Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1.
|
|
|
- Mervin Terry
- 6 years ago
- Views:
Transcription
1 Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according to transcriptome analyses. (b) 3 and 5 RACE for full length of lnc-β-catm. The length of lnc-β-catm was 2281 nucleotides verified by sequencing. Black arrowhead denotes the full length of lnc-β-catm. (c) Histogram of lnc-β-catm coding potential analyzed by CPC (left panel), CPAT (middle panel) and PhyloCSF (right panel). HOX transcript antisense RNA (Hotair), X inactivation-specific transcript (XIST) and lnctcf7 serve as control non-coding RNAs. β-actin (ACTB) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) serve as control coding genes. For CPC and phylocsf, scores above 0 indicates coding potential, whereas scores below 0 represent no coding potential. CPAT scores indicate the possibility of coding. (d) Anti-Myc and anti-β-actin Western blots. Samples were shown in left panels. Full length of lnc-β-catm was cloned into an eukaryotic expression vector pcdna4-his-myc B with transcription initiating codon ATG in three expression patterns. T, thymine. (e) Scatter plots (means±s.d.) of nomorlized lnc-β-catm expression levels. ehcc, early HCC; ahcc, advanced HCC. (f, g) Histogram of lnc-β-catm expression levels in liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) (f), or in oncospheres and non-spheres (g). Error bars, s.d. (n = 4 cell cultures). Two tailed Student s t-test was used for statistical analysis, **, P < 0.01; ***, P< (h) Western blots (upper panel) and realtime PCR (lower panel) of Nucleocytoplasmic separation fractions. Error bars, s.d. (n = 4 cell cultures). U1 RNA serves as a positive control for nuclear location. EEA1, endosome antigen 1; H3, histone 3. For b, d, h, uncropped blots and gels can be found in Supplementary Data Set 1.
2 Supplementary Figure 2 Lnc-β-Catm enhances self-renewal of liver CSCs. (a) Relative lnc-β-catm expression levels of lnc-β-catm and control cells. Error bars, s.d. (n = 3 cell cultures). Two tailed Student s t- test was used for statistical analysis, **, P < 0.01; ***, P< (b, c) Serial sphere formation (b) and tumor propagation (c) with lnc-β- Catm overexpressing and control cells. Error bars, s.d. (n = 3 cell cultures for b, n = 5 mice for c). Two tailed Student s t-test was used for statistical analysis, *, P < 0.05; **, P<0.01; ***, P< (d) Tumor-free mice ratios after 3 months tumor formation with lnc-β-catm overexpressing (oelnc) and control (oevec) cells. n = 6 mice for each group.
3 Supplementary Figure 3 Lnc-β-Catm associates with β-catenin and EZH2. (a) Relative mrna expression levels in lnc-β-catm silenced and control cells (lower panels), 16 nearby genes of lnc-β-catm locus (less
4 than 2 Mb) (upper panels) were analyzed. Error bars, s.d. (n = 4 cell cultures). Two tailed Student s t-test was used for statistical analysis, **, P < (b, c) MS-MS profiles of β-catenin (b) and EZH2 (c), corresponding peptide sequences are listed on the top of the corresponding graphs. (d) Different regions of lnc-β-catm (left panel) were labeled with biotin and incubated with PLC sphere lysates, followed by RNA pulldown assays (right panel). (e) Stem-loop structures of full length ( nt), segment #6 ( nt) and segment #9 ( nt) of lnc-β-catm. Predictions were based on minimum free energy (MFE) and partition function. Color scales denote confidence of predictions for each base with shades of red indicating strong confidence ( (f) Anti-β-catenin and anti-ezh2 Western blots. Samples were derived from co-immunoprecipitation (co-ip) with PLC spheres. (g, h) Antiβ-catenin, anti-ezh2, anti-β-actin (loading control) and anti-oct4 (serum treated control) Western blots. Samples were immunoprecipitates from spheres (S) and non-spheres (N) (g), or serum treated spheres (h). (i) Intensity profiles along the diagonal from upper left to lower right. Green profiles indicate lnc-β-catm gray value (intensity), red profiles indicate β-catenin intensity, and blue EZH2. (j) Flag-β-catenin truncates (upper panels) overexpressing spheres were established, followed by co-ip assays and Western blots (lower panels). (k) Anti-Myc and anti-his Western blots (right panels) with EZH2 truncates overepressing spheres, as in j. (l, m) Histogram of lnc-β-catm enrichment after RNA immunoprecipitation assays. β-catenin truncates (l) and EZH2 truncates (m) overexpressing PLC spheres were used. Error bars, s.d. (n = 4 independent experiments). Throughout figure, uncropped blots and gels can be found in Supplementary Data Set 1.
