Figure S1 early log mid log late log stat.
|
|
|
- Julius Heath
- 5 years ago
- Views:
Transcription
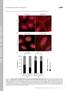
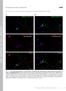
1 Figure S1 early log mid log late log stat. C D E F Fig. S1. The chromosome-expressed EI localizes to the cell poles independent of the growth phase and growth media. MG1655 (ptsi-mcherry) cells, which express the EI protein fused to mcherry from the chromosome, were grown in L () or in a minimal medium supplemented with glycerol (), glucose (C), lactose (D), mannitol (E) and ribose (F). Pictures were taken at different growth phases: early log, mid log, late log and stationary phase (stat.), as specified in the legend to Fig. 1. The EI-mCherry fusion protein was observed by fluorescence microscopy (red) and cells were observed with phase microscopy (grey). Shown are overlays of the signals from the fluorescence and phase microscopy (grey and red, merge). Scale bar corresponds to 1 µm.
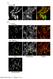
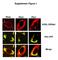
2 Figure S2 EI-mCherry EI anti mcherry b. anti His b HPr-mCherry mcherry glf-gfp C anti glf b. D anti GFP b. 1 glf * 3 4* 5 6* 2* 3 5 glg-gfp E anti glg b. F anti GFP b. 5 glg * * 4 Fig. S2. Western blot analysis of the PTS and gl proteins. Immunoblot analysis was carried out as described in Supplementary Material and Methods. The primary antibodies that were used are specified above the panels. Each panel is from a single blot and scan; removal of irrelevant lanes is marked by black separation lines. star next to a lane number denotes that it shows the expression level of the relevant protein in our experiments, as detailed below. b., antibodies. Detection of EI and HPr proteins: Panels () and (), the general PTS proteins (specified in parenthesis hereafter) were expressed from the following strains or plasmids: lanes 1, MG1655 (ptsimcherry) (chromosome-encoded EI-mCherry); lanes 2, pd18 (vector only); lanes 3, pdllmcherry (His-tagged mcherry); Lanes 4, pdllei-mcherry (His-tagged EI-mCherry); Lanes 5, pdllei-mcherry&hpr (His-tagged EI-mCherry and His-tagged HPr). Expression of the plasmidencoded EI and HPr was induced by the addition of.1 % arabinose for 1 h. rbutin (1%) was added to the last 3 min of growth. Similar amounts of cell extracts were loaded, except for lanes 1, in which two times more extract was loaded. The results confirm that the mcherry-tagged EI and HPr are stable and the tag is not liberated (compare lanes 4 and 5 to lane 3). The ratio between the chromosomeencoded EI (panel, lane 1) and the plasmid encoded EI (panel, lane 4), after normalizing to the amount of extract loaded was.33 (i.e., three times more EI encoded from the plasmid than from the chromosome). The ratio between the plasmid-encoded HPr and EI proteins (panel, lane 5) was To calculate these ratios, the intensity of the bands was quantified by the MetaMorph software and normalized to the intensity in an area in the gel lacking detectable bands.
