|
|
|
- Briana Cook
- 6 years ago
- Views:
Transcription
1 DOI: 0.038/ncb337 a RFP-Zyxin Actin b RFP Fluorescence non-swirling CCW swirling, positive tilting of radial fibers c GFP-VASP Actin d GFP Fluorescence non-swirling CCW swirling, positive tilting of radial fibers Supplementary Figure Overexpression of radial fiber components, Zyxin or VASP, is not sufficient to reverse chirality. RFP-Zyxin (a) and GFP-VASP (c) localized to radial fibers and focal adhesions. Actin was visualized either by co-transfection with LifeAct-GFP (green in a) or phalloidin staining (red in c). Scale bars, 0 μm. RFP-Zyxin (b) or GFP-VASP (d) expressing cells were filmed to follow their actin dynamics. The expression level in an individual cell was assessed by measuring either total RFP or total GFP fluorescence intensity respectively. The y-axis represents fluorescence intensity in a log scale; the cells were sorted in ascending order of their respective protein expression level along the x-axis. Each bar on the histogram represents an individual cell. Color-coding indicates the type of actin cytoskeleton dynamics for each cell. Green cells that do not demonstrate swirling during the entire period of observation (~0 hours). Blue cells demonstrating positive tilt of radial fibers and counter-clockwise swirling similar to control cells. No clockwise swirling events were observed in these experiments. 58 cells assessed for RFP-Zyxin, and 72 cells assessed for GFP-VASP Macmillan Publishers Limited. All rights reserved
2 a e b c d a a b f c d Supplementary Figure 2 Electron tomography showing 3D morphology and organization of transverse and radial fibers. (a-d) Surface representations of overlapping 3D electron tomographic reconstructions running along radial fibers. The relative spatial arrangement of the four reconstructions is shown below d. The center of the cell is towards the left of a, while the cell edge is shown in d. The height (in nm) of the features are mapped according to the color key on the right of a. The area of a reconstructed image is 2.8 µm 2. Radial (RF) and transverse (TF) fibers as well as the transversal layer (TL) are indicated in b. Nearer to the cell center (a,b), the transverse and radial fibers (blue and cyan) are above the transversal layer (orange). Towards the cell edge (c,d), the two fiber systems move closer to the substrate (color shift from blue/cyan to cyan/green and green/orange) and finally appear below the orange transversal layer plane (in d) when the focal adhesions contact the substrate. (e) Cross-section of the reconstruction shown in a. The upper panel shows a 0 nm thick cross-section along the long axis of the radial fibers to the top of a, perpendicular to the transverse fibers, as indicated in the scheme at the upper left corner of e. The transverse layer (TL) and the substrate (S) locations are marked at the sides of the figure. The location of the radial fiber (RF) and the location where the transverse fiber (TF) runs perpendicular to the image plane are also marked. Transverse fibers appear to interact with radial fibers. The height (in nm) is shown in the bar on the right. The bar is color-coded for comparison with a-d. The width of the cross-section is.9 µm. The components of the radial fibers go in and out of the cross-section, generating an alternating pattern of black and white regions. The lower panel shows the model derived from the tomographic reconstructions in a view similar to that in the upper panel for comparison. (f) XY slices through the tomographic reconstruction (each 2 nm thick) shown in b (see also Supplementary Video 4). The heights of the sections are indicated by the color of the rim, which correspond to the color-coding in a and e. Orthogonal side views are also shown for each slice on the top and to the right. The location of the XZ slices (top) within the XY view are indicated by the end of the red lines at the right of the XY view; the location of the YZ slices (right) are indicated by the end of the green lines at the top of the XY view. Radial fibers (RF), transversal layer (TL), as well as the top and bottom occurrences of the transverse fibers (TF) are marked on the slices. See also Supplementary Video 3 for comparison with fluorescence microscopy Macmillan Publishers Limited. All rights reserved
3 a b c d e cell periphery 0 sec 4 sec 8.5 sec Supplementary Figure 3 Probing actin fibers in permeabilized cells with myosin VI-coated nanoparticles. (a) Movement trajectories (green) of fluorescent nanoparticles coated with myosin VI along the actin cytoskeleton (red) of a fixed extracted cell. The white box marks the view shown in b. Scale bar, 0 μm. (b-e) High magnification view of myosin VI nanoparticle movement along the single transverse fiber. (b) A view of actin cytoskeleton (red) after extraction. White-boxed region: The transverse fiber decorated with myosin VI nanoparticles (green) is marked by the dotted line. (c-e) Sequential images from the live imaging of myosin VI nanoparticles moving along the transverse fiber. Nanoparticles # and #2 (white arrowheads) are moving away from each other. Scale bars, μm. The full-length sequence is shown in Supplementary Video Macmillan Publishers Limited. All rights reserved
4 Untreated control Nocodazole-treated Actin Microtubule Actin Microtubule Supplementary Figure 4 Self-organization of the actin cytoskeleton in cells with depolymerized microtubules. Disruption of microtubules by nocodazole was confirmed by anti-α-tubulin immunofluorescence staining. Actin was visualized with phalloidin staining. The radial and chiral patterns of the actin cytoskeleton organization in cells with an intact microtubule system and in cells lacking microtubules are shown. Scale bars, 0 μm. See also Supplementary Video Macmillan Publishers Limited. All rights reserved