5 Supplementary Figure 4 Characterization of β-catenin methylation. (a) Anti-Methylated lysine, anti-β-catenin, anti-h3 and anti-eea1 Western blots. Samples were nuclear (N) and cytoplasmic (C) fractions of the indicated spheres. EEA1, endosome antigen 1; H3, histone 3. (b) Western blots for β-catenin methylation signals in HCC tumor tissues (T) and peri-tumor tissues (P). (c) Methylation observation of β-catenin in peri-tumor and tumor tissues. β-catenin (green), methylated lysine (red), and EZH2 (blue). Scale bars, 20 μm. Uncropped blots in a and b can be found in Supplementary Data Set 1.
6 Supplementary Figure 5 β-catenin methylation promotes its stability. (a) Anti-phosphorylated β-catenin (p-β-catenin), anti-β-catenin, anti-ezh2 and anti-β-actin (control) Western blots using EZH2 overexpressing (oeezh2) and control (oevec) spheres. (b) Anti-K48 linkage ubiquitylation (K48-Ub), anti-β-catenin and anti-β-actin (control) Western blots. EZH2 inhibitors (GSK126 and GSK343) treated and control (DMSO) spheres were used for β-catenin immunoprecipitation. (c) Anti-β-catenin and anti-β-actin (control) Western blots of methylated and non-methylated β-catenin supplemented with sphere lysates (left panels). Relative β-catenin levels in the right panel. Error bars, s.d. (n = 3 cell cultures). Throughout figure, uncropped Western blot results can be found in Supplementary Data Set 1.
7 Supplementary Figure 6 Lnc-β-Catm promotes Wnt signaling by increasing β-catenin stability. (a) Relative expression levels of Wnt-β-catenin target genes in lnc-β-catm silenced and control spheres. (b) Relative β-catenin protein levels in lnc-β-catm depleted spheres and control spheres. Error bars, s.d. (n=3 cell cultures). Two tailed Student s t-test was used for statistical analysis, **, P < 0.01; ***, P < (c) Anti-methylated lysine and anti-β-catenin (immunoprecipitation control) Western blots. Lnc-β-Catm overexpressing (oelnc) and control (oevec) HCC primary spheres were used. (d, e) Realtime PCR (d) and Western blots (e) of Wnt-β-catenin target genes in lnc-β-catm KO cells (lnc-β-catm KO) and rescued cells (lnc-β-catm KO+rescueLnc). Error bars, s.d. (n=3 cell cultures). (f-i) Anti-methylated lysine, anti-β-catenin, anti-ubiquitination and anti-actin (control) Western blots for β- catenin methylation (f), phosphorylation (g), ubiquitination (h) and stability (i). Samples were lnc-β-catm KO cells, rescued cells and control cells. For i, relative β-catenin protein levels were calculated and shown in the right panel. Error bars, s.d. (n = 3 cell cultures).
8 Two tailed Student s t-test was used for statistical analysis, *, P < 0.05; **, P < 0.01; ***, P < (j, k) Sphere formation (j) and xenograft tumor growth (k). Samples were lnc-β-catm silenced cells rescued with Wnt-β-catenin target genes (c-myc, Ccnd1, and Pttg1). Scale bar, 500 μm. Error bars, s.d. (n = 3 independent experiments). Two tailed Student s t-test was used for statistical analysis, *, P < (i) Sphere formation of lnc-β-catm overexpressed spheres supplemented with Wnt-β-catenin inhibitor WIKI4. Typical images were shown in left panels and sphere formation ratios were calculated (right panels). Error bars, s.d. (n = 3 independent experiments). Two tailed Student s t-test was used for statistical analysis, *, P < 0.05; **, P < ns, not significant. Throughout figure, uncropped blots and gels can be found in Supplementary Data Set 1.
9 Supplementary Figure 7 Lnc-β-Catm plays a predominant role in HCC and liver CSCs. (a, b) Expression levels of EZH2 and Wnt-β-catenin target genes in HCC tumors (a) and metastasis patients (b) derived from Wang s cohort. Data are shown as box-and-whisker plots. Whiskers: 5th and 95th percentiles; Horizontal lines: median levels; Boxes: interquartile range (IQR); upper and lower edges: 75th and 25th percentiles. (c) Kaplan-Meier survival analysis of Wnt-β-catenin target genes. HCC samples were divided into 2 groups according to the indicated gene expression levels. (d) Sphere formation of lnc-β-catm
10 and lnctcf7 silenced and control HCC primary cells. 31 HCC primary cells were used. *, **, ***, lncrna shrna versus control shrna. #, ##, lnc-β-catm shrna versus lnctcf7 shrna. Error bars, s.d. (n = 3 cell cultures). Two tailed Student s t-test was used for statistical analysis, *, P < 0.05; **, P < 0.01; ***, P < 0.001; #, P < 0.05; ##, P < (e, f) Confocal observation with CD133 antibody (e), Oct4 antibody and c-myc antibody (f). Control, lnc-β-catm silenced and lnctcf7 silenced spheres were used. Scale bar, 20 μm.