3 Fig. S2 (cont.) Detection of glf proteins: Panel (C), the gl proteins (specified in parenthesis hereafter) were expressed from the following strains or plasmids: lane 1, M1 (bgl + ) grown with salicin that induces expression of the bgl operon (chromosome-encoded gl proteins); lanes 2 & 3, MG1655 containing pdllhpr-mcherry&ei and pllk12f-gfp (glf-gfp) grown without (lane 3) or with (lane 4) IPTG; lanes 4 & 5, MG1655 (bgl - ) containing pdllhpr-mcherry&ei, pjs185 (glg-gfp) and pllk12f (glf) grown without (lane 4) or with (lane 5) IPTG; lane 6, MG1655 containing pdllhpr-mcherry&ei, pjs185 (glg-gfp) and pzlf (glf) grown with anhydrotetracycline hydrochloride (2 ng). Expression of the plasmid-encoded EI and HPr was induced by the addition of.1% arabinose for 1 h. rbutin (1%) was added to the last 3 min of growth for cells analyzed in lanes 2-6. Similar amounts of cell extracts were loaded, except for lanes 1, in which two and a half times more extract was loaded. Panel (D) shows the reaction of lanes 2 & 3 with anti-gfp antibodies. It is obvious that the level of the plasmid encoded glf that we used in the experiment shown in Fig. 7 (analyzed here in lane 4, no IPTG) is lower (hardly detectable without IPTG induction) than the native level encoded from the chromosome upon -glucoside induction (lane 1), even after taking into consideration that two and a half times more extract was loaded in the second case. The level of the plasmid-encoded glf-gfp that we used in the experiment shown in Fig. 3 (analyzed here in lane 2, no IPTG) was comparable to the native chromosome-encoded level (1.1 times higher after normalizing to the amount of extracts loaded). The level of the plasmid-encoded glf that we used in the experiment shown in Fig. S1 and S11 (analyzed here in lane 6) was significantly higher (over 1 fold) than the native chromosome-encoded level. Detection of glg proteins: Panel (E), the gl proteins (specified in parenthesis hereafter) were expressed from the following strains or plasmids: lane 1, M1 grown without -glucosides (no gl proteins expressed); lane 2, M1 grown with salicin, an inducer of the bgl operon (chromosomeencoded gl proteins); lane 3 & 4, MG1655 containing pdllhpr-mcherry&ei and pjs185 (glg-gfp) grown without (lane 3) or with (lane 4) IPTG. Expression of the plasmid-encoded EI and HPr was induced by the addition of.1 % arabinose for 1 h (lanes 3 & 4). rbutin (1%) was added to the last 3 min of growth for cells analyzed in lanes 3 & 4. In lanes 1 & 2, two and a half times more cell extract were loaded than in lanes 3 & 4. Notably, non-specific interactions with the polyclonal anti-glg antiserum due to cross-reacting antibodies are detected in all four lanes, independent of glg or glg-gfp expression. Panel (F) shows the reaction of the extracts in (E) with anti-gfp antibodies. It is obvious that the level of the plasmid-encoded glg-gfp, which we used in the experiments shown in all figures of this manuscript (lane 3, no IPTG) is lower (detectable with anti-gfp antibodies only after IPTG induction, lane 4) than the native level encoded from chromosome upon -glucoside induction (lane 2), even after taking into consideration that two and a half times more extract was loaded in the second case.
4 Figure S3 fluorescence intensity Fluorescence intensity of HPr-mCherry of Hpr-mCherry HPr-mCherry mcherry & & EI EI. HPr-mCherry mcherry & EI-N & EI-N average intensity at the poles average intensity in the cell 5 4 Ratio poles/cell ratio pole/cell Hpr-cherry HPr-mCherry & EI & EI. Hpr-cherry HPr-mCherry & EI-N & EI-N Fig. S3. Distribution of HPr in the cell in the presence of EI or EI-N. The fluorescent intensity of HPr-mCherry near the poles (blue) and its average intensity in MG1655 pts cells (purple), which express EI () or EI-N () together with the HPr-Cherry, were quantified for 2 cells, in each case, for the experiments shown in Fig. 2F and 4E, respectively. The intensities were determined by the MetaMorph software and normalized to the intensity in regions lacking cells in the respective fields. The ratio between the two values (yellow), shown in the lower panel, was 3.3 for () and 4.1 for ().
5 Figure S4 Phase HPr-GFP EI-mCherry merge C D E Fig. S4. EI and HPr localize to the cell poles independently, but the distribution of HPr in the cell is affected by EI and by PTS sugars. MG1655 pts (, D and E) or MG1655 (ptsi-mcherry) ( and C) cells, containing a plasmid that code for HPr-GFP (-C) or for both HPr-GFP and EI-mCherry (D-E) were grown in L (, D and E) or in minimal medium containing lactose () or mannitol (C) at 37 C (-D) or at 3 C (E). The GFP and mcherry fusion proteins were observed by fluorescence microscopy (green, GFP; red, mcherry) and cells were observed with phase microscopy. Overlays of the signals from the GFP and mcherry fluorescent signal (green and red, merge) are also shown.
6 Figure S5 II glc -GFP phase II glc -GFP EI-mCherry merge phase Fig. S5. The cytoplasmic protein II glc is evenly spread throughout the cell. MG1655 () and MG1655 (ptsi-mcherry) () cells, containing a plasmid which encodes II glc fused to GFP, were grown at 3 C in L (1 and 1) or at 37 C in minimal medium supplemented with either.4% glucose (2 and 2) or 2mM ribose (3 and 3). The II glc -GFP protein was expressed from pjpk12 () or pjp12 () without induction. The GFP and mcherry fusion proteins were observed by fluorescence microscopy (green, GFP; red, mcherry; green and red, merge) and cells were observed with phase microscopy (grey, phase). Scale bar corresponds to 1 µm.