5 a α-actinin-: Actin binding (ABD) Integrin binding CH CH R R2 R3 R4 CaM Anti-parallel dimer of α-actinin- ABDdel-α-actinin-: R R2 R3 R4 CaM SR2: R R2 b GFP-ABDdel-α-actinin- Vinculin Myo-IIA Actin c Percentage of fiber (%) Angle of tilt ( ) GFP-ABDdel-α-actinin- single-ended stress-fiber Supplementary Figure 5 Long single-ended stress-fibers replace radial fibers in GFP-ABDdel-α-actinin- expressing cells. (a) Schematic diagram illustrating the molecular organization of α-actinin- and its mutants. The α-actinin- monomer has an N-terminal actin-binding domain (ABD) composed of two calponin-homology (CH) domains, a central rod consisting of four spectrin repeats (R-R4), and a C-terminal calmodulin (CaM)- like domain. α-actinin- monomers form an anti-parallel dimer with the ABD domain present at both ends. The ABDdel-α-actinin- mutant lacks the ABD domain 4. Spectrin repeats and 2 are responsible for integrin binding. The SR2 mutant consists only of these spectrin repeats (R and R2) 4. (b) Cells expressing GFP-ABDdel-α-actinin- were extracted prior to fixation to reduce background due to the presence of excess soluble GFP- ABDdel-α-actinin-. Truncated GFP-ABDdel-α-actinin- mutant protein localized to long actin fibers (purple). Unlike radial fibers, these fibers also contained myosin-iia (blue). However, unlike ventral stress-fibers, they were associated with only one focal adhesion (labeled by vinculin, red). Thus, we classified this type of actin fibers as single-ended stress-fibers. Scale bar, 0 μm. (c) Histogram showing the distribution of single-ended actin fiber tilt angle ( ) in a cell expressing GFP-ABDdel-α-actinin-. 30 actin fibers of a representative cell were analyzed. See also Supplementary Video 2. Note that unlike radial fibers in cells overexpressing the fulllength α-actinin-, the single-ended stress-fibers in cells expressing GFP-ABDdel-α-actinin- tilt in the same direction as radial fibers in control cells (see Fig. 6c) Macmillan Publishers Limited. All rights reserved
6 a b 0 time (min) 30 0 time (min) 30 Supplementary Figure 6 The effect of SR2-GFP α-actinin- fragment on actin cytoskeleton self-organization. (a,b) Actin organization (visualized by tdtomato-f-tractin) in SR2-GFP expressing cells and high magnification time-series images illustrating actin dynamics (white-boxed region). 60% of SR2-GFP transfected cells demonstrated phenotypes similar to control cells (30% counter-clockwise swirling and 30% non-swirling). The remaining 40% of the transfected cells were found to express a higher level (2-fold on average) of SR2-GFP. 60% of these higher SR2-GFP expressing cells failed to progress beyond the circular stage (a) while the remaining 40% exhibited similar behavior to GFP-ABDdel-α-actinin- mutant expressing cells, forming characteristic single-ended stress-fibers that tilt in a positive direction (b). An example of two single-ended stress-fibers are shown here (white and cyan arrowheads). Scale bars, 0 μm; 5 μm (high-magnification). n = 39 transfected cells pooled from 2 independent experiments Macmillan Publishers Limited. All rights reserved
7 Brightfield 0 min 4 min 38 min * ** * * * Actin Supplementary Figure 7 Membrane-bound beads move in a chiral motion coupled to the swirling of the actin network. Fibronectin-coated beads (4.5 μm in diameter) moved in a counter-clockwise direction on the cell surface as the actin cytoskeleton swirled in the same direction. Yellow, red and cyan asterisks (*) marks the bead position at time points 0-, 4- and 38-minute respectively. The arrow indicates the direction of the bead movement. Scale bar, 0 μm Macmillan Publishers Limited. All rights reserved
8 < < SUPPLEMENTARY INFORMATION a Immobilized formin dimer and unconstrained filament x Stair-stepping filament rotation Radial fiber elongation b Immobilized formin dimer and constrained filament screwstep screwstep x Radial fiber elongation Screw-stepping filament rotation Supplementary Figure 8 Schematic illustration depicting polymerization of individual actin filaments nucleated by formins. (a) Polarized actin filaments (green) capped by a formin dimer (blue-red). The first three actin subunits in the two actin helices are numbered -3 and -3 respectively. The formin dimer is immobilized while the opposite end of the actin filament is not. Unconstrained actin filaments rotate in the positive angular direction about -axis (magenta arrow), as experimentally shown by Mizuno et al. 50 (b) Actin filaments are prevented from rotating via immobilization of barbed end by formin and trapping of opposite end by α-actinin cross-linking. Immobile actin subunits are labeled in yellow. Torsional elastic energy accumulated during polymerization is periodically released via high amplitude rotation ( screw-step highlighted by gray background) in the negative direction (yellow arrow). See details in Shemesh et al. 49. See also Supplementary Videos 7 and 8) Macmillan Publishers Limited. All rights reserved
9 a Membrane Membrane 2 anti-actn4 anti-tubulin 250 kd 50 kd 00 kd 75 kd 50 kd 37 kd 25 kd anti-actn anti-tubulin 250 kd 50 kd 00 kd 75 kd 50 kd 37 kd 25 kd b Lower exposure Membrane Membrane kd 50 kd 00 kd 75 kd 50 kd 37 kd 250 kd 50 kd 00 kd 75 kd 50 kd 37 kd 25 kd 25 kd Region of interest M = marker = control sirna 2 = ACTN sirna c Higher exposure Membrane Membrane 2 Supplementary Figure 9 Primary immunoblotting data (a) 0 μg samples of protein cell lysates from control sirna- and ACTN sirna-transfected cells were separated on SDS-PAGE gel and transferred to PVDF membranes. Positions of molecular weight markers are indicated to the right of each PVDF membrane. (b) Immunoblotting was performed with corresponding antibodies, anti-actn, anti-actn4 and anti-α-tubulin, and developed using standard ECL procedure. Scans of uncropped immunoblot is shown here. Data from this immunoblot was used in Fig. 7c Macmillan Publishers Limited. All rights reserved