11 Supplementary Table 1. Table 1a. Frequencies of tumor initiating cells of lnc-β-catm depletion Cell CSC ratio (95% CI) P value Control shrna (A) 1/3190 (1/8046-1/1264) Lnc shrna#1 (B) 1/31905 (1/ /12651) 7.0E-5(B vs A) Lnc shrna#2 (C) 1/12241 (1/ /4483) 0.029(C vs A) Table 1b. Frequencies of tumor initiating cells of lnc-β-catm overexpression Cell CSC ratio (95% CI) P value oevec (A) 1/3190 (1/8046-1/1264) oelnc-β-catm (B) 1/318(1/804-1/126) 6.9E-5(B vs A) Supplementary Table 1. Tumor initiating cell ratios of lnc-β-catm silenced (a) and overexpression (b) cells , , , and 10 sphere cells were subcutaneously implanted into BALB/c nude mice for tumor initiation. Frequencies of tumor initiating cells were calculated using extreme limiting dilution analysis. 95% CI, 95% confidence interval of the estimation; vs, versus. P value less than 0.05 was considered significant.
12 Supplementary Table 2. Realtime PCR primers used in this study Primers 18S (Forward) 18S (Reverse) actin (Forward) actin (Reverse) Lnc-β-Catm (Forward) Lnc-β-Catm (Reverse) EZH2 (Forward) EZH2 (Reverse) CD13 (Forward) CD13 (Reverse) c-myc (Forward) c-myc (Reverse) Ccnd1 (Forward) Ccnd1 (Reverse) n-myc (Forward) Sequences 5 -AACCCGTTGAACCCCATT-3 5 -CCATCCAATCGGTAGTAGCG-3 5 -TCCATCATGAAGTGTGACGT-3 5 -GAGCAATGATCTTGATCTTCAT-3 5 -GGACAGTGAGCGGTCCAAAT-3 5 -TCCTTGTTCCAGTGAAGCGG-3 5 -AATCAGAGTACATGCGACTGAGA-3 5 -GCTGTATCCTTCGCTGTTTCC-3 5 -GACCAAAGTAAAGCGTGGAATCG-3 5 -TCTCAGCGTCACCCGGTAG-3 5 -GGCTCCTGGCAAAAGGTCA-3 5 -CTGCGTAGTTGTGCTGATGT-3 5 -GCTGCGAAGTGGAAACCATC-3 5 -CCTCCTTCTGCACACATTTGAA-3 5 -TGATCCTCAAACGATGCCTTC-3 n-myc (Reverse) 5 -GGACGCCTCGCTCTTTATCT-3 Pparγ (Forward) 5 -ACCAAAGTGCAATCAAAGTGGA -3 Pparγ (Reverse) 5 -ATGAGGGAGTTGGAAGGCTCT-3 Pttg1 (Forward) Pttg1 (Reverse) DKK1 (Forward) DKK1 (Reverse) PCNXL2 (Forward) PCNXL2 (Reverse) 5 -ACCCGTGTGGTTGCTAAGG-3 5 -ACGTGGTGTTGAAACTTGAGAT-3 5 -CCTTGAACTCGGTTCTCAATTCC-3 5 -CAATGGTCTGGTACTTATTCCCG-3 5 -CCATCCCATAGCACCAGTGTC-3 5 -GGTATTACTGACTTGGTCGTGG-3 NTPCR (Forward) 5 -GATGTCGTCACGTTGTCCG -3 NTPCR (Reverse) 5 -GGCATTCACGTTTTCCAGGT -3 KCNK1 (Forward) 5 -TTCTGGAAACCTTCTGTGAACTC-3 KCNK1 (Reverse) IRF2BP2 (Forward) IRF2BP2 (Reverse) TOMM20 (Forward) TOMM20 (Reverse) RBM34 (Forward) 5 -AGTTGGTCATGCTCTATGATGTG-3 5 -ACACCCATTTTGTGCAGTGC-3 5 -ACTGGGACAATAGACCTCTCC-3 5 -GGTACTGCATCTACTTCGACCG-3 5 -TGGTCTACGCCCTTCTCATATTC-3 5 -TACAGGCTTGGACAGGTCG-3 RBM34 (Reverse) 5 -CGTACACGGGTTGAATCTGGG-3 ARID4B (Forward) 5 -ATGAGCCTCCCTATTTGACAGT-3 ARID4B (Reverse) 5 -GGCCCTTTATGTGGTCATCCT -3 GGPS1 (Forward) GGPS1 (Reverse) TBCE (Forward) TBCE (Reverse) GNG4 (Forward) GNG4 (Reverse) LYST (Forward) LYST (Reverse) NID1 (Forward) 5 -ACAGCATCTATGGAATCCCATCT-3 5 -CAAAAGCTGGCGGGTAAAAAG-3 5 -CAGCGGATGTCATTGGTCGAA-3 5 -TCTACTCCTAACCAGGGTCCT-3 5 -GAGGGCATGTCTAATAACAGCAC-3 5 -AGACCTTGACCCTGTCCATAC-3 5 -GCCTCAGAGCATTTGAAAGCC-3 5 -TTCACATCGTCTGTGCCTTTT-3 5 -TCTACGTCACCACAAATGGCA-3 NID1 (Reverse) 5 -GCGACTGCACCGAATGTTG-3 ERO1LB (Forward) 5 -TTCTGGATGATTGCTTGTGTGA -3 ERO1LB (Reverse) 5 -GGTCGCTTCAGATTAACCTTGT-3
13 Supplementary Table 3. shrna sequences used in this study Primers shlnc-β-catm#1 shlnc-β-catm#2 shttty16#1 shttty16#2 shlinc00211#1 shlinc00211#2 shlinc00244#1 shlinc00244#2 shflvcr1-as1#1 shflvcr1-as1#2 shncrupar#1 shncrupar#2 shhotair#1 shhotair#2 shcrym-as1#1 shcrym-as1#2 shlinc00221#1 shlinc00221#2 shlinctcf7#1 shlinctcf7#2 shβ-catenin Sequences 5 -GAAGCAAGGAAGAGACATA-3 5 -GGCTGAGATATTCCGAAGA-3 5 -GAGTCACTTTGAGGAGCAT-3 5 -GTGCAAGTGGGAATCTTGA-3 5 -GGGCTATGGAACTATGAGA-3 5 -GCAGCATTTCTGTCCTAAA-3 5 -GCAGAAAGTCAGAAGCACA-3 5 -GGGACAAGCTAAGACATGA-3 5 -GCACCAACACATGTAGTTA-3 5 -GGACCACGTTTCATAAGTT-3 5 -GCAGTAGAATGGCGTAAAC-3 5 -GGATCATGAGGTCAGGAGA-3 5 -GAACGGGAGTACAGAGAGA-3 5 -GAGGAAAAGGGAAAATCTA-3 5 -GGAGCAAGCATTATAGAAC-3 5 -GGATCAGTTTCTTACCTCT-3 5 -GCAGGAGAATCACTTGAAC-3 5 -GGATGAATCTCTGGAGAAG-3 5 -AGCCAACATTGTTGGTTAT-3 5 -CACCTAGGTGCTCACTGAA-3 5 -GCAAGCTCATCATACTGGC-3