7 Figure S6 Fluorescence intensity of glg-gfp glg-gfp expressed with: EI EI & DHPr average intensity at the pole average intensity in the cell Ratio pole/cell glg-gfp expressed with: EI EI & DHPr Fig. S6. Distribution of glg-gfp in the cell in the presence of one of the general PTS proteins or both. The fluorescent intensity of glg-gfp near the poles (blue) and its average intensity (purple) in MG1655 pts cells which express, besides glg-gfp, also EI () or EI and HPr (), were quantified for 2 cells, in each case, for the experiments shown in Fig. 5 and 5D, respectively. The intensities were determined by the MetaMorph software and normalized to the intensity in regions lacking cells in the respective fields. The ratio between the two values (yellow, shown in the lower panel) was 1.7 for () and 1.5 for ().
8 Fig. S7-I glg(h16y)- GFP phase merge Fig. S7-II glg(h28r)- GFP phase merge glg(h16y)- GFP mcherry merge glg(h28r)- GFP mcherry merge C EI C EI D HPr D HPr EI-mCherry +HPr E EI EI-mCherry +HPr E EI HPr-mCherry +EI F HPr HPr-mCheery +EI F HPr G EI(H189) G EI(H189) H HPr(H15) H HPr(H15) I EI-N) I EI-N EI-C Fig. S7. The residues required for glg phosphorylation are dispensable for its co-localization with the general PTS proteins at the poles. Images of MG1655 pts cells expressing glg(h16y)-gfp (Fig. S7-I) or glg(h28r)- GFP (Fig. S7-II). () Fluorescence images (green, left), phase contrast (grey, middle) and overlay (merge, right). (-I) Fluorescence microscopy images (green, GFP; red, mcherry; red and green, overlay) of the cells in () that contain also a second plasmid encoding EI-mCherry (), HPr-mCherry (C), EI-mCherry and HPr (D), HPr-mCherry and EI (E), EI(H189)-mCherry (F), HPr(H15)-mCherry (G), EI-N (H), EI-C (I). Cells were grown in rich medium at 3 C. Expression of the glg proteins was induced with IPTG (.1mM) for 1 hour. Scale bar corresponds to 1 µm. EI-C
9 Figure S8 [min] t t5 glg-gfp glg-gfp Fig. S8. glg migrates from the cell periphery to the cell poles after addition of the stimulating sugar. Shown are larger fragments from the fields presented in Fig. 7, before (t) and 5 min after (t5) arbutin addition.
10 Figure S9 Fluorescence intensity of glg-gfp fluorescence intensity of glg-gfp t t5 t t5 exp. 1 exp. 2 average intensity at the poles average intensity in the cell ratio pole/cell t t5 t t5 exp. 1 exp. 2 Fig. S9. glg migrates from the cell periphery to the cell poles after addition of the stimulating sugar. () The fluorescent intensity of glg-gfp near the poles (blue) and its average intensity in the cytoplasm and membrane (purple) of cells expressing glg-gfp together with glf and the general PTS proteins, before (t) and 5 min fter (t5) arbutin addition. The intensities were quantified for 2 cells from two independent experiments (exp. 1 and exp. 2), the first shown in Fig. 7, by the MetaMorph software and normalized to the intensity in regions lacking cells in the respective fields. Of note, quantifications were made for snapshot images, explaining the change in the absolute cell fluorescence between the time points. () The ratios (yellow) between each pair in () are as following: 1.9 (exp.1) or.97 (exp. 2) at t and 3.25 (exp.1) or 2.88 (exp.2) at t5.