10 Supplementary Video Legends Supplementary Video A typical example of the self-organization of the actin cytoskeleton in cell on a circular adhesive island. Images were recorded at 2 minute intervals over a period of ~ hours. Display rate is 30 frames/sec and corresponds to the time-lapse series in Fig. b. Supplementary Video 2 Evolution of the actin cytoskeleton from the radial into the chiral pattern. Images were recorded at 2 minute intervals over a period of ~ 4 hours. Display rate is 7 frames/sec and corresponds to the time-lapse series in Fig. 5 (untreated). Supplementary Video 3 Organization of radial and transverse fibers seen in confocal microscopy Z-section. Cells were fixed and stained with phalloidin to visualize actin. Images of the actin structures at the cell edge of non-dehydrated cell on a circular island were captured by Z-stepping at 0.25 μm per step from the bottom to the top of the cell. Supplementary Video 4 Electron tomographic reconstruction of intersection between transverse and radial fibers. Movie showing successive 2-nm slices of the tomographic reconstruction (0 frames/sec) as shown in Supplementary Fig. 2f. The image display starts from the top, goes to the bottom, and then back up to the top. Supplementary Video S5 Movement of myosin V- or VI-coated nanoparticles (green) along filopodial cell projections (red) of fixed extracted cell. An asterisk (*) indicates a filopodium. Myosin V-coated nanoparticles moved towards filament barbed ends away from the cell body, while myosin VI-coated nanoparticles moved towards filament pointed ends in the direction of the cell body. Images were recorded at half a second intervals. Display rate is 30 frames/sec. Supplementary Video 6 Movement of myosin V- (left) and myosin VI-coated (right) nanoparticles (green) along the actin transverse fiber (red) of fixed extracted cell. Note that the movements of both myosin V- and VI-coated nanoparticles could proceed in opposite directions along the same transverse fibers. Images were recorded at half a second intervals. Display rate is 7 frames/sec and corresponds to Fig. 3d-f (myosin V) and Supplementary Fig. 3b-e (myosin VI). Supplementary Video 7 Actin dynamics upon treatment with the 0 μm formin inhibitor, SMIFH2. Images were recorded at 2 minute intervals over a period of ~ 3.5 hours. Display rate is 20 frames/sec and corresponds to the time-lapse series in Fig. 4a (lower panel). Supplementary Video 8 Growth of radial fiber is dependent on formin activity. SMIFH2-treated cell was monitored for ~ hour prior to drug washout. The recovery phase was followed for ~ 4 hours with images taken at 2 minute interval. Display rate is 30 frames/sec. Note the recovery of radial fibers growth started at ~ 3 hours after drug washout, at the cell periphery. Supplementary Video 9 Actin dynamics upon treatment with 00 μm Y27632 (left) or 50 μm blebbistatin (Bleb, right). Images were recorded at 2 minute intervals over a period of ~ 5 hours. Display rate is 20 frames/sec and corresponds to the time-lapse series in Fig. 5. Supplementary Video 0 Actin dynamics upon treatment with μm nocodazole. Images were recorded at 2 minute intervals over a period of ~ 5 hours. Note that depolymerization of microtubules does not interfere with formation of chiral pattern and does not change handedness of the actin swirl. Display rate is 20 frames/sec and corresponds to the time-lapse series in Fig. 5. Supplementary Video Handedness of the chiral pattern in cell expressing low (left) or high (right) level of GFP-α-actinin-. Radial fibers were visualized with GFP-α-actinin-. Images were recorded at 2 minute intervals over a period of ~ 6 hours. Display rate is 20 frames/sec and corresponds to Fig. 6a. Supplementary Video 2 A typical example of actin dynamics in cell expressing truncated mutant GFP-ABDdel-α-actinin-. Images were recorded at 4 minute intervals over a period of ~ 9 hours. Display rate is 6 frames/sec and corresponds to the time-lapse series in Fig. 7a. Supplementary Video 3 A typical example of actin dynamics in α-actinin- knockdown cell. Images were recorded at 3 minute intervals over a period of ~ 9 hours. Display rate is 8 frames/sec and corresponds to the time-lapse series in Fig. 7f. Supplementary Video 4 Rotation of the microtubule array in the counter-clockwise direction, together with counter-clockwise swirling of the actin network, seen in cell expressing GFP-tubulin and tdtomato-f-tractin to visualize microtubule and actin respectively. Images were recorded at 2 minute intervals over a period of ~ 2 hours. Display rate is 30 frames/sec. Supplementary Video 5 Simulation of the chiral self-organization of the actin cytoskeleton. Corresponds to Fig. 8b. Supplementary Video 6 Simulation showing polymerization of a free actin filament capped by formin-fh2 dimer. Actin monomers are shown in green (dark and light green are used to distinguish the long pitch helices). FH2 dimer is shown in blue-red. Note the rotation of the formin dimer. Supplementary Video 7 Simulation showing polymerization of actin filament capped by an immobilized formin-fh2 dimer. Actin monomers are shown in green (dark and light green are used to distinguish the long pitch helices). FH2 dimer is shown in blue-red. Note the rotation of the actin filament. Supplementary Video 8 Simulation showing polymerization of actin filament capped by an immobilized formin-fh2 dimer, and prevented from rotating at the pointed end. Actin monomers are shown in green (dark and light green are used to distinguish the long pitch helices). FH2 dimer is shown in blue-red. Immobilized monomers at the tip of the filament are shown in yellow. Note the periodic high amplitude rotation of the actin filament upon transit release of the contact of its barbed end with formin dimer ( screw-steps ; highlighted by change in background color from black to light gray) Macmillan Publishers Limited. All rights reserved
11 Supplementary Note for Cellular chirality arising from the self-organization of the actin cytoskeleton Yee Han Tee, Tom Shemesh, Visalatchi Thiagarajan, Rizal Fajar Hariadi, Karen L. Anderson, Christopher Page, Niels Volkmann, Dorit Hanein, Sivaraj Sivaramakrishnan, Michael M. Kozlov, Alexander D. Bershadsky 205 Macmillan Publishers Limited. All rights reserved