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
ENCODE RBP Antibody Characterization Guidelines
 ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
What we ll do today. Types of stem cells. Do engineered ips and ES cells have. What genes are special in stem cells?
 Do engineered ips and ES cells have similar molecular signatures? What we ll do today Research questions in stem cell biology Comparing expression and epigenetics in stem cells asuring gene expression
Do engineered ips and ES cells have similar molecular signatures? What we ll do today Research questions in stem cell biology Comparing expression and epigenetics in stem cells asuring gene expression
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Do engineered ips and ES cells have similar molecular signatures?
 Do engineered ips and ES cells have similar molecular signatures? Comparing expression and epigenetics in stem cells George Bell, Ph.D. Bioinformatics and Research Computing 2012 Spring Lecture Series
Do engineered ips and ES cells have similar molecular signatures? Comparing expression and epigenetics in stem cells George Bell, Ph.D. Bioinformatics and Research Computing 2012 Spring Lecture Series
Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA.
 Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Wnt16 smact merge VK/AB
 A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation
 1 2 3 4 5 SUPPLEMENTAL DATA Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation Magalí Nazar, Juan Pablo Nicola, María Laura
1 2 3 4 5 SUPPLEMENTAL DATA Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation Magalí Nazar, Juan Pablo Nicola, María Laura
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Int. J. Mol. Sci. 2016, 17, 1259; doi: /ijms
 S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets.
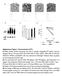 Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing
 Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing Chase L. Beisel, Yvonne Y. Chen, Stephanie J. Culler, Kevin G. Hoff, & Christina
Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing Chase L. Beisel, Yvonne Y. Chen, Stephanie J. Culler, Kevin G. Hoff, & Christina
Correction: The Leukemia-Associated Mllt10/ Af10-Dot1l Are Tcf4/β-Catenin Coactivators Essential for Intestinal Homeostasis
 CORRECTION Correction: The Leukemia-Associated Mllt10/ Af10-Dot1l Are Tcf4/β-Catenin Coactivators Essential for Intestinal Homeostasis Tokameh Mahmoudi, Sylvia F. Boj, Pantelis Hatzis, Vivian S. W. Li,
CORRECTION Correction: The Leukemia-Associated Mllt10/ Af10-Dot1l Are Tcf4/β-Catenin Coactivators Essential for Intestinal Homeostasis Tokameh Mahmoudi, Sylvia F. Boj, Pantelis Hatzis, Vivian S. W. Li,
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl
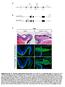 Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Supplementary Figure 1 Characterization of sirna-onv stability. (a) Fluorescence recovery curves of SQ-siRNA-ONV and SQ-ds-siRNA in 1 TAMg buffer
 Supplementary Figure 1 Characterization of sirna-onv stability. (a) Fluorescence recovery curves of SQ-siRNA-ONV and SQ-ds-siRNA in 1 TAMg buffer containing 10% serum The data error bars indicate means
Supplementary Figure 1 Characterization of sirna-onv stability. (a) Fluorescence recovery curves of SQ-siRNA-ONV and SQ-ds-siRNA in 1 TAMg buffer containing 10% serum The data error bars indicate means
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
Nature Immunology: doi: /ni Supplementary Figure 1. Zranb1 gene targeting.
 Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor
 Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2774 Figure S1 TRF2 dosage modulates the tumorigenicity of mouse and human tumor cells. (a) Left: immunoblotting with antibodies directed against the Myc tag of the transduced TRF2 forms
DOI: 10.1038/ncb2774 Figure S1 TRF2 dosage modulates the tumorigenicity of mouse and human tumor cells. (a) Left: immunoblotting with antibodies directed against the Myc tag of the transduced TRF2 forms
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
Supplementary Figure Legend
 Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Supplemental Information. The TRAIL-Induced Cancer Secretome. Promotes a Tumor-Supportive Immune. Microenvironment via CCR2
 Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
EPIGENTEK. EpiQuik In Situ Histone H3-K27 Tri-Methylation Assay Kit. Base Catalog # P-3014T PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik In Situ Histone H3-K27 Tri-Methylation Assay Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik In Situ Histone H3-K27 Tri-Methylation Assay Kit is suitable for specifically
EpiQuik In Situ Histone H3-K27 Tri-Methylation Assay Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik In Situ Histone H3-K27 Tri-Methylation Assay Kit is suitable for specifically
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well
 Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
Supplemental Information: Supplemental Methods: Cell culture To isolate single GNS 144 cell clones, cells were plated at a density of 1cell/well in 96 well Primaria plates in GNS media and incubated at
translation The building blocks of proteins are? amino acids nitrogen containing bases like A, G, T, C, and U Complementary base pairing links
 The actual process of assembling the proteins on the ribosome is called? translation The building blocks of proteins are? Complementary base pairing links Define and name the Purines amino acids nitrogen
The actual process of assembling the proteins on the ribosome is called? translation The building blocks of proteins are? Complementary base pairing links Define and name the Purines amino acids nitrogen
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on
 Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1.
 Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Fig. 1. Characteristics of transcription elongation by YonO. a. YonO forms a saltstable EC. Immobilized ECs were washed with
 Supplementary Fig. 1. Characteristics of transcription elongation by YonO. a. YonO forms a saltstable EC. Immobilized ECs were washed with transcription buffer with or without a high salt concentration
Supplementary Fig. 1. Characteristics of transcription elongation by YonO. a. YonO forms a saltstable EC. Immobilized ECs were washed with transcription buffer with or without a high salt concentration
Rat IGF-1 ELISA Kit (rigf-1-elisa)
 Rat IGF-1 ELISA Kit (rigf-1-elisa) Cat. No. EK0377 96 Tests in 8 x 12 divisible strips Background Insulin-like growth factor 1 (IGF-1), also known as somatomedin C, is a polypeptide protein hormone similar
Rat IGF-1 ELISA Kit (rigf-1-elisa) Cat. No. EK0377 96 Tests in 8 x 12 divisible strips Background Insulin-like growth factor 1 (IGF-1), also known as somatomedin C, is a polypeptide protein hormone similar
Quantitative Real Time PCR USING SYBR GREEN
 Quantitative Real Time PCR USING SYBR GREEN SYBR Green SYBR Green is a cyanine dye that binds to double stranded DNA. When it is bound to D.S. DNA it has a much greater fluorescence than when bound to
Quantitative Real Time PCR USING SYBR GREEN SYBR Green SYBR Green is a cyanine dye that binds to double stranded DNA. When it is bound to D.S. DNA it has a much greater fluorescence than when bound to
Supplementary Figure 1. shrna screen against CSD-containing proteins in hescs. (a) Brightfield images of H9 hescs. Knockdown of YBX1, YBX2 and YBX3
 Supplementary Figure 1. shrna screen against CSD-containing proteins in hescs. (a) Brightfield images of H9 hescs. Knockdown of YBX1, YBX2 and YBX3 do not change hesc colony morphology. Scale bar represents
Supplementary Figure 1. shrna screen against CSD-containing proteins in hescs. (a) Brightfield images of H9 hescs. Knockdown of YBX1, YBX2 and YBX3 do not change hesc colony morphology. Scale bar represents