11 Figure S1 [min] glg-gfp EI-mCherry t t5 t1 t2 Fig. S1. glg is observed in foci that migrate from the cell periphery to the cell poles after addition of the stimulating sugar. Fluorescence microscopy images of MG1655 pts cells grown in rich medium. Expression of glg-gfp was not induced. Expression of glf from pzlf was induced significantly by the addition of 2 ng/ml anhydrotetracycline hydrochloride (see Fig. S2C, lane 6 and figure legend), to slow glg-gfp release from glf and follow its route after sugar stimulation and before its accumulation at the poles Samples were taken before (t) and at the indicated times after the addition of 1% arbutin. The fluorescent signal of glg-gfp was twodimensional deconvolved to reveal the localization of most glg molecules.
12 Figure S11 fluorescence intensity t t2 t t2 snapshots average intensity at the poles time-lapse average intensity in the cell ratio pole/cell t t2 t t2 snapshots time-lapse Fig. S11. glg migrates from the cell periphery to the cell poles after addition of the stimulating sugar. The fluorescent intensity of glg-gfp near the poles (blue) and its average intensity (purple) in the cytoplasm and membrane of cells expressing glg-gfp together with glf and the general PTS proteins, before (t) and after (t2) arbutin addition, were quantified for 1 cells in snapshot images (see Fig. S1) and for 1 cells visualized by time-lapse microscopy. glf was overexpressed from pzlf as described in Fig. S1 (see explanation there). The intensities were determined by the MetaMorph software and normalized to the intensity in regions lacking cells in the respective fields. The ratio between the two values (yellow), shown in the lower panel, was 1.12 (snapshots) or 1.9 (time-lapse) at t compared to 2.91 (snapshots) or 1.99 (timelapse) at t2.
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Supplemental Data. Sethi et al. (2014). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Autophagy induction [h]
![Autophagy induction [h] Autophagy induction [h]](/thumbs/80/80564099.jpg) A SD-N [h]: SD-N [h]: 1 2 4 1 2 4 1 2 4 -GFP 1 2 4 percentage GFP-Atg8 [%] 1 9 8 7 6 5 4 3 2 1 1 2 4 Autophagy induction [h] -13xmyc SD-N [h]: 1 2 4 -GFP -13xmyc 1 prape1 mape1 - - GFP - 13xmyc mape1 [%]
A SD-N [h]: SD-N [h]: 1 2 4 1 2 4 1 2 4 -GFP 1 2 4 percentage GFP-Atg8 [%] 1 9 8 7 6 5 4 3 2 1 1 2 4 Autophagy induction [h] -13xmyc SD-N [h]: 1 2 4 -GFP -13xmyc 1 prape1 mape1 - - GFP - 13xmyc mape1 [%]
Aoki et al.,
 JCB: SUPPLEMENTAL MATERIAL Aoki et al., http://www.jcb.org/cgi/content/full/jcb.200609017/dc1 Supplemental materials and methods Calculation of the raw number of translocated proteins First, the average
JCB: SUPPLEMENTAL MATERIAL Aoki et al., http://www.jcb.org/cgi/content/full/jcb.200609017/dc1 Supplemental materials and methods Calculation of the raw number of translocated proteins First, the average
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supporting information
 Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Feng et al., http://www.jcb.org/cgi/content/full/jcb.201408079/dc1 Figure S1. A modest elevation of disulfide-bonded K14 in primary mouse
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Feng et al., http://www.jcb.org/cgi/content/full/jcb.201408079/dc1 Figure S1. A modest elevation of disulfide-bonded K14 in primary mouse
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide,
 Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
42 fl organelles = 34.5 fl (1) 3.5X X 0.93 = 78,000 (2)
 SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues.
 Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
SUPPLEMENTARY INFORMATION. Tolerance of a knotted near infrared fluorescent protein to random circular permutation
 SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr
 Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplementary Fig. 1. Multiple five micron sections of liver tissues of rats treated
 Supplementary Figure Legends Supplementary Fig. 1. Multiple five micron sections of liver tissues of rats treated with either vehicle (left; n=3) or CCl 4 (right; n=3) were co-immunostained for NRP-1 (green)
Supplementary Figure Legends Supplementary Fig. 1. Multiple five micron sections of liver tissues of rats treated with either vehicle (left; n=3) or CCl 4 (right; n=3) were co-immunostained for NRP-1 (green)
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Prospéri et al.,
 Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
-7 ::(teto) ::(teto) 120. GFP membranes merge. overexposed. Supplemental Figure 1 (Marquis et al.)