12 COMPUTATIONAL MODEL Computational procedure. Each discrete computational step contains the following transformations of the system:. A new TF is formed between each pair of mature FAs with a probability r!"# Δt, where Δt is the time corresponding to one computational step and r!"# is the rate of TF generation. 2. The internal stress within all the TFs including the newly generated ones is set to be equal γ = γ. The length L of each TF is then determined by the position of its endpoints bound to two RFs. The effective relaxed length L! of each TF, is set such that!!!!!! k! = γ. 3. The force on each TF endpoint, f!, is calculated by taking the component of the internal stress γ parallel to the local direction of the RF to which it is bound. 4. The total force, F!, acting on each RF is calculated as the sum of the forces applied by all the TFs that are bound along its length, F! =!!"#$%!"! f!. 5. The endpoints of each TF advance along the RFs to a distance Δs = βf! Δt meaning that the velocity of each endpoint is proportional to the force f! applied to it, according to the linear friction law. 6. The new relaxed length L! of each TF is adjusted to the new distance between its ends such that the TF internal stress remains constant and equal γ. 7. The total elastic energy of the system is calculated as the sum of the stretching energies of all the TFs and of the bending energies of the RFs: ε!"! =!!!!!! k!!!!!!!!"!!",!!!!!"# + k!"#$ c! (t)! dt where the integration in the second terms is performed along the RF length. 8. The change of configurations of all RFs is determined numerically by minimizing the total energy ε!"! with respect to the RF shapes. 205 Macmillan Publishers Limited. All rights reserved
13 9. Each FA is grown or disintegrated, depending on whether the force applied to it by the attached RFs exceeds or is lower than a threshold growth force, f, according to the model suggested in Shemesh et al., 2, r!" = r!"#$ Γ 0 > f!!!"#!!"# 0 Γ 0 = f!!!"#!!"# r!!!"#$ Γ 0 < f!!!"#!!"# 0. A pair of RFs whose ends are connected by a TF, and are closer than a minimal distance!"#$, are fused with a probability related to the angle between them, φ : where p! is the rate of fusion attempts p!"#$ = p! Δt e!!!! Un-fused RFs are elongated by polymerization at a rate r!"# = 0 Γ 0 < Γ!"# a Γ 0 Γ!"# Γ!"# < Γ 0 < Γ!"# r!"# Γ 0 > Γ!"# and contract locally at rate r!!!"# (s) = b Γ s Γ where a, b are fitting parameters in the model. Detailed description of chiral asymmetry model. In the rack and pinion picture, rotation of individual filaments in RF about their axis (local direction x, see Supplementary Figure 8) generates a frictional force on the TF in the local direction y. Since RF are bound to TF at points x > 0, a torque develops about their anchoring to the FA at x = 0, corresponding to a CCW rotation pattern for f! < 0; and a CW rotation pattern for f! > 0. A general friction mechanism coupling the rotation of the filaments to the TF may be expressed as f(t) = β ω(t) (az)!!! ω(t) (az) () 205 Macmillan Publishers Limited. All rights reserved
14 where a is the radius of the filament, β is a friction coefficient and α is the exponent describing the velocity dependence of the friction force. Formin processive cappers continuously track the growing barbed ends of actin filaments. Due to the helical structure of the F-actin filaments, the orientation of formins relative to the bulk of the filaments must rotate with the addition of each new actin monomer. When not constrained, a formin capper tracks the right-handed long-pitch actin helix in a stairstepping mode, so that addition of each new actin monomer results in ~ 4 rotation of the formin about x (Supplementary Video 6). Conversely, if the formin dimer is immobilized while the pointed end of the filament is free to rotate, the bulk of the polymerizing actin filament will rotate along its axis in opposite direction (Supplementary Figure 8a and Supplementary Video 7). The angular velocity of a filament under such conditions is given by ω!! =!"!!! x (2) where τ!! is the time associated with a single stair-step motion of the formin. The frictional force exerted on the tsf, f!"##, is then f!"## = β ω!! a! y (3), thereby promoting CCW patterns. We next consider a filament that, in addition to being capped by an immobilized formin, is prevented from rotating at its pointed end. Under these conditions, continuous polymerization of the filament results in accumulation of torsional strain in the bulk of the filament. Moreover, the zero-rotation constraint may be applied at any distance from the growing barbed end and not necessarily at the pointed tip, resulting in strain being accrued at the region between the formin and the non-rotating region. We propose that 205 Macmillan Publishers Limited. All rights reserved
15 cross-linking between actin filaments within a bundle can effectively prevent the rotation of the individual filaments. A possible mechanism for relieving the torsional strain is suggested from the symmetry of the formin dimer. The configuration of the formin-actin complex after a stair-stepping polymerization step in which formin and actin rotate by 4 relative to each other is identical to that resulting from a polymerization step with a relative rotation of 66 in the opposite direction. We have previously suggested that employing such inverse rotations, termed screw-steps can limit the strain in the filaments (Supplementary Figure 8b and Supplementary Video 8). The optimal regime for processive capping under these constraints was found to consist of ~2 stair-steps followed by one screwstep 3. In this regime, the rotation velocity of a bound filament alternates according to: ω!"#$% t = ω!! 0 < t < 2 τ!! ω!" 2 τ!! < t < 2 τ!! + τ!" (4) where τ!" is the time required for a single screw-step of formin, and ω!" is the angular velocity of the filament bulk during a screw-step, given by ω!" =!""!!" x (5) The time average friction force produced by an actin filament under such conditions is given by:!"! f!"#$% =!!!!!" f!"#$% t dt! (2τ!! + τ!" ) (6) Using (Eq. -5) in (Eq. 6), we get f!"#$% =!!!!!!!!!"!!!!!!"!!!"!"!!!!!!" y (7) 205 Macmillan Publishers Limited. All rights reserved
16 Assuming the time for each polymerization step to be constant, τ!! = τ!", then for any α > we find that f!"#$! > 0. While, in fact, a stair-step may require less time than a screw-step due to energetic barriers to the latter, this has only a weak effect on the minimal force-velocity exponent. Moreover - while we treat here a specific mechanism for relaxation of the stresses within a bound, polymerizing filament, the friction force produced by such filaments is expected, in general, to promote CW swirl patterns in the system. The fraction of the filaments in the RF that are immobilized by cross-linkers, λ, is expected to correspond to the expression levels of α-actinin. The mean force, F, exerted on the TF in the system by all the rotating filaments is then given by: F = λ f!"#$% + ( λ) f!"## (8) As expression level of α-actinin cross-linkers increases, this force changes sign, and the system transitions from favoring counter-clockwise to clockwise patterns. 205 Macmillan Publishers Limited. All rights reserved
