Xu et al., Supplementary Figures 1-7
|
|
|
- Morris Baker
- 5 years ago
- Views:
Transcription
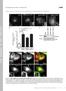
1 Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to human PIPKI (sipipki -O1, sipipki -O2) or negative control sirna (sinc) were serum starved to induce primary ciliogenesis and stained with appropriate antibodies. (b) Knockdown of PIPKI was validated by immunoblotting. (c) PIPKI exhibits basal body localization in multiple types of ciliated cells. (d, e) PIPKI and PIPKI antibodies yielded no cilium-related signal. Serum starved RPE-1 cells were fixed with methanol (d) or 1 P a g e
2 paraformaldehyde (PFA) (e) and stained with appropriate antibodies. (f) PIPKIγ forms a ring-like structure at the ciliary base. RPE-1 cells co-stained with anti-pipki and anti-plk4 antibodies were examined by 3D-SIM microscopy. (g-i) PIPKI depletion substantially inhibits primary ciliogenesis in RPE-1, NIH3T3, and RCTE cells. (g) RPE-1 cells treated with negative control (sinc) or sirnas against PIPKI (sipipki -O1 and sipipki -O2), PIPKI (sipipki ), or PIPKI (sipipki ) were serum starved to induce ciliogenesis and stained with anti-acetylated -tubulin antibody (Ac-tub). (h) NIH3T3 cells were treated with sinc or mouse PIPKI -specific (sipipki -O3) sirnas. (i) RCTE cells treated with sinc or two PIPKI -specific sirnas were analyzed as described in (a). The percentage of ciliated cells (n > 200) in each group was quantified from at least three independent experiments and plotted. Error bars represent s.d. ***P < (j) Severe ciliary defect was observed in ppk-1-null C. elegans. With dye-filling assay, ppk-1-null worms exhibited complete dye-filling minus phenotype. Brackets indicate ciliated Amphid neurons. (k) Genotyping results of ppk-1-null worms. Actin was used as a loading control. Nuclei were visualized by DAPI staining (blue). Insets show magnified images of primary cilia. Ac-tub, acetylated -tubulin. Glu-tub, polyglutamylated tubulin. -tub, -tubulin. Scale bars are 0.5 μm in (f) and 5 μm in other panels. 2 P a g e
3 Supplementary Figure 2. PIPKI functions downstream of basal-body membrane docking and upstream of TZ formation. RPE-1 cells treated with negative control (sinc) or PIPKI (sipipki ) sirna were serum starved and stained with the appropriate antibodies. Percentage of cells (n > 200) showing positive staining of indicated proteins was quantified from at least three experiments. Error bars represent standard deviation (s.d.). N.S., no statistically significant difference. -tub, -tubulin. Scale bars, 0.5 μm. 3 P a g e
4 Supplementary Figure 3. PIPKI promotes the recruitment of TTBK2 to the M-centriole/basal body. (a) CP110 retained at the M-centriole in serum-starved, PIPKI -depleted RPE-1 cells. Arrowheads point to the basal body. (b) TTBK2 recruitment to the M-centriole induced by serum starvation was impaired in RPE-1 cells when PIPKI was knocked down. Percentage of cells (n > 200) exhibiting CP110 on both centrioles (a), or TTBK2 at the M-centriol/basal body (b) was quantified from at least three independent experiments. Glu-tub, polyglutamylated tubulin. -tub, -tubulin. (c) PIPKI overexpression promotes TTBK2 recruitment to the centrosome in a PIPKI kinase activity dependent manner. RCTE cells stably expressing GFP-TTBK2 were transfected with empty vector 4 P a g e
5 (Control), wild-type (WT), or kinase-dead (KD) HA-PIPKI i3 for 8 h without serum starvation. Cep83 was used as a centrosome marker. (d) RCTE cells were transfected with empty vector (Control), HA- PIPKI, and HA-PIPKI CT. The percentage of ciliated cells (n > 20) in each group was quantified from at least three independent experiments and plotted. Error bars represent standard deviation (s.d.). ***P < N.S., no statistically significant difference. Scale bars, 0.5 μm. 5 P a g e
6 Supplementary Figure 4. INPP5E inhibits ciliogenesis by suppressing CP110 removal from the basal body. IMCD3 Cells transfected with empty vector (Control), or Flag-INPP5E were serum starved and stained with indicated antibodies. Percentage of cells with CP110 on both centrioles (a, n > 100) and ciliated cells (b, n > 200) was quantified from at least three independent experiments. Error bars represent s.d. ***P< P a g e