 P spoiie -gfp -7 ::(teto) 120-91 ::(teto) 120 GFP membranes merge overexposed Supplemental Figure 1 (Marquis et al.) Figure S1 TetR-GFP is stripped off the teto array in sporulating cells that contain
P spoiie -gfp -7 ::(teto) 120-91 ::(teto) 120 GFP membranes merge overexposed Supplemental Figure 1 (Marquis et al.) Figure S1 TetR-GFP is stripped off the teto array in sporulating cells that contain
Supplementary Figure 1 Autoaggregation of E. coli W3110 in presence of native
 Supplementary Figure 1 Autoaggregation of E. coli W3110 in presence of native Ag43 phase variation depends on Ag43, motility, chemotaxis and AI-2 sensing. Cells were grown to OD600 of 0.6 at 37 C and aggregation
Supplementary Figure 1 Autoaggregation of E. coli W3110 in presence of native Ag43 phase variation depends on Ag43, motility, chemotaxis and AI-2 sensing. Cells were grown to OD600 of 0.6 at 37 C and aggregation
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Supporting Information
 Supporting Information Üstün and Börnke, 2015 Figure S1: XopJ does not display acetyltransferase activity. Autoacetylation activity in vitro. Acetylation reactions using MBP, MBP-XopJ, GST and GST-HopZ1a
Supporting Information Üstün and Börnke, 2015 Figure S1: XopJ does not display acetyltransferase activity. Autoacetylation activity in vitro. Acetylation reactions using MBP, MBP-XopJ, GST and GST-HopZ1a
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Your Protein Manufacturer
 Expression of Toxic Proteins in E. coli Your Protein Manufacturer Contents 1. Definition of Toxic Protein 2. Mechanisms of Protein Toxicity 3. Percentage of Protein Toxicity 4. Phenotypes of Protein Toxicity
Expression of Toxic Proteins in E. coli Your Protein Manufacturer Contents 1. Definition of Toxic Protein 2. Mechanisms of Protein Toxicity 3. Percentage of Protein Toxicity 4. Phenotypes of Protein Toxicity
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Table S1. List of DNA constructs and primers, part 1 Construct
 SUPPLEMENTARY TABLES: Table S1. List of DNA constructs and primers, part 1 Construct Comment (Expressed protein) Vector, PCR primers and template or source of Restriction Sites name Resistance construct
SUPPLEMENTARY TABLES: Table S1. List of DNA constructs and primers, part 1 Construct Comment (Expressed protein) Vector, PCR primers and template or source of Restriction Sites name Resistance construct
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Craft et al., http://www.jcb.org/cgi/content/full/jcb.201409036/dc1 Figure S1. GFP -tubulin interacts with endogenous -tubulin. Ciliary
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Craft et al., http://www.jcb.org/cgi/content/full/jcb.201409036/dc1 Figure S1. GFP -tubulin interacts with endogenous -tubulin. Ciliary
Spatial focalization of pheromone/mapk signaling triggers commitment to cell-cell fusion
 Supplemental data for: Spatial focalization of pheromone/mapk signaling triggers commitment to cell-cell fusion Omaya Dudin, Laura Merlini and Sophie G Martin Figure S1 (related to Figure 1): Myo51 and
Supplemental data for: Spatial focalization of pheromone/mapk signaling triggers commitment to cell-cell fusion Omaya Dudin, Laura Merlini and Sophie G Martin Figure S1 (related to Figure 1): Myo51 and
The fusion used in most of these studies (MinC4-GFP) contains GFP and
 Gregory, Becker and Pogliano, Supplemental Materials page 1 Supplemental Methods Description of the MinC and MinD GFP insertions The fusion used in most of these studies (MinC4-GFP) contains GFP and flanking
Gregory, Becker and Pogliano, Supplemental Materials page 1 Supplemental Methods Description of the MinC and MinD GFP insertions The fusion used in most of these studies (MinC4-GFP) contains GFP and flanking
Figure S1. USP-46 is expressed in several tissues including the nervous system
 Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Single cell resolution in vivo imaging of DNA damage following PARP inhibition. Supplementary Data
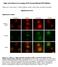 Single cell resolution in vivo imaging of DNA damage following PARP inhibition Katherine S. Yang, Rainer H. Kohler, Matthieu Landon, Randy Giedt, and Ralph Weissleder Supplementary Data Supplementary Figures
Single cell resolution in vivo imaging of DNA damage following PARP inhibition Katherine S. Yang, Rainer H. Kohler, Matthieu Landon, Randy Giedt, and Ralph Weissleder Supplementary Data Supplementary Figures
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1.
 Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Laloux and Jacobs-Wagner, http://www.jcb.org/cgi/content/full/jcb.201303036/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Supporting data on the PopZ variants. Related
Supplemental material Laloux and Jacobs-Wagner, http://www.jcb.org/cgi/content/full/jcb.201303036/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Supporting data on the PopZ variants. Related
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
over time using live cell microscopy. The time post infection is indicated in the lower left corner.
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Supplementary Movie 1 Description: Fusion of NBs. BSR cells were infected
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Supplementary Movie 1 Description: Fusion of NBs. BSR cells were infected
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION 1 Supplementary Figure S1. Proportion of HEK293T transfectants showing fluorescent foci after heat sho Blinded slides were counted for the appearance of fluorescent foci of tagrfp-t
SUPPLEMENTARY INFORMATION 1 Supplementary Figure S1. Proportion of HEK293T transfectants showing fluorescent foci after heat sho Blinded slides were counted for the appearance of fluorescent foci of tagrfp-t
T H E J O U R N A L O F C E L L B I O L O G Y
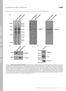 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Bays et al., http://www.jcb.org/cgi/content/full/jcb.201309092/dc1 Figure S1. Specificity of the phospho-y822 antibody. (A) Total cell
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Bays et al., http://www.jcb.org/cgi/content/full/jcb.201309092/dc1 Figure S1. Specificity of the phospho-y822 antibody. (A) Total cell
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplementary information
 Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Engineering splicing factors with designed specificities
 nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
* ** ** * IB: p-p90rsk. p90rsk (Ser380) (arbitrary units) (Ser380) p90rsk. IB: p90rsk. Tubulin. IB: Tubulin. Ang II (200 nm) Ang II (200 nm)
 I: p-p9rsk I: p9rsk I: C I: p-p9rsk I: p9rsk 5 (ka) 5 5 (min) Ang II ( nm) p-p9rsk (Ser8) p9rsk p-p9rsk (Ser8) p9rsk (h) Mannitol 5 mm -Glucose 5 mm p9rsk (Ser8) (arbitrary units) p-p9rsk (Ser8) (arbitrary
I: p-p9rsk I: p9rsk I: C I: p-p9rsk I: p9rsk 5 (ka) 5 5 (min) Ang II ( nm) p-p9rsk (Ser8) p9rsk p-p9rsk (Ser8) p9rsk (h) Mannitol 5 mm -Glucose 5 mm p9rsk (Ser8) (arbitrary units) p-p9rsk (Ser8) (arbitrary
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kanaani et al., http://www.jcb.org/cgi/content/full/jcb.200912101/dc1 Figure S1. The K2 rabbit polyclonal antibody is specific for GAD67,
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kanaani et al., http://www.jcb.org/cgi/content/full/jcb.200912101/dc1 Figure S1. The K2 rabbit polyclonal antibody is specific for GAD67,
Supplemental Materials
 Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Supplementary Figure 1.
 Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplement Figure 1. Plin5 Plin2 Plin1. KDEL-DSRed. Plin-YFP. Merge
 Supplement Figure 1 Plin5 Plin2 Plin1 KDEL-DSRed Plin-YFP Merge Supplement Figure 2 A. Plin5-Ab MitoTracker Merge AML12 B. Plin5-YFP Cytochrome c-cfp merge Supplement Figure 3 Ad.GFP Ad.Plin5 Supplement
Supplement Figure 1 Plin5 Plin2 Plin1 KDEL-DSRed Plin-YFP Merge Supplement Figure 2 A. Plin5-Ab MitoTracker Merge AML12 B. Plin5-YFP Cytochrome c-cfp merge Supplement Figure 3 Ad.GFP Ad.Plin5 Supplement
Supporting Information
 Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
J. Cell Sci. 128: doi: /jcs : Supplementary Material. Supplemental Figures. Journal of Cell Science Supplementary Material
 Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material JCB Tanaka et al., http://www.jcb.org/cgi/content/full/jcb.201402128/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nonselective autophagy is normal in Hrr25-depleted
Supplemental material JCB Tanaka et al., http://www.jcb.org/cgi/content/full/jcb.201402128/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nonselective autophagy is normal in Hrr25-depleted
Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction
 Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
VERIFY Tagged Antigen. Validation Data
 VERIFY Tagged Antigen Validation Data Antibody Validation Figure 1. Over-expression cell lysate for human STAT3 (NM_139276) was used to test 3 commercial antibodies. Antibody A shows strong antigen binding.