Multiple choice questions (numbers in brackets indicate the number of correct answers)
 1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
1 Multiple choice questions (numbers in brackets indicate the number of correct answers) February 1, 2013 1. Ribose is found in Nucleic acids Proteins Lipids RNA DNA (2) 2. Most RNA in cells is transfer
REAL TIME PCR USING SYBR GREEN
 REAL TIME PCR USING SYBR GREEN 1 THE PROBLEM NEED TO QUANTITATE DIFFERENCES IN mrna EXPRESSION SMALL AMOUNTS OF mrna LASER CAPTURE SMALL AMOUNTS OF TISSUE PRIMARY CELLS PRECIOUS REAGENTS 2 THE PROBLEM
REAL TIME PCR USING SYBR GREEN 1 THE PROBLEM NEED TO QUANTITATE DIFFERENCES IN mrna EXPRESSION SMALL AMOUNTS OF mrna LASER CAPTURE SMALL AMOUNTS OF TISSUE PRIMARY CELLS PRECIOUS REAGENTS 2 THE PROBLEM
pgbkt7 Anti- Myc AH109 strain (KDa) 50
 pgbkt7 (KDa) 50 37 Anti- Myc AH109 strain Supplementary Figure 1. Protein expression of CRN and TDR in yeast. To analyse the protein expression of CRNKD and TDRKD, total proteins extracted from yeast culture
pgbkt7 (KDa) 50 37 Anti- Myc AH109 strain Supplementary Figure 1. Protein expression of CRN and TDR in yeast. To analyse the protein expression of CRNKD and TDRKD, total proteins extracted from yeast culture
Roche Molecular Biochemicals Technical Note No. LC 10/2000
 Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Technical Review. Real time PCR
 Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Supplementary Information
 Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplemental Materials and Methods
 Supplemental Materials and Methods 125 I-CXCL12 binding assay KG1 cells (2 10 6 ) were preincubated on ice with cold CXCL12 (1.6µg/mL corresponding to 200nM), CXCL11 (1.66µg/mL corresponding to 200nM),
Supplemental Materials and Methods 125 I-CXCL12 binding assay KG1 cells (2 10 6 ) were preincubated on ice with cold CXCL12 (1.6µg/mL corresponding to 200nM), CXCL11 (1.66µg/mL corresponding to 200nM),
Learning Objectives. Define RNA interference. Define basic terminology. Describe molecular mechanism. Define VSP and relevance
 Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination.
 Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Nature Structural and Molecular Biology: doi: /nsmb.2959
 Supplementary Figure 1 EIciNAs were pulled down with an antibody to Pol II. (a) Western blot showing that pol II was efficiently pulled down with a pol II antibody in HeLa cell lysates. (b) The enrichment
Supplementary Figure 1 EIciNAs were pulled down with an antibody to Pol II. (a) Western blot showing that pol II was efficiently pulled down with a pol II antibody in HeLa cell lysates. (b) The enrichment
The Genetic Code and Transcription. Chapter 12 Honors Genetics Ms. Susan Chabot
 The Genetic Code and Transcription Chapter 12 Honors Genetics Ms. Susan Chabot TRANSCRIPTION Copy SAME language DNA to RNA Nucleic Acid to Nucleic Acid TRANSLATION Copy DIFFERENT language RNA to Amino
The Genetic Code and Transcription Chapter 12 Honors Genetics Ms. Susan Chabot TRANSCRIPTION Copy SAME language DNA to RNA Nucleic Acid to Nucleic Acid TRANSLATION Copy DIFFERENT language RNA to Amino
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and
 Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts. (A) Shoot morphology of 19-day-old nla mutants under Pi-sufficient conditions. (B) [ 33 P]Pi uptake
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts. (A) Shoot morphology of 19-day-old nla mutants under Pi-sufficient conditions. (B) [ 33 P]Pi uptake
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
Supplementary Figure 1 Activated B cells are subdivided into three groups
 Supplementary Figure 1 Activated B cells are subdivided into three groups according to mitochondrial status (a) Flow cytometric analysis of mitochondrial status monitored by MitoTracker staining or differentiation
Supplementary Figure 1 Activated B cells are subdivided into three groups according to mitochondrial status (a) Flow cytometric analysis of mitochondrial status monitored by MitoTracker staining or differentiation
- NaCr. + NaCr. α H3K4me2 α H3K4me3 α H3K9me3 α H3K27me3 α H3K36me3 H3 H2A-2B H4 H3 H2A-2B H4 H3 H2A-2B H4. α Kcr. (rabbit) α Kac.
