Supplementary Figure 1. Lnc-β-Catm characterization. (A, B) LncBRM silenced
|
|
|
- Norman Bryant
- 6 years ago
- Views:
Transcription
1 Supplementary Figure 1. Lnc-β-Catm characterization. (A, B) LncBRM silenced Hep3B (A) and Huh7 (B) cells were established using psicor lentivirus, followed by sphere formation assays. (C, D) LncBRM deleted Hep3B (C) and Huh7 (D) cells were established using a CRISPR/Cas9 approach, followed by serial sphere formation assays. (E) 12 genes nearby lncbrm locus (less than 2 Mb) were shown. Location information was obtained from UCSC dataset ( (F) LncBRM silenced and shctrl spheres were collected, followed by examination of gene expression levels by realtime PCR. (G) Schematic annotation of lncbrm genomic locus on chromasome 5. Purple rectangles represent exons (upper panel), sequence conservation was analyzed by Phylop software (lower panel). (H) Full length of lncbrm was examined by 3 and 5 rapid-amplification of cdna ends (RACE) assays. The length of lncbrm was 1321 nucleotides. Black arrowhead indicates the full length of lncbrm. (I) Coding potential of lncbrm was analyzed using CPC (left panel) and CPAT (right panel). HOX transcript antisense RNA (Hotair), X inactivation-specific transcript (XIST) and
2 lnc-tcf7 served as control non-coding RNAs. β-actin (ACTB) and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as control coding genes. CPC scores represent the coding ability; CPAT scores indicate the possibility of coding. (J) The indicated genes were cloned into PCDNA4-HisMycB vector and transfected into Huh7 cells. 2 days later, cells were lyzed with RIPA buffer, followed by Western blot with Myc antibody. ZIC2 served as a coding control. Data are shown as means ± SD. Two tailed Student s t-test was used for statistical analysis, **, p < 0.01; ***, p<0.001; ns, not significant. Data are representative of three independent experiments.
3 Supplementary Figure 2. LncBRM promotes self-renewal of liver CSCs. (A) LncBrm silenced primary cells were established using psicor lentivirus and confirmed by realtime PCR. shctrl, Control shrna. (B) LncBRM knockout cells were established using a CRISPR/Cas9 approach (left panel), followed by sphere formation. Representative sphere images were shown in the right panel and statistic ratios were shown in the lower right panel. (C) Self-renewal of lncbrm knockout and rescued HCC cells were detected by serial sphere formation assays. (D) LncBRM overexpressing primary cells were established using lentivirus, and the overexpressing efficiency was examined by realtime PCR. oevec, overexpressing Vector control; oelncbrm, overexpressing lncbrm. (E) LncBRM overexpressing and control primary cells were established with psicor lentivirus and used for subcutaneous injection. Xenograt tumors were used for serial tumor implantation. (F) LncBrm overexpressing and control cells (10, 10 2, 10 3, 10 4 and 10 5 ) were subcutaneously injected into BALB/c nude mice. Tumor-free mice ratios are calculated three month later. n=6 for each group. Data are shown as means ± SD. Two tailed Student s t-test was used for statistical analysis, *, p < 0.05; **, p < 0.01; ***, p < Data are representative of four independent experiments.
4 Supplementary Figure 3. LncBRM interacts with BRM. (A) MS/MS profiles of BRM. Corresponding peptide sequences are listed on top of the corresponding graphs. (B) LncBRM interacts with free BRM but not other BAF components. LncBRM was labeled with biotin using in vitro transcription and incubated with oncosphere lysates for RNA pulldown assay. Precipitates were subjected to SDS-PAGE for separation, and detected by Western blot with the indicated antibodies. (C) Full length ( nt) and segment #3 ( nt) of lncbrm were predicted to harbor stable stem-loop structures. Predictions were based on minimum free energy (MFE) and partition function. Color scales denote confidence of predictions for each base with shades of red indicating strong confidence ( (D) LncBRM has no impact on BRM stability. LncBRM
5 silenced and shctrl spheres were lyzed for immunoprecipitation with BRM antibody, followed by immunoblotting using anti-ubiquitin (Ub) antibody. (E) LncBRM has no effect on BRM ATPase activity. Precipitated BRM from lncbrm silenced or shctrl spheres was measured for ATPase activity by ATPase/GTPase Activity Assay Kit (Sigma). Data are shown as means ± SD. ns, not significant. Data are representative of three independent experiments.
6 Supplementary Figure 4. KLF4 plays a critical role in activation of YAP1 signaling. (A) Full length (FL) and C terminal truncate ( C) KLF4 were cloned into pbplv vector and transfected into KLF4 knockout cells, followed by sphere formation assay. YAP1 signaling target genes were examined by realtime PCR. PR, proline-rich domain; AD, acidic domain; PEST, PEST domain, ZF, Zinc finger domain. (B) KLF4 binding sequence (GGGCGGGG) was replaced with mutant sequence (ATAATATT) (left panels) by a CRISPR/Cas9 approach. WT and mutant Huh7 cells (Mut) were collected for ChIP assays with KLF4, BRG1 and H3K4me3 antibodies, and YAP1 promoter enrichment was examined using realtime PCR. (C, D) BRG1, BAF170, SNF5 and ARID1A ChIP assays were performed using lncbrm silenced (C) and overexpressing (D) cells. (E, F) Activation of YAP1 promoter was examined by luciferase assay (E) and H3K4me3 ChIP assay (F) in lncbrm silenced spheres. (G) LncBRM silenced cells were established (upper panels), followed by Western blot (lower panels). 18S rrna served as a loading control for Northern blot, and β-actin served as a loading control for Western blot. **, p < 0.01; ***, p < Data are representative of three independent experiments.