VERIFY Tagged Antigen Validation Data Antibody Validation Figure 1. Over-expression cell lysate for human STAT3 (NM_139276) was used to test 3 commercial antibodies. Antibody A shows strong antigen binding.
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplementary Information (Ha, et. al) Supplementary Figures Supplementary Fig. S1
 Supplementary Information (Ha, et. al) Supplementary Figures Supplementary Fig. S1 a His-ORMDL3 ~ 17 His-ORMDL3 GST-ORMDL3 - + - + IPTG GST-ORMDL3 ~ b Integrated Density (ORMDL3/ -actin) 0.4 0.3 0.2 0.1
Supplementary Information (Ha, et. al) Supplementary Figures Supplementary Fig. S1 a His-ORMDL3 ~ 17 His-ORMDL3 GST-ORMDL3 - + - + IPTG GST-ORMDL3 ~ b Integrated Density (ORMDL3/ -actin) 0.4 0.3 0.2 0.1
7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV
 1 Fall 2006 7.03 7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that
1 Fall 2006 7.03 7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that
7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV
 7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand
7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
SUPPLEMENTARY INFORMATION FIGURE LEGENDS
 SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Nature Structural & Molecular Biology: doi: /nsmb.3308
 Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum. Blood-stage Development
 Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
SUPPLEMENTARY INFORMATION
 a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy
 Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Supplemental Information. A Versatile Tool for Live-Cell Imaging. and Super-Resolution Nanoscopy Studies. of HIV-1 Env Distribution and Mobility
 Cell Chemical Biology, Volume 24 Supplemental Information A Versatile Tool for Live-Cell Imaging and Super-Resolution Nanoscopy Studies of HIV-1 Env Distribution and Mobility Volkan Sakin, Janina Hanne,
Cell Chemical Biology, Volume 24 Supplemental Information A Versatile Tool for Live-Cell Imaging and Super-Resolution Nanoscopy Studies of HIV-1 Env Distribution and Mobility Volkan Sakin, Janina Hanne,
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
A novel mechanism of post-translational modulation of HMGA functions by the histone chaperone nucleophosmin.
 A novel mechanism of post-translational modulation of HMGA functions by the histone chaperone nucleophosmin. Laura Arnoldo 1,#, Riccardo Sgarra 1,#, Eusebio Chiefari 2, Stefania Iiritano 2, Biagio Arcidiacono
A novel mechanism of post-translational modulation of HMGA functions by the histone chaperone nucleophosmin. Laura Arnoldo 1,#, Riccardo Sgarra 1,#, Eusebio Chiefari 2, Stefania Iiritano 2, Biagio Arcidiacono
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Impaired ITS2 processing results in mislocalization of foot-factors to the cytoplasm. (a) Schematic representation of ITS2 processing. The endonuclease Las1 initiates ITS2 processing
Supplementary Figure 1 Impaired ITS2 processing results in mislocalization of foot-factors to the cytoplasm. (a) Schematic representation of ITS2 processing. The endonuclease Las1 initiates ITS2 processing
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Supplementary Fig. 1 Kinetics of appearence of the faster migrating form of Bcl-10.
