei Wang 1, Dandan Niu 1, Liyun Lai 1, & Ee Chee Ren 1,2 1 Singapore Immunology
Supplemental Material
 Supplemental Material Figure S1.(A) Immuno-staining of freshly isolated Myf5-Cre:ROSA-YFP fiber (B) Satellite cell enumeration in WT and cko mice from resting hind limb muscles. Bulk hind limb muscles
Supplemental Material Figure S1.(A) Immuno-staining of freshly isolated Myf5-Cre:ROSA-YFP fiber (B) Satellite cell enumeration in WT and cko mice from resting hind limb muscles. Bulk hind limb muscles
Nature Biotechnology doi: /nbt Supplementary Figure 1. Substrate-linked protein evolution components.
 Supplementary Figure 1 Substrate-linked protein evolution components. (a) loxbtr subsites used for the combinational directed-evolution process to generate Brec1. Sequence differences (compared to the
Supplementary Figure 1 Substrate-linked protein evolution components. (a) loxbtr subsites used for the combinational directed-evolution process to generate Brec1. Sequence differences (compared to the
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
PrimePCR Assay Validation Report
 Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID Glyceraldehyde-3-phosphate dehydrogenase Gapdh Rat This gene encodes a member of
Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID Glyceraldehyde-3-phosphate dehydrogenase Gapdh Rat This gene encodes a member of
gacgacgaggagaccaccgctttg aggcacattgaaggtctcaaacatg
 Supplementary information Supplementary table 1: primers for cloning and sequencing cloning for E- Ras ggg aat tcc ctt gag ctg ctg ggg aat ggc ttt gcc ggt cta gag tat aaa gga agc ttt gaa tcc Tpbp Oct3/4
Supplementary information Supplementary table 1: primers for cloning and sequencing cloning for E- Ras ggg aat tcc ctt gag ctg ctg ggg aat ggc ttt gcc ggt cta gag tat aaa gga agc ttt gaa tcc Tpbp Oct3/4
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
hnrnp C promotes APP translation by competing with FMRP for APP mrna recruitment to P bodies
 hnrnp C promotes APP translation by competing with for APP mrna recruitment to P bodies Eun Kyung Lee 1, Hyeon Ho Kim 1, Yuki Kuwano 1, Kotb Abdelmohsen 1, Subramanya Srikantan 1, Sarah S. Subaran 2, Marc
hnrnp C promotes APP translation by competing with for APP mrna recruitment to P bodies Eun Kyung Lee 1, Hyeon Ho Kim 1, Yuki Kuwano 1, Kotb Abdelmohsen 1, Subramanya Srikantan 1, Sarah S. Subaran 2, Marc
Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by. Regulating Occludin
 Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by Regulating Occludin Chaithanya Chelakkot 1,ǂ, Jaewang Ghim 2,3,ǂ, Nirmal Rajasekaran 4, Jong-Sun Choi 5, Jung-Hwan
Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by Regulating Occludin Chaithanya Chelakkot 1,ǂ, Jaewang Ghim 2,3,ǂ, Nirmal Rajasekaran 4, Jong-Sun Choi 5, Jung-Hwan
Inhibition of HSV-1 Replication by Gene Editing Strategy. Running Title: Targeting HSV-1 Infection by CRISPR/Cas9
 SUPPLEMENTARY MATERIALS 2/4/16 Inhibition of HSV-1 Replication by Gene Editing Strategy Running Title: Targeting HSV-1 Infection by CRISPR/Cas9 Pamela C. Roehm 1,2,3*, Masoud Shekarabi 1, Hassen Wollebo
SUPPLEMENTARY MATERIALS 2/4/16 Inhibition of HSV-1 Replication by Gene Editing Strategy Running Title: Targeting HSV-1 Infection by CRISPR/Cas9 Pamela C. Roehm 1,2,3*, Masoud Shekarabi 1, Hassen Wollebo
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3164 Supplementary Figure 1 Validation of effective Gnas deletion and epithelial thickness. a, Representative genotyping in mice treated or not with tamoxifen to show Gnas deletion. To
DOI: 10.1038/ncb3164 Supplementary Figure 1 Validation of effective Gnas deletion and epithelial thickness. a, Representative genotyping in mice treated or not with tamoxifen to show Gnas deletion. To
Electrophoretic Mobility Shift Assay (EMSA). Nuclear extracts were. oligonucleotide spanning the NF-kB site (5 -GATCC-
 SUPPLEMENTARY MATERIALS AND METHODS Electrophoretic Mobility Shift Assay (EMSA). Nuclear extracts were prepared as previously described. (1) A [ 32 P] datp-labeled doublestranded oligonucleotide spanning
SUPPLEMENTARY MATERIALS AND METHODS Electrophoretic Mobility Shift Assay (EMSA). Nuclear extracts were prepared as previously described. (1) A [ 32 P] datp-labeled doublestranded oligonucleotide spanning
Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death
 SUPPLEMENTAL DATA Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death Huajun Jin 1, Arthi Kanthasamy 1, Vellareddy Anantharam