7 Supplementary Figure 5. YAP1 signaling is required for self-renewal of liver CSCs. (A) Liver CSCs and non-cscs were treated with Verteportin and YAP1 overexpression, respectively. Liver CSC self-renewal was detected using the indicated CSC markers and stem factors. (B) YAP1 deficient cells were established using a CRISPR/Cas9 approach, and validated with Western blot. WT, wild type; YAP1KO, YAP1 knockout. (C) YAP1 overexpressing cells were established and followed by sphere formation assays. (D, E) Birc5 or Foxm1 silenced cells were established (left panels), followed by sphere formation assays. (F) LncBRM overexpressing cells were treated with YAP1 inhibitor Verteporfin, followed by sphere formation assays. oevec, overexpression vector; oelnc, overexpression lncbrm. For C, D, E, F, scale bars, 500 μm. Data are shown as means ± SD. *, p < 0.05; ***, p < ns, not significant. Data are representative of four independent experiments.
8 Supplementary Figure 6. Expression levels of BRG1 and YAP1 targets are positively related to cancer severity of HCC. (A, B) Decreased BRM expression (left panels) and increased BRG1 expression (right panels) in HCC tumor tissues derived from Zhang s cohort (GSE25097, A) and Wang s cohort (GSE54238, B). R language was used for gene expression analysis. P, peri-tumor; T, tumor; NL, normal liver; CL, cirrhosis liver; ehcc, early HCC; ahcc, advanced HCC. (C) Expression levels of BRG1 and BRM were analyzed using Zhang s cohort (GSE25097) and Wang s cohort (GSE14520) datasets using R language, and shown as heat map. r, Pearson correlation coefficient. (D) Higher expression levels of YAP1 target genes in HCC tumors were validated using Zhang s cohort (GSE25097). There were no probes for Lats1, YAP1 and other YAP1 target genes. NL, normal liver; PL, peri-tumor liver tissues. (E) Elevated Foxm1 and Sox9 expression appeared in advanced HCC patients derived from Wang s cohort. NL, normal liver; IL, inflammatory liver; CL, cirrhosis liver; ehcc, early HCC; ahcc, advanced HCC. (F) Expression levels of YAP1 and its target genes were related to clinical prognosis of HCC patients. HCC samples were divided into two groups according to the indicated gene expression levels, followed by Kaplan Meier survival analysis. For A, D: data are shown as box and whisker plot. Box: interquartile range (IQR); horizontal line within box: median;
9 whiskers: 5 95 percentile. For B, E, data are shown as means ± SD. Two tailed Student s t-test was used for statistical analysis.
10 Supplementary Figure 7. LncBRM drives YAP1 activation in liver CSCs. (A, B) LncBRM and lnctcf7 knockout cells were established using a CRISPR/Cas9 approach, followed by mrna detection by realtime PCR. (C) LncBRM silenced, lnctcf7 silenced or double knockdown cells were established using PSiCoR lentivirus, followed by sphere formation assays. (D) CD90 +, EpCAM +, CD24 + or CD44 + Cells were enriched by FACS, followed by realtime PCR for lncbrm expression. (E) CD90 +, EPCAM +, CD24 + or CD44 + cells were isolated using FACS and infected with shlncbrm virus or shctrl control virus, followed by sphere formation assays. Representative images were shown in the left panel and sphere ratios shown in the right panel. (F) CD13 + CD133 + cells were detected using FACS, and ratios were shown as histogram. (G) CD13 + CD133 + cells were enriched and seeded onto cover slips for 2 days in incubator, and then cells were fixed, penetrated and
11 stained with the indicated antibodies. Representative images were shown in the middle panel and statistic ratios were shown in the right panel. Asymmetric division (%) = asymmetric division / (asymmetric division + symmetric division). (H) CD133 + and CD133 - cells were sorted with FACS, followed by lncbrm knockdown and subsequent long-term culture for differentiation assays. Ratios of CD133 - cell differentiation were analyzed by FACS. Statistic ratios of CD133 - cells on day 20 were shown in the right panel. (I) In non-cscs, lncbrm is much lowly expressed, the BRM-embedded BAF complex exists, which blocks YAP1 signaling activation. By contrast, in liver CSCs, lncbrm is much highly expressed, lncbrm interacts with BRM to sequester BRM away from the BAF complex, facilitating the formation of BRG1-biased BAF complex, leading to YAP1 signaling activation. Consequently, lncbrm-mediated YAP1 signaling activation regulates the self-renewal of liver CSCs and their tumorigenesis. Scale bars, E, 500 μm; G, 10 μm. Data are shown as means ± SD. Two tailed Student s t-test was used for statistical analysis, *, p < 0.05; **, p < 0.01; ns, not significant. Data are representative of three independent experiments.
12 Supplementary Figure 8. Uncropped blots of figures. The red sections indicate blot results shown in the indicated figures.
13 Supplementary Figure 8. Uncropped blots of figures. The red sections indicate blot results shown in the indicated figures.
14 Supplementary Figure 8. Uncropped blots of figures. The red sections indicate blot results shown in the indicated figures.
15 Supplementary Figure 8. Uncropped blots of figures. The red sections indicate blot results shown in the indicated figures.