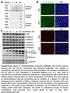 + NaCr NaCr + NaCr NaCr Peptides 10ng 50ng 250ng K α Pan (mouse) Pan (mouse) 10ng 50ng 250ng α Pan (rabbit) C 10ng 50ng 250ng α Pan (mouse) 0 1.25 2.5 5 10 20 40 (mm) NaCr 24h α Pan (rabbit) α K4me2 α
+ NaCr NaCr + NaCr NaCr Peptides 10ng 50ng 250ng K α Pan (mouse) Pan (mouse) 10ng 50ng 250ng α Pan (rabbit) C 10ng 50ng 250ng α Pan (mouse) 0 1.25 2.5 5 10 20 40 (mm) NaCr 24h α Pan (rabbit) α K4me2 α
Small-Molecule Drug Target Identification/Deconvolution Technologies
 Small-Molecule Drug Target Identification/Deconvolution Technologies Case-Studies Shantani Target ID Technology Tool Box Target Deconvolution is not Trivial = A single Tool / Technology May Not necessarily
Small-Molecule Drug Target Identification/Deconvolution Technologies Case-Studies Shantani Target ID Technology Tool Box Target Deconvolution is not Trivial = A single Tool / Technology May Not necessarily
Supplemental Movie Legend.
 Supplemental Movie Legend. Transfected T cells were dropped onto SEE superantigen-pulsed Raji B cells (approximate location indicated by circle). Maximum-intensity projections from Z-stacks (17 slices,
Supplemental Movie Legend. Transfected T cells were dropped onto SEE superantigen-pulsed Raji B cells (approximate location indicated by circle). Maximum-intensity projections from Z-stacks (17 slices,
Regulation of Gene Expression
 Slide 1 Chapter 18 Regulation of Gene Expression PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions
Slide 1 Chapter 18 Regulation of Gene Expression PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions
3.1.4 DNA Microarray Technology
 3.1.4 DNA Microarray Technology Scientists have discovered that one of the differences between healthy and cancer is which genes are turned on in each. Scientists can compare the gene expression patterns
3.1.4 DNA Microarray Technology Scientists have discovered that one of the differences between healthy and cancer is which genes are turned on in each. Scientists can compare the gene expression patterns
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
Supplemental Data. LMO4 Controls the Balance between Excitatory. and Inhibitory Spinal V2 Interneurons
 Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Galina Gabriely, Ph.D. BWH/HMS
 Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Bio 101 Sample questions: Chapter 10
 Bio 101 Sample questions: Chapter 10 1. Which of the following is NOT needed for DNA replication? A. nucleotides B. ribosomes C. Enzymes (like polymerases) D. DNA E. all of the above are needed 2 The information
Bio 101 Sample questions: Chapter 10 1. Which of the following is NOT needed for DNA replication? A. nucleotides B. ribosomes C. Enzymes (like polymerases) D. DNA E. all of the above are needed 2 The information
Jae Myoung Suh, Daniel Zeve, Renee McKay, Jin Seo, Zack Salo, Robert Li, Michael Wang, and Jonathan M. Graff
 Cell Metabolism, Volume 6 Supplemental Data Adipose Is a Conserved, Dosage-Sensitive Antiobesity Gene Jae Myoung Suh, Daniel Zeve, Renee McKay, Jin Seo, Zack Salo, Robert Li, Michael Wang, and Jonathan
Cell Metabolism, Volume 6 Supplemental Data Adipose Is a Conserved, Dosage-Sensitive Antiobesity Gene Jae Myoung Suh, Daniel Zeve, Renee McKay, Jin Seo, Zack Salo, Robert Li, Michael Wang, and Jonathan
Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo
 Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo Rampyari Raja Walia and Bakhos A. Tannous 1 2 1 Pluristem Innovations, 1453
Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo Rampyari Raja Walia and Bakhos A. Tannous 1 2 1 Pluristem Innovations, 1453
Metabolic collateral vulnerabilities of MTAP-deleted cancers as therapeutic opportunities Keystone on Tumor Metabolism 2017
 Metabolic collateral vulnerabilities of MTAP-deleted cancers as therapeutic opportunities Keystone on Tumor Metabolism 2017 5 9 March 2017 Whistler, Canada The Challenge: Identifying Precision Medicine
Metabolic collateral vulnerabilities of MTAP-deleted cancers as therapeutic opportunities Keystone on Tumor Metabolism 2017 5 9 March 2017 Whistler, Canada The Challenge: Identifying Precision Medicine
Functional characterisation
 Me Me Ac Ac Functional characterisation How can we know if measured changes in DNA methylation and function (phenotype) and linked, and in what way? DNMT Dianne Ford Professor of Molecular Nutritional
Me Me Ac Ac Functional characterisation How can we know if measured changes in DNA methylation and function (phenotype) and linked, and in what way? DNMT Dianne Ford Professor of Molecular Nutritional
Strep-tag detection in Western blots
 Strep-tag detection in Western blots General protocol for the detection of Strep-tag fusion proteins Last date of revision April 2012 Version PR07-0010 www.strep-tag.com For research use only Important