 α-cd3 + α-cd28: Time (min): + + + + + + + + + 0 5 15 30 60 120 180 240 300 360 360 n.s. Supplementary Fig. 1 Kinetics of appearence of the faster migrating form of. Immunoblot of lysates from Jurkat cells
α-cd3 + α-cd28: Time (min): + + + + + + + + + 0 5 15 30 60 120 180 240 300 360 360 n.s. Supplementary Fig. 1 Kinetics of appearence of the faster migrating form of. Immunoblot of lysates from Jurkat cells
Supplementary Information
 Supplementary Information Figure S1. Transiently overexpressed ECFP-TIAF1/EYFP-TIAF1 causes apoptosis of Mv1Lu cells. Mv1Lu cells were transfected by electroporation with ECFP, EYFP, ECFP-TIAF1, EYFP-
Supplementary Information Figure S1. Transiently overexpressed ECFP-TIAF1/EYFP-TIAF1 causes apoptosis of Mv1Lu cells. Mv1Lu cells were transfected by electroporation with ECFP, EYFP, ECFP-TIAF1, EYFP-
Methanol fixation allows better visualization of Kal7. To compare methods for
 Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Supplementary Figure 1. Drawing of spinal cord open-book preparations and DiI tracing. Nature Neuroscience: doi: /nn.3893
 Supplementary Figure 1 Drawing of spinal cord open-book preparations and DiI tracing. Supplementary Figure 2 In ovo electroporation of dominant-negative PlexinA1 in commissural neurons induces midline
Supplementary Figure 1 Drawing of spinal cord open-book preparations and DiI tracing. Supplementary Figure 2 In ovo electroporation of dominant-negative PlexinA1 in commissural neurons induces midline
Supplementary material for: Materials and Methods:
 Supplementary material for: Iron-responsive degradation of iron regulatory protein 1 does not require the Fe-S cluster: S.L. Clarke, et al. Materials and Methods: Fe-S Cluster Reconstitution: Cells treated
Supplementary material for: Iron-responsive degradation of iron regulatory protein 1 does not require the Fe-S cluster: S.L. Clarke, et al. Materials and Methods: Fe-S Cluster Reconstitution: Cells treated
Slope: Y-Intercept: Amount of DNA (ng)
 Cp A 30 25 20 Landing pad Slope: -3.522 Y-Intercept: 11.17 R 2 : 0.99 15 10 5 hgh Slope: -3.314 Y-Intercept: 9.983 R 2 : 0.99 IFNa-2b Slope: -3.527 Y-Intercept: 9.910 R 2 : 0.99 0 2.0 10-5 2.0 10-4 2.0
Cp A 30 25 20 Landing pad Slope: -3.522 Y-Intercept: 11.17 R 2 : 0.99 15 10 5 hgh Slope: -3.314 Y-Intercept: 9.983 R 2 : 0.99 IFNa-2b Slope: -3.527 Y-Intercept: 9.910 R 2 : 0.99 0 2.0 10-5 2.0 10-4 2.0
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Supplementary Information
 Supplementary Information Fluorescent protein choice The dual fluorescence channel ratiometric promoter characterization plasmid we designed for this study required a pair of fluorescent proteins with
Supplementary Information Fluorescent protein choice The dual fluorescence channel ratiometric promoter characterization plasmid we designed for this study required a pair of fluorescent proteins with
Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1.
 Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
Resveratrol inhibits epithelial-mesenchymal transition of retinal. pigment epithelium and development of proliferative vitreoretinopathy
 Resveratrol inhibits epithelial-mesenchymal transition of retinal pigment epithelium and development of proliferative vitreoretinopathy Keijiro Ishikawa, 1,2 Shikun He, 2, 3 Hiroto Terasaki, 1 Hossein
Resveratrol inhibits epithelial-mesenchymal transition of retinal pigment epithelium and development of proliferative vitreoretinopathy Keijiro Ishikawa, 1,2 Shikun He, 2, 3 Hiroto Terasaki, 1 Hossein
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
mcherry Monoclonal Antibody (16D7) Catalog Number M11217 Product data sheet
 Website: thermofisher.com Customer Service (US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 mcherry Monoclonal Antibody (16D7) Catalog Number M11217 Product data sheet Details
Website: thermofisher.com Customer Service (US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 mcherry Monoclonal Antibody (16D7) Catalog Number M11217 Product data sheet Details
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators.
 Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
14_integrins_EGFR
 α1 Integrin α1 -/- fibroblasts from integrin α1 knockout animals Figure S1. Serum-starved fibroblasts from α -/- 1 and +/+ mice were stimulated with 10% FBS for 30 min. (FBS) or plated on collagen I (CI)
α1 Integrin α1 -/- fibroblasts from integrin α1 knockout animals Figure S1. Serum-starved fibroblasts from α -/- 1 and +/+ mice were stimulated with 10% FBS for 30 min. (FBS) or plated on collagen I (CI)
Nature Structural & Molecular Biology: doi: /nsmb.1583
 Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