16 Supplementary Table 1. Clinical information of HCC patients we studied Sample Diagnosis Differentiation Metastasis Size (cm) Stage Tumor location 1# HCC low no III left lobe 8# HCC low no III right anterior lobe 9# HCC medium Yes II Left lateral lobe 13# HCC low Yes III left lobe 17# HCC medium yes II right lobe 20# HCC low yes II-III right lobe
17 Supplementary Table 2. CSC ratios of the indicated cells. A Cell CSC ratio (95% CI) P value shctrl (A) 1/3190 (1/8046-1/1264) shlncbrm #1 (B) 1/12241 (1/ /4483) (B vs A) shlncbrm #2 (C) 1/27311 (1/ /10472) (C vs A) B Cell CSC ratio (95% CI) P value oevec (A) 1/3190 (1/8046-1/1264) oelncbrm (B) 1/459 (1/1109-1/190) 0.002(A vs B) C Cell CSC ratio (95% CI) P value WT(A) 1/3190 (1/8046-1/1264) YAP1KO#1(B) 1/26597 (1/ /10135) (B vs A) YAP1KO#2(C) 1/12241 (1/ /4483) 0.029(C vs A) 10, , , and indicated cells were subcutaneously injected into BALB/c nude mice on the back and tumor formation was observed three months later. Liver CSC ratios were calculated using extreme limiting dilution analysis. CI, Confidence interval; vs, versus.
18 Supplementary Table 3. Realtime PCR primers used in this study Primers 18S (Forward) 18S (Reverse) actin (Forward) actin (Reverse) LncBRM (Forward) LncBRM (Reverse) BRG1 (Forward) BRG1 (Reverse) BRM (Forward) BRM (Reverse) Sequences 5 -AACCCGTTGAACCCCATT-3 5 -CCATCCAATCGGTAGTAGCG-3 5 -TCCATCATGAAGTGTGACGT-3 5 -GAGCAATGATCTTGATCTTCAT-3 5 -GGTCAAGAGGCCAGGAAGAG-3 5 -TTCTCACTTCAGCCCAATGCT-3 5 -CAGATCCGTCACAGGCAAAAT-3 5 -TCTCGATCCGCTCGTTCTCTT-3 5 -AGGGGATTGTAGAAGACATCCA-3 5 -TTGGCTGTGTTGATCCATTGG-3 Ctgf (Forward) 5 -CAGCATGGACGTTCGTCTG-3 Ctgf (Reverse) 5 -AACCACGGTTTGGTCCTTGG-3 YAP1 (Forward) 5 -TAGCCCTGCGTAGCCAGTTA -3 YAP1 (Reverse) 5 -TCATGCTTAGTCCACTGTCTGT-3 Ankrd1 (Forward) 5 -AGTAGAGGAACTGGTCACTGG -3 Ankrd1 (Reverse) 5 -TGTTTCTCGCTTTTCCACTGTT -3 Birc5 (Forward) 5 -CCAGATGACGACCCCATAGAG -3 Birc5 (Reverse) 5 -TTGTTGGTTTCCTTTGCAATTTT-3 Foxm1 (Forward) 5 -CGTCGGCCACTGATTCTCAAA-3 Foxm1 (Reverse) 5 -GGCAGGGGATCTCTTAGGTTC-3 Sox9 (Forward) 5 -AGCGAACGCACATCAAGAC-3 Sox9 (Reverse) 5 -CTGTAGGCGATCTGTTGGGG-3 DDX4 (Forward) DDX4 (Reverse) IL31RA (Forward) IL31RA (Reverse) IL6ST (Forward) IL6ST (Reverse) MAP3K1 (Forward) MAP3K1 (Reverse) SETD9 (Forward) SETD9 (Reverse) GPBP1 (Forward) GPBP1 (Reverse) ACTBL2 (Forward) ACTBL2 (Reverse) PLK2 (Forward) PLK2 (Reverse) GAPT (Forward) GAPT (Reverse) RAB3C (Forward) RAB3C (Reverse) PDE4D (Forward) PDE4D (Reverse) VEGF (Forward) VEGF (Reverse) Bcl2l1 (Forward) Bcl2l1 (Reverse) Hif1a (Forward) 5 -TCATACTTGCAGGACGAGATTTG AACGACTGGCAGTTATTCCATC-3 5 -CACACTTCGATTCAGGACAGTC-3 5 -CACATCGCAGAGCTATGACAT-3 5 -GTGAGTGGGATGGTGGAAGG-3 5 -CAAACTTGTGTGTTGCCCATTC-3 5 -CATCAGGTCGCACAGTGAAAT-3 5 -TCAGGGCTATATGGTGAGAAGC-3 5 -GCCGTTACAAGTACCGCTTC-3 5 -CTCTGGAACATATCGGAGGGT-3 5 -GCAGCCTCGTCTAACCAAACT-3 5 -CAGCACGGCTTTCATCTTCAT-3 5 -CTCGACACCAGGGCGTTATG-3 5 -CCACTCCATGCTCGATAGGAT-3 5 -CTACGCCGCAAAAATTATTCCTC-3 5 -TCTTTGTCCTCGAAGTAGTGGT-3 5 -AAGCAGGTCCTCCCAAAGCTA-3 5 -CCTCGAAATTAGACTGCCCTGT-3 5 -GGAAGACGAGCGGGTCATC-3 5 -CTCTCTGACATTTTGTCGCAGAT-3 5 -ACGGACCGGATAATGGAGGAG-3 5 -ATTTTTCCACGGAAGCATTGTG-3 5 -ATCACGAAGTGGTGAAGTTC-3 5 -TGCTGTAGGAAGCTCATCTC-3 5 -GAGCTGGTGGTTGACTTTCTC-3 5 -TCCATCTCCGATTCAGTCCCT-3 5 -GAACGTCGAAAAGAAAAGTCTCG-3