7 Supplementary Figure 5. Localization of PI variants in cell. (a, b) IMCD3 cells were stained with indicated antibodies, and nuclei were visualized by DAPI staining. Anti-PtdIns(4)P (PI(4)P) (a) and anti-ptdins(4,5)p 2 (PIP2) (b) antibodies were pre-absorbed with control PolyPIPosomes (Control), or PolyPIPosomes containing 5% of PI(4)P or PIP2 before adding onto samples for immunostaining. (c, d) IMCD3 cells transfected with the PI(3)P probe GFP-FYVE, or the PI(5)P probe GFP-PHD were processed for immunofluorescence microscopy. Scale bars: green, 5 μm; white, 0.5 μm. 7 P a g e
8 Supplementary Figure 6. CEP164 and TTBK2 contain evolutionarily conserved PI binding motifs. The protein sequences of Cep164 (a) and TTBK2 (b) orthologs were analyzed by MULTALIN alignment software. Evolutionarily conserved PI-binding motifs with lysine/arginine-rich patches were highlighted. (c, d) PolyPIPosome pull-down assay. HEK293T Cells transfected with 8 P a g e
9 wild-type (WT) Myc-CEP164 or GFP-TTBK2, or mutated Myc-CEP164 (9A) and GFP-TTBK2 (ΔPI) were subjected to pull-down assay with PolyPIPosomes containing PI(4)P. The resulting precipitates and cell lysates (Input) were immunoblotted with indicated antibodies. 9 P a g e
10 Supplementary Figure 7. Uncropped images of immunoblotting results for Figs P a g e
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Supplemental Material
 Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of
 Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
1 Supplementary Figure 1. Kif13b expression and localization in serum starved
 1 2 3 4 5 6 Supplementary Figure 1. Kif13b expression and localization in serum starved cells. (a) Immunoblot of NIH3T3 cell lysates after 24 hrs of serum deprivation, using rabbit polyclonal KIF13B antiserum.
1 2 3 4 5 6 Supplementary Figure 1. Kif13b expression and localization in serum starved cells. (a) Immunoblot of NIH3T3 cell lysates after 24 hrs of serum deprivation, using rabbit polyclonal KIF13B antiserum.
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Nature Structural & Molecular Biology: doi: /nsmb.3308
 Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein 22 mutant that causes type 1A Charcot-Marie-Tooth disease
 Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein mutant that causes type 1A Charcot-Marie-Tooth disease Taichi Hara, Yukiko Hashimoto, Tomoko Akuzawa, Rika Hirai,
Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein mutant that causes type 1A Charcot-Marie-Tooth disease Taichi Hara, Yukiko Hashimoto, Tomoko Akuzawa, Rika Hirai,
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions.
 Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions. 1 (a) Etoposide treatment gradually changes acetylation level and co-chaperone
Supplementary Figure 1. The Hsp70 acetylation level is related to the co-chaperone binding of Hsp70 under various stress conditions. 1 (a) Etoposide treatment gradually changes acetylation level and co-chaperone
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplementary Figure 1 Structural modeling and purification of V. cholerae ABH. (a) The migration of the purified rabh and catalytically inactive
 Supplementary Figure 1 Structural modeling and purification of V. cholerae ABH. (a) The migration of the purified rabh and catalytically inactive variants rabhs, rabhd, and rabhh are shown on 12% SDS-PAGE
Supplementary Figure 1 Structural modeling and purification of V. cholerae ABH. (a) The migration of the purified rabh and catalytically inactive variants rabhs, rabhd, and rabhh are shown on 12% SDS-PAGE
Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr
 Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various
 Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
supplementary information
 DOI: 10.1038/ncb2156 Figure S1 Depletion of p114rhogef with different sirnas. Caco-2 (a) and HCE (b) cells were transfected with individual sirnas, pools of the two sirnas or the On Target (OnT) sirna
DOI: 10.1038/ncb2156 Figure S1 Depletion of p114rhogef with different sirnas. Caco-2 (a) and HCE (b) cells were transfected with individual sirnas, pools of the two sirnas or the On Target (OnT) sirna
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
Supplemental Information
 Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
Hossain_Supplemental Figure 1
 Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2880 Supplementary Figure 1 Sequence alignment of Deup1 and Cep63. The protein sequence alignment was generated by the Clustal X 2.0 multiple sequence alignment program using default parameters.