SUPPLEMENTAL DATA Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death Huajun Jin 1, Arthi Kanthasamy 1, Vellareddy Anantharam
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Information
 Supplementary Information Supplementary Fig. 1 Morphology of NP69 and CNE1 cells. Phase contrast images of normal nasopharyngeal epithelial NP69 cells (left) and the well differentiated NPC CNE1 cells
Supplementary Information Supplementary Fig. 1 Morphology of NP69 and CNE1 cells. Phase contrast images of normal nasopharyngeal epithelial NP69 cells (left) and the well differentiated NPC CNE1 cells
HCT116 SW48 Nutlin: p53
 Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
Figure S1 Proteasome inhibition leads to formation of aggregates in human cells and tissues. (a)
 SUPPLEMENTARY MATERIAL Figure S1 Proteasome inhibition leads to formation of aggregates in human cells and tissues. (a) Flow cytometry. Cells were treated with MG132 for the indicated times. Cells were
SUPPLEMENTARY MATERIAL Figure S1 Proteasome inhibition leads to formation of aggregates in human cells and tissues. (a) Flow cytometry. Cells were treated with MG132 for the indicated times. Cells were
Supplementary Figure 2
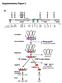 Supplementary Figure 2 a SBS-C1 SBS-C2 SBS-C3 SBS-C4 SBS-C5 SBS-C6 SBS-C7 SBS-C8 SBS-C9 LCR CNS-1 CNS-2 Il5 Rad50 Il13 Il4 Kif3a Sept8 0 50 100 150 200kb Sau3AI 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17
Supplementary Figure 2 a SBS-C1 SBS-C2 SBS-C3 SBS-C4 SBS-C5 SBS-C6 SBS-C7 SBS-C8 SBS-C9 LCR CNS-1 CNS-2 Il5 Rad50 Il13 Il4 Kif3a Sept8 0 50 100 150 200kb Sau3AI 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17
Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative
 Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative RT-PCR For CD8-1 transcript, forward primer CD8-1F 5 -TAGTAACCAGAGGCCGCAAGA-3 reverse primer CD8-1R 5 -TCTACTAAGGTGTCCCATAGCATGAT-3
Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative RT-PCR For CD8-1 transcript, forward primer CD8-1F 5 -TAGTAACCAGAGGCCGCAAGA-3 reverse primer CD8-1R 5 -TCTACTAAGGTGTCCCATAGCATGAT-3
Supplement 1: Sequences of Capture Probes. Capture probes were /5AmMC6/CTG TAG GTG CGG GTG GAC GTA GTC
 Supplementary Appendixes Supplement 1: Sequences of Capture Probes. Capture probes were /5AmMC6/CTG TAG GTG CGG GTG GAC GTA GTC ACG TAG CTC CGG CTG GA-3 for vimentin, /5AmMC6/TCC CTC GCG CGT GGC TTC CGC
Supplementary Appendixes Supplement 1: Sequences of Capture Probes. Capture probes were /5AmMC6/CTG TAG GTG CGG GTG GAC GTA GTC ACG TAG CTC CGG CTG GA-3 for vimentin, /5AmMC6/TCC CTC GCG CGT GGC TTC CGC
H3K36me3 polyclonal antibody
 H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
H3K36me3 polyclonal antibody Cat. No. C15410192 Type: Polyclonal ChIP-grade/ChIP-seq grade Source: Rabbit Lot #: A1845P Size: 50 µg/32 µl Concentration: 1.6 μg/μl Specificity: Human, mouse, Arabidopsis,
B Western blot of the p150caf-1 complex
 Loyola et al., Supplementary Figure 1 A MS data of the HP1α complex RIF1 SPT6 SMARCA4 POGZ CAF-1 p150 KAP-1 HP1α 200 116 97.1 CAF-1 p150 tnasp Importin4 B Western blot of the p150caf-1 complex Input mock
Loyola et al., Supplementary Figure 1 A MS data of the HP1α complex RIF1 SPT6 SMARCA4 POGZ CAF-1 p150 KAP-1 HP1α 200 116 97.1 CAF-1 p150 tnasp Importin4 B Western blot of the p150caf-1 complex Input mock
Eli Hefner, Ph.D. Sr. Scientist Gene Expression Division
 Multi-Discipline Analysis of Gene Silencing: Complementary use of Multiplex RT-qPCR, 2-D electrophoresis and Western blotting for RNAi based pathway analysis. Eli Hefner, Ph.D. Sr. Scientist Gene Expression
Multi-Discipline Analysis of Gene Silencing: Complementary use of Multiplex RT-qPCR, 2-D electrophoresis and Western blotting for RNAi based pathway analysis. Eli Hefner, Ph.D. Sr. Scientist Gene Expression
Supplementary Figure 1: MYCER protein expressed from the transgene can enhance
 Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1 Characterization of GSCs. a. Immunostaining of primary GSC spheres from GSC lines. Nestin (neural progenitor marker, red), TLX (green). Merged images of nestin,
Supplementary Figures Supplementary Fig. 1 Characterization of GSCs. a. Immunostaining of primary GSC spheres from GSC lines. Nestin (neural progenitor marker, red), TLX (green). Merged images of nestin,