19 Hif1a (Reverse) CD74 (Forward) CD74 (Reverse) Cox2 (Forward) Cox2 (Reverse) MMP2 (Forward) MMP2 (Reverse) c-myc (Forward) c-myc (Reverse) Tiam (Forward) Tiam (Reverse) Kiaa (Forward) Kiaa (Reverse) Ccnd2 (Forward) Ccnd2 (Reverse) Sox4 (Forward) Sox4 (Reverse) Fn14 (Forward) Fn14 (Reverse) Sox2 (Forward) Sox2 (Reverse) Hes6 (Forward) Hes6 (Reverse) Hey1 (Forward) Hey1 (Reverse) Hes1 (Forward) Hes1 (Reverse) Gli1 (Forward) Gli1 (Reverse) Ptch1 (Forward) Ptch1 (Reverse) Gli3 (Forward) Gli3 (Reverse) JunB (Forward) JunB (Reverse) Cbfa1 (Forward) Cbfa1 (Reverse) Msx2 (Forward) Msx2 (Reverse) Smad7 (Forward) Smad7 (Reverse) Nanog (Forward) Nanog (Reverse) 5 -CCTTATCAAGATGCGAACTCACA-3 5 -GACGAGAACGGCAACTATCTG-3 5 -GTTGGGGAAGACACACCAGC-3 5 -ATGCTGACTATGGCTACAAAAGC-3 5 -TCGGGCAATCATCAGGCAC-3 5 -CCCACTGCGGTTTTCTCGAAT-3 5 -CAAAGGGGTATCCATCGCCAT-3 5 -GGCTCCTGGCAAAAGGTCA-3 5 -CTGCGTAGTTGTGCTGATGT-3 5 -CCTGTGTCTTACACTGACTCTTC-3 5 -CATCCCCGTAAAGCCTGCTC-3 5 -GAACGCCACTCAGCTTTTGC-3 5 -GAAGCACTTATGTTGGGGTCTT-3 5 -TTTGCCATGTACCCACCGTC-3 5 -AGGGCATCACAAGTGAGCG-3 5 -AGCGACAAGATCCCTTTCATTC-3 5 -CGTTGCCGGACTTCACCTT-3 5 -CTGCTAACTGTTCCTCGGCT-3 5 -GCCTTGGCAATCCTTTCACA-3 5 -GCCGAGTGGAAACTTTTGTCG-3 5 -GGCAGCGTGTACTTATCCTTCT-3 5 -AGCAGGAGCCTGACTCAGTT-3 5 -AGCTCCTGAACCATCTGCTC-3 5 -GTTCGGCTCTAGGTTCCATGT-3 5 -CGTCGGCGCTTCTCAATTATTC-3 5 -TCAACACGACACCGGATAAAC-3 5 -GCCGCGAGCTATCTTTCTTCA-3 5 -TGGATATGATGGTTGGCAAGTG-3 5 -ACAGACTCAGGCTCAGGCTTCT-3 5 -CCACAGAAGCGCTCCTACA-3 5 -CTGTAATTTCGCCCCTTCC-3 5 -GAAGTGCTCCACTCGAACAGA-3 5 -GTGGCTGCATAGTGATTGCG-3 5 -ACAAACTCCTGAAACCGAGCC-3 5 -CGAGCCCTGACCAGAAAAGTA-3 5 -TGGTTACTGTCATGGCGGGTA-3 5 -TCTCAGATCGTTGAACCTTGCTA-3 5 -ATGGCTTCTCCGTCCAAAGG-3 5 -CGGCTTCTTGTCGGACATGA-3 5 -TTCCTCCGCTGAAACAGGG-3 5 -CCTCCCAGTATGCCACCAC-3 5 -TTTGTGGGCCTGAAGAAAACT-3 5 -AGGGCTGTCCTGAATAAGCAG-3 Runx1 (Forward) 5 -CTGCCCATCGCTTTCAAGGT-3 Runx1 (Reverse) 5 -GCCGAGTAGTTTTCATCATTGCC (Forward) (YAP1p) 5 -GCGAACTGGAAGCGCCTTTC (Reverse) (YAP1p) 5 -GAAGCGTGCCCCGTATTCT (Forward) (YAP1p) 5 -GTTGCGGCTTCCAGTGACTA (Reverse) (YAP1p) 5 -AAGCCGCGAGGATAGATTGG (Forward) (YAP1p) 5 -GGATGACCTCCTTGCCCATT (Reverse) (YAP1p) 5 -CCTACCCCTGCTATCCCTGT (Forward) (YAP1p) 5 -ACCTTTTTGCTCACGGACTT (Reverse) (YAP1p) 5 -GCACAAGCCCATTCTAAGGAAAT-3
20 -1486 (Forward) (YAP1p) 5 -GACAGGAAACTGGGGGTTCA (Reverse) (YAP1p) 5 -GCAGGCCTCAGACACTCAAA (Forward) (YAP1p) 5 -GCATAGTTTCTGCCCAAAGGTT (Reverse) (YAP1p) 5 -AGGGACCCTGTTGGTAGTCC (Forward) (YAP1p) 5 -ATCCCCTACTACACCGTGGTT (Reverse) (YAP1p) 5 -TGGGATTGGAATAGAAGCTGTCC (Forward) (YAP1p) 5 -GAAGGGAGTGATAGGTTGCAGA (Reverse) (YAP1p) 5 -TCGTCTGTACTCTGCATCATCC (Forward) (YAP1p) 5 -ACAATGAATTACGATTTCCA (Reverse) (YAP1p) 5 -GTGAGAAATAAAATTCTAG (Forward) (YAP1p) 5 -AGCTTAGGCGTTGGAATAAAACA (Reverse) (YAP1p) 5 -TCAATCCCGTTCTCTCATGGATT (Forward) (YAP1p) 5 -CACCATGTTGGCCAGGCTGGTG (Reverse) (YAP1p) 5 -GATGGAGCCATTGCACTCCAGC (Forward) (YAP1p) 5 -AAATGTGGAAGGACACTAATGC (Reverse) (YAP1p) 5 -TAACACCTTGGTTTCTGGGGC (Forward) (LncBRMp) 5 -GGGAATTGGTGTGGCAGGGGGA (Reverse) (LncBRMp) 5 -ATAGTCTACACAGGTCAGCCTC-3 YAP1p: YAP1 promoter LncBRMp: LncBRM promoter.