Strep-tag detection in Western blots General protocol for the detection of Strep-tag fusion proteins Last date of revision April 2012 Version PR07-0010 www.strep-tag.com For research use only Important
Transcriptional Regulation in Eukaryotes
 Transcriptional Regulation in Eukaryotes Concepts, Strategies, and Techniques Michael Carey Stephen T. Smale COLD SPRING HARBOR LABORATORY PRESS NEW YORK 2000 Cold Spring Harbor Laboratory Press, 0-87969-537-4
Transcriptional Regulation in Eukaryotes Concepts, Strategies, and Techniques Michael Carey Stephen T. Smale COLD SPRING HARBOR LABORATORY PRESS NEW YORK 2000 Cold Spring Harbor Laboratory Press, 0-87969-537-4
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles
 Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
Different Potential of Extracellular Vesicles to Support Thrombin Generation: Contributions of Phosphatidylserine, Tissue Factor, and Cellular Origin
 Different Potential of Extracellular Vesicles to Support Thrombin Generation: Contributions of Phosphatidylserine, Tissue Factor, and Cellular Origin Carla Tripisciano 1, René Weiss 1, Tanja Eichhorn 1,
Different Potential of Extracellular Vesicles to Support Thrombin Generation: Contributions of Phosphatidylserine, Tissue Factor, and Cellular Origin Carla Tripisciano 1, René Weiss 1, Tanja Eichhorn 1,
Chapter 18: Regulation of Gene Expression. 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer
 Chapter 18: Regulation of Gene Expression 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer Gene Regulation Gene regulation refers to all aspects of controlling
Chapter 18: Regulation of Gene Expression 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer Gene Regulation Gene regulation refers to all aspects of controlling
Gene Expression and Heritable Phenotype. CBS520 Eric Nabity
 Gene Expression and Heritable Phenotype CBS520 Eric Nabity DNA is Just the Beginning DNA was determined to be the genetic material, and the structure was identified as a (double stranded) double helix.
Gene Expression and Heritable Phenotype CBS520 Eric Nabity DNA is Just the Beginning DNA was determined to be the genetic material, and the structure was identified as a (double stranded) double helix.
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications
![MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications](/thumbs/74/71368870.jpg) MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna
 Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Custom Antibodies Services. GeneCust Europe. GeneCust Europe
 GeneCust Europe Laboratoire de Biotechnologie du Luxembourg S.A. 2 route de Remich L-5690 Ellange Luxembourg Tél. : +352 27620411 Fax : +352 27620412 Email : info@genecust.com Web : www.genecust.com Custom
GeneCust Europe Laboratoire de Biotechnologie du Luxembourg S.A. 2 route de Remich L-5690 Ellange Luxembourg Tél. : +352 27620411 Fax : +352 27620412 Email : info@genecust.com Web : www.genecust.com Custom
Supplemental Materials. Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans
 Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Supplemental Information. Lysine-5 Acetylation Negatively Regulates. Lactate Dehydrogenase A and Is Decreased. in Pancreatic Cancer
 Cancer Cell, Volume 23 Supplemental Information Lysine-5 Acetylation Negatively Regulates Lactate Dehydrogenase A and Is Decreased in Pancreatic Cancer Di Zhao, Shao-Wu Zou, Ying Liu, Xin Zhou, Yan Mo,
Cancer Cell, Volume 23 Supplemental Information Lysine-5 Acetylation Negatively Regulates Lactate Dehydrogenase A and Is Decreased in Pancreatic Cancer Di Zhao, Shao-Wu Zou, Ying Liu, Xin Zhou, Yan Mo,
Identification of Microprotein-Protein Interactions via APEX Tagging
 Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian
Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian
Year III Pharm.D Dr. V. Chitra
 Year III Pharm.D Dr. V. Chitra 1 Genome entire genetic material of an individual Transcriptome set of transcribed sequences Proteome set of proteins encoded by the genome 2 Only one strand of DNA serves
Year III Pharm.D Dr. V. Chitra 1 Genome entire genetic material of an individual Transcriptome set of transcribed sequences Proteome set of proteins encoded by the genome 2 Only one strand of DNA serves
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
GUGAUAAUGGAGCGAGAUUUUCUGUUGUGCUUGAUCUAACCAUGUGCUUGCGAGGUAUGA GAAAAACAUGGUUCCGUCAAGCACCAUGGAACGUCACGCAGCUUUCUACA
 Precursor mmu-mir-8- GUGAUAAUGGAGCGAGAUUUUCUGUUGUGCUUGAUCUAACCAUGUGCUUGCGAGGUAUGA GAAAAACAUGGUUCCGUCAAGCACCAUGGAACGUCACGCAGCUUUCUACA Precursor mmu-mir-8- GACCAGUUGCCGCGGGGCUUUCCUUUGUGCUUGAUCUAACCAUGUGGUGGAACGAUGGAA
Precursor mmu-mir-8- GUGAUAAUGGAGCGAGAUUUUCUGUUGUGCUUGAUCUAACCAUGUGCUUGCGAGGUAUGA GAAAAACAUGGUUCCGUCAAGCACCAUGGAACGUCACGCAGCUUUCUACA Precursor mmu-mir-8- GACCAGUUGCCGCGGGGCUUUCCUUUGUGCUUGAUCUAACCAUGUGGUGGAACGAUGGAA