17 REFERENCES. Shemesh, T., Verkhovsky, A.B., Svitkina, T.M., Bershadsky, A.D. & Kozlov, M.M. Role of focal adhesions and mechanical stresses in the formation and progression of the lamellipodium-lamellum interface [corrected]. Biophysical journal 97, (2009). 2. Shemesh, T., Bershadsky, A.D. & Kozlov, M.M. Physical model for self-organization of actin cytoskeleton and adhesion complexes at the cell front. Biophysical journal 02, (202). 3. Shemesh, T., Otomo, T., Rosen, M.K., Bershadsky, A.D. & Kozlov, M.M. A novel mechanism of actin filament processive capping by formin: solution of the rotation paradox. The Journal of cell biology 70, (2005). 205 Macmillan Publishers Limited. All rights reserved
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing
 Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Cellular chirality arising from the self-organization of the actin cytoskeleton
 A RT I C L E S Cellular chirality arising from the self-organization of the actin cytoskeleton Yee Han Tee,7, Tom Shemesh 2,7,8, Visalatchi Thiagarajan, Rizal Fajar Hariadi 3, Karen L. Anderson 4, Christopher
A RT I C L E S Cellular chirality arising from the self-organization of the actin cytoskeleton Yee Han Tee,7, Tom Shemesh 2,7,8, Visalatchi Thiagarajan, Rizal Fajar Hariadi 3, Karen L. Anderson 4, Christopher
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Prospéri et al.,
 Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 )
 Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Lecture 13. Motor Proteins I
 Lecture 13 Motor Proteins I Introduction: The study of motor proteins has become a major focus in cell and molecular biology. Motor proteins are very interesting because they do what no man-made engines
Lecture 13 Motor Proteins I Introduction: The study of motor proteins has become a major focus in cell and molecular biology. Motor proteins are very interesting because they do what no man-made engines
T H E J O U R N A L O F C E L L B I O L O G Y
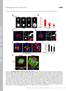 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
SUPPLEMENTARY INFORMATION
 DOI: 1.18/ncb46 a Damage Fracture s s 7 5 16 1 9 26 5 µm 5 µm b Position of photo-damage site in cells 15 Counts 1 5-14 -12-1 -8-6 -4-2 Distance from damage site to tip (µm) Supplementary Figure 1 Effect
DOI: 1.18/ncb46 a Damage Fracture s s 7 5 16 1 9 26 5 µm 5 µm b Position of photo-damage site in cells 15 Counts 1 5-14 -12-1 -8-6 -4-2 Distance from damage site to tip (µm) Supplementary Figure 1 Effect
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supporting Information
 Supporting Information Shao et al. 10.1073/pnas.1504837112 SI Materials and Methods Immunofluorescence and Immunoblotting. For immunofluorescence, cells were fixed with 4% paraformaldehyde and permeabilized
Supporting Information Shao et al. 10.1073/pnas.1504837112 SI Materials and Methods Immunofluorescence and Immunoblotting. For immunofluorescence, cells were fixed with 4% paraformaldehyde and permeabilized
J. Cell Sci. 128: doi: /jcs : Supplementary Material. Supplemental Figures. Journal of Cell Science Supplementary Material
 Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
SUPPLEMENTARY INFORMATION
 a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
supplementary information
 DOI: 10.1038/ncb1977 Figure S1 a. Immunofluorescence analysis of IFT20 localization in PBL costained with anti-β-tubulin antibodies. b. Immunofluorescence analysis of IFT20 localization in Jurkat cells,
DOI: 10.1038/ncb1977 Figure S1 a. Immunofluorescence analysis of IFT20 localization in PBL costained with anti-β-tubulin antibodies. b. Immunofluorescence analysis of IFT20 localization in Jurkat cells,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Supplementary Materials for
 www.sciencemag.org/cgi/content/full/science.1245533/dc1 Supplementary Materials for A Mechanism for Reorientation of Cortical Microtubule Arrays Driven by Microtubule Severing Jelmer J. Lindeboom, Masayoshi
www.sciencemag.org/cgi/content/full/science.1245533/dc1 Supplementary Materials for A Mechanism for Reorientation of Cortical Microtubule Arrays Driven by Microtubule Severing Jelmer J. Lindeboom, Masayoshi
supplementary information
 DOI: 10.1038/ncb1992 Figure S1 (a-b) Verification of actin flow patter using photobleaching of actin:gfp and Lifeact:RFP in migrating dendritic cells (DCs). (a) Actin:GFP and Lifeact:RFP double-trafected
DOI: 10.1038/ncb1992 Figure S1 (a-b) Verification of actin flow patter using photobleaching of actin:gfp and Lifeact:RFP in migrating dendritic cells (DCs). (a) Actin:GFP and Lifeact:RFP double-trafected
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplemental material JCB. Pitaval et al., Cell shape and ciliogenesis Pitaval et al.
 Supplemental material THE JOURNAL OF CELL BIOLOGY Pitaval et al., http://www.jcb.org/cgi/content/full/jcb.201004003/dc1 S1 Figure S1. Cell cycle exit analysis. (A) Experimental procedure. RPE1 cells were
Supplemental material THE JOURNAL OF CELL BIOLOGY Pitaval et al., http://www.jcb.org/cgi/content/full/jcb.201004003/dc1 S1 Figure S1. Cell cycle exit analysis. (A) Experimental procedure. RPE1 cells were
T H E J O U R N A L O F C E L L B I O L O G Y
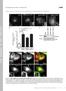 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
supplementary information
 DOI: 10.1038/ncb2156 Figure S1 Depletion of p114rhogef with different sirnas. Caco-2 (a) and HCE (b) cells were transfected with individual sirnas, pools of the two sirnas or the On Target (OnT) sirna
DOI: 10.1038/ncb2156 Figure S1 Depletion of p114rhogef with different sirnas. Caco-2 (a) and HCE (b) cells were transfected with individual sirnas, pools of the two sirnas or the On Target (OnT) sirna
Ac(n cytoskeleton 12/7/11. The Cytoskeleton. Func0ons of the Cytoskeleton. B Visegrady. Elements of the Cytoskeleton
 The Cytoskeleton Ac(n cytoskeleton B Visegrady The cytoskeleton is an intricate network of protein filaments that extends throughout the cytoplasm. A skin cell (fibroblast) stained with Coomassie blue.
The Cytoskeleton Ac(n cytoskeleton B Visegrady The cytoskeleton is an intricate network of protein filaments that extends throughout the cytoplasm. A skin cell (fibroblast) stained with Coomassie blue.