21 Supplementary Table 4. shrna sequences used in this study shrna shlncbrm#1 shlncbrm#2 sh sh sh sh sh sh sh sh sh shlnctcf7 Sequence 5 -GGAATATTGTCGACTTAAC-3 5 -GGACCACTAGGTTTCATAT-3 5 -GGAATATGATGAATGATCA-3 5 -GGAGGAAGTTGTTGGCTGT-3 5 -GCACCAAGACTCCTGATCA-3 5 -GGATGAATGAGTGCAGAGA-3 5 -GCATCACCATAGCCATAAG-3 5 -GCAGAAGAGTGAGGAGTTA-3 5 -GCCTGAAGTCATAACCAGA-3 5 -GGACCACACTGAACTCTTG-3 5 -GGTTTATGATGCCAGCAGA-3 5 -AGCCAACATTGTTGGTTAT-3
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Nature Immunology: doi: /ni Supplementary Figure 1. Zranb1 gene targeting.
 Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Int. J. Mol. Sci. 2016, 17, 1259; doi: /ijms
 S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
ENCODE RBP Antibody Characterization Guidelines
 ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2774 Figure S1 TRF2 dosage modulates the tumorigenicity of mouse and human tumor cells. (a) Left: immunoblotting with antibodies directed against the Myc tag of the transduced TRF2 forms
DOI: 10.1038/ncb2774 Figure S1 TRF2 dosage modulates the tumorigenicity of mouse and human tumor cells. (a) Left: immunoblotting with antibodies directed against the Myc tag of the transduced TRF2 forms
Supplemental Information. The TRAIL-Induced Cancer Secretome. Promotes a Tumor-Supportive Immune. Microenvironment via CCR2
 Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
REAL TIME PCR USING SYBR GREEN
 REAL TIME PCR USING SYBR GREEN 1 THE PROBLEM NEED TO QUANTITATE DIFFERENCES IN mrna EXPRESSION SMALL AMOUNTS OF mrna LASER CAPTURE SMALL AMOUNTS OF TISSUE PRIMARY CELLS PRECIOUS REAGENTS 2 THE PROBLEM
REAL TIME PCR USING SYBR GREEN 1 THE PROBLEM NEED TO QUANTITATE DIFFERENCES IN mrna EXPRESSION SMALL AMOUNTS OF mrna LASER CAPTURE SMALL AMOUNTS OF TISSUE PRIMARY CELLS PRECIOUS REAGENTS 2 THE PROBLEM
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA.
 Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
Galina Gabriely, Ph.D. BWH/HMS
 Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets.
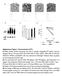 Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Quantitative Real Time PCR USING SYBR GREEN
 Quantitative Real Time PCR USING SYBR GREEN SYBR Green SYBR Green is a cyanine dye that binds to double stranded DNA. When it is bound to D.S. DNA it has a much greater fluorescence than when bound to
Quantitative Real Time PCR USING SYBR GREEN SYBR Green SYBR Green is a cyanine dye that binds to double stranded DNA. When it is bound to D.S. DNA it has a much greater fluorescence than when bound to
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl
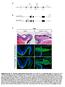 Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Endoglin Is Essential for the Maintenance of Self-Renewal and Chemoresistance
 Stem Cell Reports, Volume 9 Supplemental Information Endoglin Is Essential for the Maintenance of Self-Renewal and Chemoresistance in Renal Cancer Stem Cells Junhui Hu, Wei Guan, Peijun Liu, Jin Dai, Kun
Stem Cell Reports, Volume 9 Supplemental Information Endoglin Is Essential for the Maintenance of Self-Renewal and Chemoresistance in Renal Cancer Stem Cells Junhui Hu, Wei Guan, Peijun Liu, Jin Dai, Kun
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Figure Legend
 Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit
 Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Wnt16 smact merge VK/AB
 A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX
 Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
Supplementary Information
 Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Supplementary Information Supplementary Figure 1. ZBTB20 expression in the developing DRG. ZBTB20 expression in the developing DRG was detected by immunohistochemistry using anti-zbtb20 antibody 9A10 on
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor
 Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination.
 Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplemental Materials. Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans
 Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna
 Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
GUGAUAAUGGAGCGAGAUUUUCUGUUGUGCUUGAUCUAACCAUGUGCUUGCGAGGUAUGA GAAAAACAUGGUUCCGUCAAGCACCAUGGAACGUCACGCAGCUUUCUACA
 Precursor mmu-mir-8- GUGAUAAUGGAGCGAGAUUUUCUGUUGUGCUUGAUCUAACCAUGUGCUUGCGAGGUAUGA GAAAAACAUGGUUCCGUCAAGCACCAUGGAACGUCACGCAGCUUUCUACA Precursor mmu-mir-8- GACCAGUUGCCGCGGGGCUUUCCUUUGUGCUUGAUCUAACCAUGUGGUGGAACGAUGGAA
Precursor mmu-mir-8- GUGAUAAUGGAGCGAGAUUUUCUGUUGUGCUUGAUCUAACCAUGUGCUUGCGAGGUAUGA GAAAAACAUGGUUCCGUCAAGCACCAUGGAACGUCACGCAGCUUUCUACA Precursor mmu-mir-8- GACCAGUUGCCGCGGGGCUUUCCUUUGUGCUUGAUCUAACCAUGUGGUGGAACGAUGGAA
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure 1: MYCER protein expressed from the transgene can enhance
 Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1 Activated B cells are subdivided into three groups
 Supplementary Figure 1 Activated B cells are subdivided into three groups according to mitochondrial status (a) Flow cytometric analysis of mitochondrial status monitored by MitoTracker staining or differentiation
Supplementary Figure 1 Activated B cells are subdivided into three groups according to mitochondrial status (a) Flow cytometric analysis of mitochondrial status monitored by MitoTracker staining or differentiation
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
SureSilencing sirna Array Technology Overview
 SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
3.1.4 DNA Microarray Technology
 3.1.4 DNA Microarray Technology Scientists have discovered that one of the differences between healthy and cancer is which genes are turned on in each. Scientists can compare the gene expression patterns
3.1.4 DNA Microarray Technology Scientists have discovered that one of the differences between healthy and cancer is which genes are turned on in each. Scientists can compare the gene expression patterns
Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo
 Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo Rampyari Raja Walia and Bakhos A. Tannous 1 2 1 Pluristem Innovations, 1453
Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo Rampyari Raja Walia and Bakhos A. Tannous 1 2 1 Pluristem Innovations, 1453
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
Supplementary Figure 1
 Supplementary Figure A Name Location Start position End position Oct4P Chr 3 29433673 29435 Annotation Mus musculus predicted gene 572 (Gm572), noncoding RNA RefSeq accession number NR_33594. Oct4P2 Chr
Supplementary Figure A Name Location Start position End position Oct4P Chr 3 29433673 29435 Annotation Mus musculus predicted gene 572 (Gm572), noncoding RNA RefSeq accession number NR_33594. Oct4P2 Chr
Supplementary Data: Fig. 1S Detailed description of In vivo experimental design
 1 2 Supplementary Data: Fig. 1S Detailed description of In vivo experimental design 3 4 5 6 7 8 9 Relative Expression Studies 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 The
1 2 Supplementary Data: Fig. 1S Detailed description of In vivo experimental design 3 4 5 6 7 8 9 Relative Expression Studies 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 The
Roche Molecular Biochemicals Technical Note No. LC 10/2000
 Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
mir-24-mediated down-regulation of H2AX suppresses DNA repair
 Supplemental Online Material mir-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells Ashish Lal 1,4, Yunfeng Pan 2,4, Francisco Navarro 1,4, Derek M. Dykxhoorn
Supplemental Online Material mir-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells Ashish Lal 1,4, Yunfeng Pan 2,4, Francisco Navarro 1,4, Derek M. Dykxhoorn
Nature Structural and Molecular Biology: doi: /nsmb.2959
 Supplementary Figure 1 EIciNAs were pulled down with an antibody to Pol II. (a) Western blot showing that pol II was efficiently pulled down with a pol II antibody in HeLa cell lysates. (b) The enrichment
Supplementary Figure 1 EIciNAs were pulled down with an antibody to Pol II. (a) Western blot showing that pol II was efficiently pulled down with a pol II antibody in HeLa cell lysates. (b) The enrichment
The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns
 The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns Kenneth J. Oestreich, 1 Albert C. Huang, 1,2 and Amy S. Weinmann 1,2 1 Department of Immunology and 2 Molecular
The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns Kenneth J. Oestreich, 1 Albert C. Huang, 1,2 and Amy S. Weinmann 1,2 1 Department of Immunology and 2 Molecular
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/323/5910/124/dc1 Supporting Online Material for Regulation of Neuronal Survival Factor MEF2D by Chaperone-Mediated Autophagy Qian Yang, Hua She, Marla Gearing, Emanuela
www.sciencemag.org/cgi/content/full/323/5910/124/dc1 Supporting Online Material for Regulation of Neuronal Survival Factor MEF2D by Chaperone-Mediated Autophagy Qian Yang, Hua She, Marla Gearing, Emanuela
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles
 Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles Long-term gene silencing shrna-specific design algorithm High titer, purified particles Thermo Scientific Dharmacon SMARTvector shrna
Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles Long-term gene silencing shrna-specific design algorithm High titer, purified particles Thermo Scientific Dharmacon SMARTvector shrna
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing
 Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing Chase L. Beisel, Yvonne Y. Chen, Stephanie J. Culler, Kevin G. Hoff, & Christina
Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing Chase L. Beisel, Yvonne Y. Chen, Stephanie J. Culler, Kevin G. Hoff, & Christina
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
What we ll do today. Types of stem cells. Do engineered ips and ES cells have. What genes are special in stem cells?