Supplementary Figure 1. Gating strategy for flow cytometry analysis of mouse aorta. Cell suspensions from mouse aorta digested with enzyme cocktail
 Supplementary Figure 1. Gating strategy for flow cytometry analysis of mouse aorta. Cell suspensions from mouse aorta digested with enzyme cocktail were stained with propidium iodide (PI), anti-cd45 (FITC),
Supplementary Figure 1. Gating strategy for flow cytometry analysis of mouse aorta. Cell suspensions from mouse aorta digested with enzyme cocktail were stained with propidium iodide (PI), anti-cd45 (FITC),
Control of Eukaryotic Genes. AP Biology
 Control of Eukaryotic Genes The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different, specialized functions? Evolution
Control of Eukaryotic Genes The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different, specialized functions? Evolution
Chromatin immunoprecipitation (ChIP) ES and FACS-sorted GFP+/Flk1+ cells were fixed in 1% formaldehyde and sonicated until fragments of an average
 Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted
Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted
Supplementary information; Mungamuri et al., 2006
 Supplementary information; Mungamuri et al., 6 Antibodies used for western blotting: The following antibodies were used for western blotting: antiser473 Akt (#4), antiakt (#97), antiser9 Gsk 3b (#9336),
Supplementary information; Mungamuri et al., 6 Antibodies used for western blotting: The following antibodies were used for western blotting: antiser473 Akt (#4), antiakt (#97), antiser9 Gsk 3b (#9336),
PrimePCR Assay Validation Report
 Gene Information Gene Name laminin, beta 3 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID LAMB3 Human The product encoded by this gene is a laminin that
Gene Information Gene Name laminin, beta 3 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID LAMB3 Human The product encoded by this gene is a laminin that
Hypoxia-induced carbonic anhydrase IX facilitates lactate flux in human breast cancer cells by non-catalytic function
 Hypoxia-induced carbonic anhydrase IX facilitates lactate flux in human breast cancer cells by non-catalytic function Somayeh Jamali 1, Michael Klier 1,2, Samantha Ames 1,2, L. Felipe Barros 3, Robert
Hypoxia-induced carbonic anhydrase IX facilitates lactate flux in human breast cancer cells by non-catalytic function Somayeh Jamali 1, Michael Klier 1,2, Samantha Ames 1,2, L. Felipe Barros 3, Robert
SUPPLEMENTARY INFORMATION
 doi:.38/nature08267 Figure S: Karyotype analysis of ips cell line One hundred individual chromosomal spreads were counted for each of the ips samples. Shown here is an spread at passage 5, predominantly
doi:.38/nature08267 Figure S: Karyotype analysis of ips cell line One hundred individual chromosomal spreads were counted for each of the ips samples. Shown here is an spread at passage 5, predominantly
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in
 Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary information for: Ten-Eleven Translocation-2 (Tet2) Is Involved in Myogenic Differentiation of Skeletal Myoblast Cells in Vitro Xia Zhong*, Qian-Qian Wang*, Jian-Wei Li, Yu-Mei Zhang, Xiao-Rong
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Fig S1 Nutlin-3a treatment does not affect cell cycle progression in the absence
 Supplementary Figure Legends Supplementary Fig S1 Nutlin-3a treatment does not affect cell cycle progression in the absence of p53 or p21. HCT116 cells which were null for either p53 (A) or p21 (B) were
Supplementary Figure Legends Supplementary Fig S1 Nutlin-3a treatment does not affect cell cycle progression in the absence of p53 or p21. HCT116 cells which were null for either p53 (A) or p21 (B) were
(A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: WT; lower
 Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figure 1 Characterization of sirna-onv stability. (a) Fluorescence recovery curves of SQ-siRNA-ONV and SQ-ds-siRNA in 1 TAMg buffer
 Supplementary Figure 1 Characterization of sirna-onv stability. (a) Fluorescence recovery curves of SQ-siRNA-ONV and SQ-ds-siRNA in 1 TAMg buffer containing 10% serum The data error bars indicate means
Supplementary Figure 1 Characterization of sirna-onv stability. (a) Fluorescence recovery curves of SQ-siRNA-ONV and SQ-ds-siRNA in 1 TAMg buffer containing 10% serum The data error bars indicate means
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification.
 Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Table 1. Primers, annealing temperatures, and product sizes for PCR amplification. Gene Direction Primer sequence (5 3 ) Annealing Temperature Size (bp) BRCA1 Forward TTGCGGGAGGAAAATGGGTAGTTA 50 o C 292
Supplementary Information: Materials and Methods. Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered
 Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Supplementary Information
 Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
Supplementary Information MED18 interaction with distinct transcription factors regulates plant immunity, flowering time and responses to hormones Supplementary Figure 1. Diagram showing T-DNA insertion
SUPPLEMENTARY FIG. S9. Morphology and DNA staining of cipsc-differentiated trophoblast cell-like cell. Scale bar: 100 mm. SUPPLEMENTARY FIG. S10.
 Supplementary Data SUPPLEMENTARY FIG. S1. Lentivirus-infected canine fibroblasts show YFP expression. (A, B) Uninfected canine skin fibroblasts (CSFs); (C, D) infected CSFs; (E, F) uninfected CTFs; (G,
Supplementary Data SUPPLEMENTARY FIG. S1. Lentivirus-infected canine fibroblasts show YFP expression. (A, B) Uninfected canine skin fibroblasts (CSFs); (C, D) infected CSFs; (E, F) uninfected CTFs; (G,
EPIGENTEK. EpiQuik Methylated DNA Immunoprecipitation Kit. Base Catalog # P-2019 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Methylated DNA Immunoprecipitation Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik MeDIP Kit can be used for immunoprecipitating the methylated DNA from a broad range
EpiQuik Methylated DNA Immunoprecipitation Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik MeDIP Kit can be used for immunoprecipitating the methylated DNA from a broad range
Supplemental Table/Figure Legends
 MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/4/198/ra74/dc1 Supplementary Materials for Short RNA Duplexes Elicit RIG-I Mediated Apoptosis in a Cell Type and Length-Dependent Manner Osamu Ishibashi, Md. Moksed
www.sciencesignaling.org/cgi/content/full/4/198/ra74/dc1 Supplementary Materials for Short RNA Duplexes Elicit RIG-I Mediated Apoptosis in a Cell Type and Length-Dependent Manner Osamu Ishibashi, Md. Moksed
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
A RRM1 H2AX DAPI. RRM1 H2AX DAPI Merge. Cont. sirna RRM1
 A H2AX DAPI H2AX DAPI Merge Cont sirna Figure S1: Accumulation of RRM1 at DNA damage sites (A) HeLa cells were subjected to in situ detergent extraction without IR irradiation, and immunostained with the
A H2AX DAPI H2AX DAPI Merge Cont sirna Figure S1: Accumulation of RRM1 at DNA damage sites (A) HeLa cells were subjected to in situ detergent extraction without IR irradiation, and immunostained with the
Apoptosis assay: Apoptotic cells were identified by Annexin V-Alexa Fluor 488 and Propidium
 Apoptosis assay: Apoptotic cells were identified by Annexin V-Alexa Fluor 488 and Propidium Iodide (Invitrogen, Carlsbad, CA) staining. Briefly, 2x10 5 cells were washed once in cold PBS and resuspended
Apoptosis assay: Apoptotic cells were identified by Annexin V-Alexa Fluor 488 and Propidium Iodide (Invitrogen, Carlsbad, CA) staining. Briefly, 2x10 5 cells were washed once in cold PBS and resuspended
Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like
 Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like morphology in co-culture with mouse embryonic feeder fibroblasts
Supplementary Figure 1. Characterization of hipscs derived from primary human fibroblasts. a,b. Morphology of hipscs. hipscs exhibit hesc-like morphology in co-culture with mouse embryonic feeder fibroblasts
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Table S1. Primer sequences
 Table S1. Primer sequences Primers for quantitative PCR Tgf 1 Forward Tgf 1 Reverse Tgf 2Forward Tgf 2Reverse Tgf 3 Forward Tgf 3 Reverse Tgf r1 Forward Tgf r1 Reverse Tgf r2 Forward Tgf r2 Reverse Thbs1
Table S1. Primer sequences Primers for quantitative PCR Tgf 1 Forward Tgf 1 Reverse Tgf 2Forward Tgf 2Reverse Tgf 3 Forward Tgf 3 Reverse Tgf r1 Forward Tgf r1 Reverse Tgf r2 Forward Tgf r2 Reverse Thbs1
Nature Genetics: doi: /ng.3556 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE. Supplementary Figure 1
 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