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Prashar et al., http://www.jcb.org/cgi/content/full/jcb.201304095/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. FBT phagocytosis in cells expressing PM-GFP. (A) T-PC
Supplemental material Prashar et al., http://www.jcb.org/cgi/content/full/jcb.201304095/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. FBT phagocytosis in cells expressing PM-GFP. (A) T-PC
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Paul et al.,
 Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Biophysics of contractile ring assembly
 Biophysics of contractile ring assembly Dimitrios Vavylonis Department of Physics, Lehigh University October 1, 2007 Physical biology of the cell Physical processes in cell organization and function: Transport
Biophysics of contractile ring assembly Dimitrios Vavylonis Department of Physics, Lehigh University October 1, 2007 Physical biology of the cell Physical processes in cell organization and function: Transport
Supplementary Figure 1. Two activation pathways and four conformations of β 2 integrins. KIM127 (red) can specifically detect
 Supplementary Figure 1 Two activation pathways and four conformations of β 2 integrins. KIM127 (red) can specifically detect integrin extension (E + ) and mab24 (green) can specifically detect headpiece-opening
Supplementary Figure 1 Two activation pathways and four conformations of β 2 integrins. KIM127 (red) can specifically detect integrin extension (E + ) and mab24 (green) can specifically detect headpiece-opening
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in
 Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking
 Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Sequence of C-terminal tail regions of myosin heavy chains in class XI of Nicotiana benthamiana (Nb
 Fig. S1 Sequence of C-terminal tail regions of myosin heavy chains in class XI of Nicotiana benthamiana (Nb myosin XI-2, -F and K) and BY-2 cell (Nt 170-kD myosin and Nt 175-kD myosin). Amino acids identical
Fig. S1 Sequence of C-terminal tail regions of myosin heavy chains in class XI of Nicotiana benthamiana (Nb myosin XI-2, -F and K) and BY-2 cell (Nt 170-kD myosin and Nt 175-kD myosin). Amino acids identical
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Mechanism for signal-induced opening of the DNA box. a, An atomic model of the DNA box held closed by locks (orange and blue) that are double helices formed by two short strands
Supplementary Figure 1. Mechanism for signal-induced opening of the DNA box. a, An atomic model of the DNA box held closed by locks (orange and blue) that are double helices formed by two short strands
Fast, three-dimensional super-resolution imaging of live cells
 Nature Methods Fast, three-dimensional super-resolution imaging of live cells Sara A Jones, Sang-Hee Shim, Jiang He & Xiaowei Zhuang Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3
Nature Methods Fast, three-dimensional super-resolution imaging of live cells Sara A Jones, Sang-Hee Shim, Jiang He & Xiaowei Zhuang Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3
SUPPLEMENTARY INFORMATION
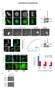 SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
b number of motif occurrences
 supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
BME Engineering Molecular Cell Biology. The Cytoskeleton (I): Actin The Cytoskeleton (II): Microtubule & Intermediate Filament
 BME 42-620 Engineering Molecular Cell Biology Lecture 09: The Cytoskeleton (I): Actin The Cytoskeleton (II): Microtubule & Intermediate Filament BME42-620 Lecture 09, September 27, 2011 1 Outline Overviewofcytoskeletal
BME 42-620 Engineering Molecular Cell Biology Lecture 09: The Cytoskeleton (I): Actin The Cytoskeleton (II): Microtubule & Intermediate Filament BME42-620 Lecture 09, September 27, 2011 1 Outline Overviewofcytoskeletal
Plutoni et al., http :// /cgi /content /full /jcb /DC1
 Supplemental material JCB Plutoni et al., http ://www.jcb.org /cgi /content /full /jcb.201505105 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. Cell morphology during migration. (a) Protein extracts (20
Supplemental material JCB Plutoni et al., http ://www.jcb.org /cgi /content /full /jcb.201505105 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. Cell morphology during migration. (a) Protein extracts (20
Supplementary Figure S1. Hoechst. cells/field. Myogenin. % of Myogenin+ cells. 0 h chase CONTROL. BrdU. 72 h chase. CONTROL BrdU CONTROL BrdU
 Hoechst Myogenin amyhc 24 h 48 h 72 h b cells/field 50 40 30 20 10 0 24h 48h 72h h after plating (4 days growth+ t) c e CONTROL 0 h chase % of Myogenin+ cells h after plating (4 days growth+ t) CONTROL
Hoechst Myogenin amyhc 24 h 48 h 72 h b cells/field 50 40 30 20 10 0 24h 48h 72h h after plating (4 days growth+ t) c e CONTROL 0 h chase % of Myogenin+ cells h after plating (4 days growth+ t) CONTROL
Cytoskeleton. B. Balen
 Cytoskeleton B. Balen structural backbone of the cell supports the fragile plasma membrane provides mechanical linkages that let the cell bear stresses without being ripped apart Importance and function
Cytoskeleton B. Balen structural backbone of the cell supports the fragile plasma membrane provides mechanical linkages that let the cell bear stresses without being ripped apart Importance and function
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Asymmetric endocytosis and remodeling of β1-integrin adhesions during growth cone chemorepulsion by MAG
 Asymmetric endocytosis and remodeling of β1-integrin adhesions during growth cone chemorepulsion by MAG Jacob H. Hines, Mohammad Abu-Rub and John R. Henley Supplementary Figure 1. Validation of FM 5-95
Asymmetric endocytosis and remodeling of β1-integrin adhesions during growth cone chemorepulsion by MAG Jacob H. Hines, Mohammad Abu-Rub and John R. Henley Supplementary Figure 1. Validation of FM 5-95
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Hong et al.,
 Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2880 Supplementary Figure 1 Sequence alignment of Deup1 and Cep63. The protein sequence alignment was generated by the Clustal X 2.0 multiple sequence alignment program using default parameters.
DOI: 10.1038/ncb2880 Supplementary Figure 1 Sequence alignment of Deup1 and Cep63. The protein sequence alignment was generated by the Clustal X 2.0 multiple sequence alignment program using default parameters.
Inverted formin 2 in focal adhesions promotes dorsal stress fiber and fibrillar adhesion formation to drive extracellular matrix assembly
 Inverted formin 2 in focal adhesions promotes dorsal stress fiber and fibrillar adhesion formation to drive extracellular matrix assembly Colleen T. Skau a, Sergey V. Plotnikov b, Andrew. oyle c, and Clare
Inverted formin 2 in focal adhesions promotes dorsal stress fiber and fibrillar adhesion formation to drive extracellular matrix assembly Colleen T. Skau a, Sergey V. Plotnikov b, Andrew. oyle c, and Clare
me239 mechanics of the cell homework biopolymers - motivation 3.1 biopolymers - motivation 3. biopolymers biopolymers biopolymers
 3. biopolymers the inner life of a cell, viel & lue, harvard [2006] me239 mechanics of the cell 1 2 biopolymers Figure 3.1. Biopolymers. Characteristic length scales on the cellular and sucellular level..
3. biopolymers the inner life of a cell, viel & lue, harvard [2006] me239 mechanics of the cell 1 2 biopolymers Figure 3.1. Biopolymers. Characteristic length scales on the cellular and sucellular level..
Dimeric WH2 domains in Vibrio VopF promote actin filament barbed end uncapping and assisted elongation
 Dimeric WH2 domains in Vibrio VopF promote actin filament barbed end uncapping and assisted elongation Julien Pernier, Jozsef Orban 1, Balendu Sankara Avvaru, Antoine Jégou, Guillaume Romet- Lemonne, Bérengère
Dimeric WH2 domains in Vibrio VopF promote actin filament barbed end uncapping and assisted elongation Julien Pernier, Jozsef Orban 1, Balendu Sankara Avvaru, Antoine Jégou, Guillaume Romet- Lemonne, Bérengère
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
Supplementary Figures
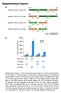 Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
Supplementary Figure 1: Geometry of the in situ tensile substrate. The dotted rectangle indicates the location where the TEM sample was placed.