 Do engineered ips and ES cells have similar molecular signatures? What we ll do today Research questions in stem cell biology Comparing expression and epigenetics in stem cells asuring gene expression
Do engineered ips and ES cells have similar molecular signatures? What we ll do today Research questions in stem cell biology Comparing expression and epigenetics in stem cells asuring gene expression
Supporting Information
 Supporting Information Horie et al. 10.1073/pnas.1008499107 SI Materials and Methods ell ulture and Reagents. THP-1 cells were obtained from the American Type ell ollection. THP-1 cells were transformed
Supporting Information Horie et al. 10.1073/pnas.1008499107 SI Materials and Methods ell ulture and Reagents. THP-1 cells were obtained from the American Type ell ollection. THP-1 cells were transformed
Plasma EGFR T790M ctdna status is associated with clinical outcome in. advanced NSCLC patients with acquired EGFR-TKI resistance
 Plasma EGFR T790M ctdna status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance 1# D Zheng; 2# X Ye; 3 MZ Zhang; 2 Y Sun; 1 JY Wang; 1 J Ni; 1 HP Zhang;
Plasma EGFR T790M ctdna status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance 1# D Zheng; 2# X Ye; 3 MZ Zhang; 2 Y Sun; 1 JY Wang; 1 J Ni; 1 HP Zhang;
mmu-mir-200a-3p Real-time RT-PCR Detection and U6 Calibration Kit User Manual MyBioSource.com Catalog # MBS826230
 mmu-mir-200a-3p Real-time RT-PCR Detection and U6 Calibration Kit User Manual Catalog # MBS826230 For the detection and quantification of mirnas mmu-mir-200a-3p normalized by U6 snrna using Real-time RT-PCR
mmu-mir-200a-3p Real-time RT-PCR Detection and U6 Calibration Kit User Manual Catalog # MBS826230 For the detection and quantification of mirnas mmu-mir-200a-3p normalized by U6 snrna using Real-time RT-PCR
Do engineered ips and ES cells have similar molecular signatures?
 Do engineered ips and ES cells have similar molecular signatures? Comparing expression and epigenetics in stem cells George Bell, Ph.D. Bioinformatics and Research Computing 2012 Spring Lecture Series
Do engineered ips and ES cells have similar molecular signatures? Comparing expression and epigenetics in stem cells George Bell, Ph.D. Bioinformatics and Research Computing 2012 Spring Lecture Series
Transcriptional Regulation in Eukaryotes
 Transcriptional Regulation in Eukaryotes Concepts, Strategies, and Techniques Michael Carey Stephen T. Smale COLD SPRING HARBOR LABORATORY PRESS NEW YORK 2000 Cold Spring Harbor Laboratory Press, 0-87969-537-4
Transcriptional Regulation in Eukaryotes Concepts, Strategies, and Techniques Michael Carey Stephen T. Smale COLD SPRING HARBOR LABORATORY PRESS NEW YORK 2000 Cold Spring Harbor Laboratory Press, 0-87969-537-4
microrna-122 stimulates translation of Hepatitis C Virus RNA
 Supplementary information for: microrna-122 stimulates translation of Hepatitis C Virus RNA Jura Inga Henke 1,5, Dagmar Goergen 1,5, Junfeng Zheng 3, Yutong Song 4, Christian G. Schüttler 2, Carmen Fehr
Supplementary information for: microrna-122 stimulates translation of Hepatitis C Virus RNA Jura Inga Henke 1,5, Dagmar Goergen 1,5, Junfeng Zheng 3, Yutong Song 4, Christian G. Schüttler 2, Carmen Fehr
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Genome Engineering. Brian Petuch
 Genome Engineering Brian Petuch Guiding Principles An understanding of the details of gene engineering is essential for understanding protocols when making recommendations on: Risk assessment Containment
Genome Engineering Brian Petuch Guiding Principles An understanding of the details of gene engineering is essential for understanding protocols when making recommendations on: Risk assessment Containment
Technical Review. Real time PCR
 Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
Sensitivity vs Specificity
 Viral Detection Animal Inoculation Culturing the Virus Definitive Length of time Serology Detecting antibodies to the infectious agent Detecting Viral Proteins Western Blot ELISA Detecting the Viral Genome
Viral Detection Animal Inoculation Culturing the Virus Definitive Length of time Serology Detecting antibodies to the infectious agent Detecting Viral Proteins Western Blot ELISA Detecting the Viral Genome
Figure S1. sporulation frequency (%) 80. sme2-5. sme2-3. sme2
 Figure S1 N WT -5-3 - DSRless mei4 100 ssm4 MS2loop () (kb) 1.0 0.5 sporulation frequency (%) 80 60 40 20 rrna 1 2 3 4 5 6 7 8 0 WT -5-3 -DSRless Figure S1. The DSR motifs in are crucial for its function.
Figure S1 N WT -5-3 - DSRless mei4 100 ssm4 MS2loop () (kb) 1.0 0.5 sporulation frequency (%) 80 60 40 20 rrna 1 2 3 4 5 6 7 8 0 WT -5-3 -DSRless Figure S1. The DSR motifs in are crucial for its function.