 Supplementary Figures Supplementary Figure 1: Geometry of the in situ tensile substrate. The dotted rectangle indicates the location where the TEM sample was placed. Supplementary Figure 2: The original
Supplementary Figures Supplementary Figure 1: Geometry of the in situ tensile substrate. The dotted rectangle indicates the location where the TEM sample was placed. Supplementary Figure 2: The original
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
Journal of Cell Science Supplementary Material
 Figure S4 A His-(RBCPS)T15 His-(_BCPS) T15 His-PXN His-PXN+T15 His-PXN+ (RB_PS)T15 Myc-Ub IP:Ni-NTA 6M GuHCL WB: anti-myc WB: anti-his (Ub)n His-PXN His-(RBCPS)T15 His-(_BCPS)T15 + + + + + + + + + -100
Figure S4 A His-(RBCPS)T15 His-(_BCPS) T15 His-PXN His-PXN+T15 His-PXN+ (RB_PS)T15 Myc-Ub IP:Ni-NTA 6M GuHCL WB: anti-myc WB: anti-his (Ub)n His-PXN His-(RBCPS)T15 His-(_BCPS)T15 + + + + + + + + + -100
Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction
 Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain
 Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain architecture. Various C-terminal fragments were cloned and
Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain architecture. Various C-terminal fragments were cloned and
Quantitative analysis of Bidirectional Signaling (qbids)
 Quantitative analysis of Bidirectional Signaling (qbids) EphB2 + cells were labeled independently with light (C 12 N 14 ) arginine and lysine or heavy (C 13 N 15 ) arginine and lysine ephrin-b1 + cells
Quantitative analysis of Bidirectional Signaling (qbids) EphB2 + cells were labeled independently with light (C 12 N 14 ) arginine and lysine or heavy (C 13 N 15 ) arginine and lysine ephrin-b1 + cells
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/8/eaat4712/dc1 Supplementary Materials for In situ manipulation and switching of dislocations in bilayer graphene Peter Schweizer, Christian Dolle, Erdmann Spiecker*
advances.sciencemag.org/cgi/content/full/4/8/eaat4712/dc1 Supplementary Materials for In situ manipulation and switching of dislocations in bilayer graphene Peter Schweizer, Christian Dolle, Erdmann Spiecker*
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
CYTOSKELETON, MOTORPROTEINS.
 CYTOSKELETON, MOTORPROTEINS. SCIENCE PHOTO LIBRARY Tamás Huber 15. 10. 2012. CYTOSKELETON: Dynamic scaffold in eukaryotic cells Three main filament systems: A. Intermediate filaments B. Microtubules C.
CYTOSKELETON, MOTORPROTEINS. SCIENCE PHOTO LIBRARY Tamás Huber 15. 10. 2012. CYTOSKELETON: Dynamic scaffold in eukaryotic cells Three main filament systems: A. Intermediate filaments B. Microtubules C.
HEK293T. Fig. 1 in the
 Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Materials for
 www.advances.sciencemag.org/cgi/content/full/1/1/e1400205/dc1 Supplementary Materials for Myosin-binding protein C corrects an intrinsic inhomogeneity in cardiac excitation-contraction coupling Michael
www.advances.sciencemag.org/cgi/content/full/1/1/e1400205/dc1 Supplementary Materials for Myosin-binding protein C corrects an intrinsic inhomogeneity in cardiac excitation-contraction coupling Michael
ANAT Cell Biology Lecture 11 School of Medical Sciences The University of New South Wales. UNSW Copyright Notice
 ANAT3231 - Cell Biology Lecture 11 School of Medical Sciences The University of New South Wales The actin cytoskeleton Prof Peter Gunning Oncology Research Unit Room 502A Wallace Wurth Building Email:
ANAT3231 - Cell Biology Lecture 11 School of Medical Sciences The University of New South Wales The actin cytoskeleton Prof Peter Gunning Oncology Research Unit Room 502A Wallace Wurth Building Email:
Supplementary Figure 1. Serial deletion mutants of BLITz.
 Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Chapter 15. Cytoskeletal Systems. Lectures by Kathleen Fitzpatrick Simon Fraser University Pearson Education, Inc.
 Chapter 15 Cytoskeletal Systems Lectures by Kathleen Fitzpatrick Simon Fraser University Table 15-1 - Microtubules Table 15-1 - Microfilaments Table 15-1 Intermediate Filaments Table 15-3 Microtubules
Chapter 15 Cytoskeletal Systems Lectures by Kathleen Fitzpatrick Simon Fraser University Table 15-1 - Microtubules Table 15-1 - Microfilaments Table 15-1 Intermediate Filaments Table 15-3 Microtubules
SUPPLEMENTARY INFORMATION
 Transcription upregulation via force-induced direct stretching of chromatin Arash Tajik 1, 2#, Yuejin Zhang 1#, Fuxiang Wei 1#, Jian Sun 2#, Qiong Jia 1, Wenwen Zhou 1, Rishi Singh 2, Nimish Khanna 3,
Transcription upregulation via force-induced direct stretching of chromatin Arash Tajik 1, 2#, Yuejin Zhang 1#, Fuxiang Wei 1#, Jian Sun 2#, Qiong Jia 1, Wenwen Zhou 1, Rishi Singh 2, Nimish Khanna 3,
System of protein filaments in the cytoplasm of. eukaryotic cell that gives the cell shape and capacity for
 Cytoskeleton System of protein filaments in the cytoplasm of eukaryotic cell that gives the cell shape and capacity for directed movement. The protein filaments are responsible for the shaping, moving,
Cytoskeleton System of protein filaments in the cytoplasm of eukaryotic cell that gives the cell shape and capacity for directed movement. The protein filaments are responsible for the shaping, moving,
Supplemental Data. Sethi et al. (2014). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12116 Supplementary Figure 1 Theoretical prediction of temperature profiles. a, sketch of sample, immersed into X-ray beam, its environment (agarose and buffer solution are not distinguished),
doi:10.1038/nature12116 Supplementary Figure 1 Theoretical prediction of temperature profiles. a, sketch of sample, immersed into X-ray beam, its environment (agarose and buffer solution are not distinguished),
Supplementary Information
 Supplementary Information Figure S1. Transiently overexpressed ECFP-TIAF1/EYFP-TIAF1 causes apoptosis of Mv1Lu cells. Mv1Lu cells were transfected by electroporation with ECFP, EYFP, ECFP-TIAF1, EYFP-
Supplementary Information Figure S1. Transiently overexpressed ECFP-TIAF1/EYFP-TIAF1 causes apoptosis of Mv1Lu cells. Mv1Lu cells were transfected by electroporation with ECFP, EYFP, ECFP-TIAF1, EYFP-
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Microtubule Assembly Dynamics at the Nanoscale
 Microtubule Assembly Dynamics at the Nanoscale Melissa K. Gardner, Henry T. Schek III*, Alan J. Hunt, and David J. Odde Department of Biomedical Engineering, University of Minnesota, Minneapolis, MN 55455
Microtubule Assembly Dynamics at the Nanoscale Melissa K. Gardner, Henry T. Schek III*, Alan J. Hunt, and David J. Odde Department of Biomedical Engineering, University of Minnesota, Minneapolis, MN 55455
Guidance of microtubule growth and organization by F-actin
 Chapter 4 Guidance of microtubule growth and organization by F-actin In this chapter we study interactions between dynamic microtubules and actin filament bundles. We find that microtubule growth can be
Chapter 4 Guidance of microtubule growth and organization by F-actin In this chapter we study interactions between dynamic microtubules and actin filament bundles. We find that microtubule growth can be
Biosensors. DNA Microarrays (for chemical analysis) Protein Sensors (for identifying viruses)
 Biosensors DNA Microarrays (for chemical analysis) Protein Sensors (for identifying viruses) DNA Microarrays 40 000 detectors in parallel, each detecting a specific DNA sequence. Combinatorial Chemistry
Biosensors DNA Microarrays (for chemical analysis) Protein Sensors (for identifying viruses) DNA Microarrays 40 000 detectors in parallel, each detecting a specific DNA sequence. Combinatorial Chemistry
me239 mechanics of the cell me239 mechanics of the cell 3.1 biopolymers - motivation 3.2 biopolymers - polymerization
 4. the cytoskeleton - fiber bundle models bathe, heussinger, claessens, bausch, frey [2008] me239 mechanics of the cell 1 me239 mechanics of the cell 2 biopolymers polymerization of actin and tubulin Figure
4. the cytoskeleton - fiber bundle models bathe, heussinger, claessens, bausch, frey [2008] me239 mechanics of the cell 1 me239 mechanics of the cell 2 biopolymers polymerization of actin and tubulin Figure
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Modeling cytoskeleton self-assembly
 Modeling cytoskeleton self-assembly Dimitrios Vavylonis Department of Physics, Lehigh University BIOS 10 Lehigh University Oct 30, 2009 Cell organization How does the cell achieve internal organization?