The Effect of cdna on Tumor Cells Growth on Nude Mice
 The Effect of cdna on Tumor Cells Growth on Nude Mice Yiming Ding Department of Electric and Computer Engineering Portland State University, OR Email: dym@cecs.pdx.edu This project is assigned in the class
The Effect of cdna on Tumor Cells Growth on Nude Mice Yiming Ding Department of Electric and Computer Engineering Portland State University, OR Email: dym@cecs.pdx.edu This project is assigned in the class
Real-time PCR. TaqMan Protein Assays. Unlock the power of real-time PCR for protein analysis
 Real-time PCR TaqMan Protein Assays Unlock the power of real-time PCR for protein analysis I can use my real-time PCR instrument to quantitate protein? Do protein levels correlate with related mrna levels
Real-time PCR TaqMan Protein Assays Unlock the power of real-time PCR for protein analysis I can use my real-time PCR instrument to quantitate protein? Do protein levels correlate with related mrna levels
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 BALB/c LYVE1-deficient mice exhibited reduced lymphatic trafficking of all DC subsets after oxazolone-induced sensitization. (a) Schematic overview of the mouse skin oxazolone contact
Supplementary Figure 1 BALB/c LYVE1-deficient mice exhibited reduced lymphatic trafficking of all DC subsets after oxazolone-induced sensitization. (a) Schematic overview of the mouse skin oxazolone contact
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
Supplemental Data Supplementary Figure Legends and Scheme Figure S1.
 Supplemental Data Supplementary Figure Legends and Scheme Figure S1. UTK1 inhibits the second EGF-induced wave of lamellipodia formation in TT cells. A and B, EGF-induced lamellipodia formation in TT cells,
Supplemental Data Supplementary Figure Legends and Scheme Figure S1. UTK1 inhibits the second EGF-induced wave of lamellipodia formation in TT cells. A and B, EGF-induced lamellipodia formation in TT cells,
TOOLS sirna and mirna. User guide
 TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation
 1 2 3 4 5 SUPPLEMENTAL DATA Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation Magalí Nazar, Juan Pablo Nicola, María Laura
1 2 3 4 5 SUPPLEMENTAL DATA Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation Magalí Nazar, Juan Pablo Nicola, María Laura
7.06 Problem Set #3, Spring 2005
 7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
Bronchial epithelium and its associated tissues act as a
 The Journal of Immunology A JNK-Independent Signaling Pathway Regulates TNF -Stimulated, c-jun-driven FRA-1 Protooncogene Transcription in Pulmonary Epithelial Cells 1 Pavan Adiseshaiah,* Dhananjaya V.
The Journal of Immunology A JNK-Independent Signaling Pathway Regulates TNF -Stimulated, c-jun-driven FRA-1 Protooncogene Transcription in Pulmonary Epithelial Cells 1 Pavan Adiseshaiah,* Dhananjaya V.
Recent technology allow production of microarrays composed of 70-mers (essentially a hybrid of the two techniques)
 Microarrays and Transcript Profiling Gene expression patterns are traditionally studied using Northern blots (DNA-RNA hybridization assays). This approach involves separation of total or polya + RNA on
Microarrays and Transcript Profiling Gene expression patterns are traditionally studied using Northern blots (DNA-RNA hybridization assays). This approach involves separation of total or polya + RNA on
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms
 Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on
 Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
Supporting Information
 Supporting Information Ho et al. 1.173/pnas.81288816 SI Methods Sequences of shrna hairpins: Brg shrna #1: ccggcggctcaagaaggaagttgaactcgagttcaacttccttcttgacgnttttg (TRCN71383; Open Biosystems). Brg shrna
Supporting Information Ho et al. 1.173/pnas.81288816 SI Methods Sequences of shrna hairpins: Brg shrna #1: ccggcggctcaagaaggaagttgaactcgagttcaacttccttcttgacgnttttg (TRCN71383; Open Biosystems). Brg shrna
LATE-PCR. Linear-After-The-Exponential
 LATE-PCR Linear-After-The-Exponential A Patented Invention of the Laboratory of Human Genetics and Reproductive Biology Lab. Director: Lawrence J. Wangh, Ph.D. Department of Biology, Brandeis University,
LATE-PCR Linear-After-The-Exponential A Patented Invention of the Laboratory of Human Genetics and Reproductive Biology Lab. Director: Lawrence J. Wangh, Ph.D. Department of Biology, Brandeis University,
Theoretical cloning project
 Theoretical cloning project Needed to get credits Make it up yourself, don't copy Possible to do in groups of 2-4 students If you need help or an idea, ask! If you have no idea what to clone, I can give
Theoretical cloning project Needed to get credits Make it up yourself, don't copy Possible to do in groups of 2-4 students If you need help or an idea, ask! If you have no idea what to clone, I can give
Supplemental Figure 1. Overview of the ΔSHAPE analysis workflow used to compare SHAPE- MaP datasets.
 Supporting Information for: Detection of RN-protein interactions in living cells with SHPE Matthew J. Smola 1, J. Mauro alabrese, and Kevin M. Weeks 1,3 1 Department of hemistry, niversity of North arolina,
Supporting Information for: Detection of RN-protein interactions in living cells with SHPE Matthew J. Smola 1, J. Mauro alabrese, and Kevin M. Weeks 1,3 1 Department of hemistry, niversity of North arolina,