Modeling cytoskeleton self-assembly Dimitrios Vavylonis Department of Physics, Lehigh University BIOS 10 Lehigh University Oct 30, 2009 Cell organization How does the cell achieve internal organization?
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Conformational changes in IgE contribute to its. uniquely slow dissociation rate from receptor FcεRI
 Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI M.D. Holdom, A.M. Davies, J.E. Nettleship, S.C. Bagby, B. Dhaliwal, E. Girardi, J. Hunt, H.J. Gould,
Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI M.D. Holdom, A.M. Davies, J.E. Nettleship, S.C. Bagby, B. Dhaliwal, E. Girardi, J. Hunt, H.J. Gould,
SUPPLEMENTARY INFORMATION
 Nanomechanical mapping of first binding steps of a virus to animal cells David Alsteens, Richard Newton, Rajib Schubert, David Martinez-Martin, Martin Delguste, Botond Roska and Daniel J. Müller This PDF
Nanomechanical mapping of first binding steps of a virus to animal cells David Alsteens, Richard Newton, Rajib Schubert, David Martinez-Martin, Martin Delguste, Botond Roska and Daniel J. Müller This PDF
No wash 2 Washes 2 Days ** ** IgG-bead phagocytosis (%)
 Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
MEMS based sensors for cellular studies
 MEMS based sensors for cellular studies Taher Saif Mechanical Science and Engineering University of Illinois at Urbana-Champaign Part of GEM4 Summer School lectures on instruments for cell mechanics studies
MEMS based sensors for cellular studies Taher Saif Mechanical Science and Engineering University of Illinois at Urbana-Champaign Part of GEM4 Summer School lectures on instruments for cell mechanics studies
Acetylated Microtubules Are Preferentially Bundled Leading to Enhanced
 Biophysical Journal, Volume 113 Supplemental Information Acetylated Microtubules Are Preferentially Bundled Leading to Enhanced Kinesin-1 Motility Linda Balabanian, Christopher L. Berger, and Adam G. Hendricks
Biophysical Journal, Volume 113 Supplemental Information Acetylated Microtubules Are Preferentially Bundled Leading to Enhanced Kinesin-1 Motility Linda Balabanian, Christopher L. Berger, and Adam G. Hendricks
SUPPLEMENTARY INFORMATION
 Normalized dorsal trunk length (µm) Normalized dorsal trunk length (TC1-5) (µm) DOI: 1.138/ncb2467 a b c d btl>src64 Src42, btl>src64 e Src64 f g h DT Src64 DT Midgut Src64 Midgut i Src64 Atpα j k l DT
Normalized dorsal trunk length (µm) Normalized dorsal trunk length (TC1-5) (µm) DOI: 1.138/ncb2467 a b c d btl>src64 Src42, btl>src64 e Src64 f g h DT Src64 DT Midgut Src64 Midgut i Src64 Atpα j k l DT
Specificity: Induced Fit
 Specificity: Induced Fit Conformational changes may occur upon ligand binding (Daniel Koshland in 1958) This adaptation is called the induced fit Induced fit allows for tighter binding of the ligand Induced
Specificity: Induced Fit Conformational changes may occur upon ligand binding (Daniel Koshland in 1958) This adaptation is called the induced fit Induced fit allows for tighter binding of the ligand Induced
Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity
 Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity Supplemental figure 1. The quantification method to determine the
Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity Supplemental figure 1. The quantification method to determine the
CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration
 /, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
/, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid
 Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Mitotic cell rounding and epithelial thinning regulate lumen
 Mitotic cell rounding and epithelial thinning regulate lumen growth and shape Esteban Hoijman, Davide Rubbini, Julien Colombelli and Berta Alsina SUPPLEMENTARY FIGURES Supplementary Figure 1 (a-c) Localization
Mitotic cell rounding and epithelial thinning regulate lumen growth and shape Esteban Hoijman, Davide Rubbini, Julien Colombelli and Berta Alsina SUPPLEMENTARY FIGURES Supplementary Figure 1 (a-c) Localization
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1
 Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
MT minus end catastrophe
 DOI: 1.138/ncb3241 MT minus end catastrophe frequency [s -1 ].2.15.1.5 control mgfp- TPX2 * * mgfp- TPX2 mini Supplementary Figure 1. TPX2 reduces catastrophes at the microtubule minus ends. Modified box-and-whiskers
DOI: 1.138/ncb3241 MT minus end catastrophe frequency [s -1 ].2.15.1.5 control mgfp- TPX2 * * mgfp- TPX2 mini Supplementary Figure 1. TPX2 reduces catastrophes at the microtubule minus ends. Modified box-and-whiskers