DOI: 10.1038/ncb2880 Supplementary Figure 1 Sequence alignment of Deup1 and Cep63. The protein sequence alignment was generated by the Clustal X 2.0 multiple sequence alignment program using default parameters.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplemental material JCB. Pitaval et al., Cell shape and ciliogenesis Pitaval et al.
 Supplemental material THE JOURNAL OF CELL BIOLOGY Pitaval et al., http://www.jcb.org/cgi/content/full/jcb.201004003/dc1 S1 Figure S1. Cell cycle exit analysis. (A) Experimental procedure. RPE1 cells were
Supplemental material THE JOURNAL OF CELL BIOLOGY Pitaval et al., http://www.jcb.org/cgi/content/full/jcb.201004003/dc1 S1 Figure S1. Cell cycle exit analysis. (A) Experimental procedure. RPE1 cells were
Supplementary Figure 1. Nur77 and leptin-controlled obesity. (A) (B) (C)
 Supplementary Figure 1. Nur77 and leptin-controlled obesity. (A) Effect of leptin on body weight and food intake between WT and KO mice at the age of 12 weeks (n=7). Mice were i.c.v. injected with saline
Supplementary Figure 1. Nur77 and leptin-controlled obesity. (A) Effect of leptin on body weight and food intake between WT and KO mice at the age of 12 weeks (n=7). Mice were i.c.v. injected with saline
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Figure 1. Drawing of spinal cord open-book preparations and DiI tracing. Nature Neuroscience: doi: /nn.3893
 Supplementary Figure 1 Drawing of spinal cord open-book preparations and DiI tracing. Supplementary Figure 2 In ovo electroporation of dominant-negative PlexinA1 in commissural neurons induces midline
Supplementary Figure 1 Drawing of spinal cord open-book preparations and DiI tracing. Supplementary Figure 2 In ovo electroporation of dominant-negative PlexinA1 in commissural neurons induces midline
SUPPLEMENTARY INFORMATION FIGURE LEGENDS
 SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
A RRM1 H2AX DAPI. RRM1 H2AX DAPI Merge. Cont. sirna RRM1
 A H2AX DAPI H2AX DAPI Merge Cont sirna Figure S1: Accumulation of RRM1 at DNA damage sites (A) HeLa cells were subjected to in situ detergent extraction without IR irradiation, and immunostained with the
A H2AX DAPI H2AX DAPI Merge Cont sirna Figure S1: Accumulation of RRM1 at DNA damage sites (A) HeLa cells were subjected to in situ detergent extraction without IR irradiation, and immunostained with the
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/4/182/ra46/dc1 Supplementary Materials for The Deacetylase Promotes Membrane Localization and Activation of and PDK1 During Tumorigenesis and Cardiac Hypertrophy
www.sciencesignaling.org/cgi/content/full/4/182/ra46/dc1 Supplementary Materials for The Deacetylase Promotes Membrane Localization and Activation of and PDK1 During Tumorigenesis and Cardiac Hypertrophy
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Regulation of autophagic activity by ζ proteins associated with class III. phosphatidylinositol-3 kinase. Mercedes Pozuelo Rubio
 ONLINE SUPPORTING INFORMATION Regulation of autophagic activity by 14-3-3ζ proteins associated with class III phosphatidylinositol-3 kinase Mercedes Pozuelo Rubio entro Andaluz de Biología Molecular y
ONLINE SUPPORTING INFORMATION Regulation of autophagic activity by 14-3-3ζ proteins associated with class III phosphatidylinositol-3 kinase Mercedes Pozuelo Rubio entro Andaluz de Biología Molecular y
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated
 Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy
 Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Information
 Supplementary Information stability is regulated by CK2-dependent interaction with R2TP complex Patrick von Morgen 1,2, Kamila Burdova 1, Thomas G. Flower 3, Nicola J. O'Reilly 4, Simon J. Boulton 5, Stephen
Supplementary Information stability is regulated by CK2-dependent interaction with R2TP complex Patrick von Morgen 1,2, Kamila Burdova 1, Thomas G. Flower 3, Nicola J. O'Reilly 4, Simon J. Boulton 5, Stephen
Supplementary Fig. 1
 a FL (1-2266) NL (1-1190) CL (1191-2266) HA-ICE1: - HA-ICE1: - - - FLAG-ICE2: + + + + FLAG-ELL: + + + + + + IP: anti-ha FLAG-ICE2 HA-ICE1-FL HA-ICE1-NL HA-ICE1-CL FLAG-ICE2 b IP: anti-ha FL (1-2266) NL
a FL (1-2266) NL (1-1190) CL (1191-2266) HA-ICE1: - HA-ICE1: - - - FLAG-ICE2: + + + + FLAG-ELL: + + + + + + IP: anti-ha FLAG-ICE2 HA-ICE1-FL HA-ICE1-NL HA-ICE1-CL FLAG-ICE2 b IP: anti-ha FL (1-2266) NL
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Flag-Rac Vector V12 V12 N17 C40. Vector C40 pakt (T308) Akt1. Myc-DN-PAK1 (N-SP)
 a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
THE JOURNAL OF CELL BIOLOGY
 Supplemental Material THE JOURNAL OF CELL BIOLOGY Toso et al., http://www.jcb.org/cgi/content/full/jcb.200809055/dc1 Figure S1. Control experiments for sirna depletions used in Figs. 1 3. The phenotype
Supplemental Material THE JOURNAL OF CELL BIOLOGY Toso et al., http://www.jcb.org/cgi/content/full/jcb.200809055/dc1 Figure S1. Control experiments for sirna depletions used in Figs. 1 3. The phenotype
Supplementary Information
 Supplementary Information Supplementary Figures Supplementary Figure 1. MLK1-4 phosphorylate MEK in the presence of RAF inhibitors. (a) H157 cells were transiently transfected with Flag- or HA-tagged MLK1-4
Supplementary Information Supplementary Figures Supplementary Figure 1. MLK1-4 phosphorylate MEK in the presence of RAF inhibitors. (a) H157 cells were transiently transfected with Flag- or HA-tagged MLK1-4
supplementary information
 DOI: 10.1038/ncb1977 Figure S1 a. Immunofluorescence analysis of IFT20 localization in PBL costained with anti-β-tubulin antibodies. b. Immunofluorescence analysis of IFT20 localization in Jurkat cells,
DOI: 10.1038/ncb1977 Figure S1 a. Immunofluorescence analysis of IFT20 localization in PBL costained with anti-β-tubulin antibodies. b. Immunofluorescence analysis of IFT20 localization in Jurkat cells,
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
SUPPLEMENTAL FIGURES AND TABLES
 SUPPLEMENTAL FIGURES AND TABLES A B Flag-ALDH1A1 IP: α-ac HEK293T WT 91R 128R 252Q 367R 41/ 419R 435R 495R 412R C Flag-ALDH1A1 NAM IP: HEK293T + + - + D NAM - + + E Relative ALDH1A1 activity 1..8.6.4.2
SUPPLEMENTAL FIGURES AND TABLES A B Flag-ALDH1A1 IP: α-ac HEK293T WT 91R 128R 252Q 367R 41/ 419R 435R 495R 412R C Flag-ALDH1A1 NAM IP: HEK293T + + - + D NAM - + + E Relative ALDH1A1 activity 1..8.6.4.2
SUPPLEMENTARY INFORMATION
 Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/4/167/ra20/dc1 Supplementary Materials for Poly(ADP-Ribose) (PAR) Binding to Apoptosis-Inducing Factor Is Critical for PAR Polymerase-1 Dependent Cell Death (Parthanatos)
www.sciencesignaling.org/cgi/content/full/4/167/ra20/dc1 Supplementary Materials for Poly(ADP-Ribose) (PAR) Binding to Apoptosis-Inducing Factor Is Critical for PAR Polymerase-1 Dependent Cell Death (Parthanatos)
SUPPLEMENTARY INFORMATION. Small molecule activation of the TRAIL receptor DR5 in human cancer cells
 SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2.
 Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Hong et al.,
 Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
TRIM31 is recruited to mitochondria after infection with SeV.
 Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplemental Data. Sethi et al. (2014). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators.
 Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Supplementary Figure 1 PARP1 is involved in regulating the stability of mrnas from pro-inflammatory cytokine/chemokine mediators. (a) A graphic depiction of the approach to determining the stability of
Supplementary Information
 Supplementary Information Supplementary Figure S1 (a) P-cRAF colocalizes with LC3 puncta. Immunofluorescence (IF) depicting colocalization of P-cRAF (green) and LC3 puncta (red) in NIH/3T3 cells treated
Supplementary Information Supplementary Figure S1 (a) P-cRAF colocalizes with LC3 puncta. Immunofluorescence (IF) depicting colocalization of P-cRAF (green) and LC3 puncta (red) in NIH/3T3 cells treated
Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates
 Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates vascular calcification (VC). (a) Von Kossa staining shows that TSA potentiated the Pi-induced VC. Scale bar, 100
Supplementary Figure 1. TSA (10 nmol/l), non-class-selective HDAC inhibitor, potentiates vascular calcification (VC). (a) Von Kossa staining shows that TSA potentiated the Pi-induced VC. Scale bar, 100
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1 sirna and shrna mediated depletion of ATP7A results in loss of melanosomal ATP7A staining. a-h, sirna mediated ATP7A depletion. Immunofluorescence microscopy (IFM) analysis of ATP7A
Supplementary Figure 1 sirna and shrna mediated depletion of ATP7A results in loss of melanosomal ATP7A staining. a-h, sirna mediated ATP7A depletion. Immunofluorescence microscopy (IFM) analysis of ATP7A
SUPPLEMENTARY INFORMATION
 DOI: 1.1/ncb2918 Supplementary Figure 1 (a) Biotin-dUTP labelling does not affect S phase progression. Cells synchronized in mid-s phase were labelled with biotindutp during a 5 min hypotonic shift (red)
DOI: 1.1/ncb2918 Supplementary Figure 1 (a) Biotin-dUTP labelling does not affect S phase progression. Cells synchronized in mid-s phase were labelled with biotindutp during a 5 min hypotonic shift (red)
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Kimura et al.,
 Supplemental material JCB Kimura et al., http://www.jcb.org/cgi/content/full/jcb.201503023/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. TRIMs regulate IFN-γ induced autophagy. (A and B) HC image analysis
Supplemental material JCB Kimura et al., http://www.jcb.org/cgi/content/full/jcb.201503023/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. TRIMs regulate IFN-γ induced autophagy. (A and B) HC image analysis
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
Supplementary Information
 Supplementary Information Supplementary Figure 1: Xenopus model of missense mutations. A mutation equivalent to MCIDAS R381H/G355D was introduced in the Xenopus mcidas (R370H, G355D) by PCR, sequenced
Supplementary Information Supplementary Figure 1: Xenopus model of missense mutations. A mutation equivalent to MCIDAS R381H/G355D was introduced in the Xenopus mcidas (R370H, G355D) by PCR, sequenced
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in
 Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Control of cortex development by ULK4, a rare risk gene for mental disorders including schizophrenia
 Control of cortex development by ULK4, a rare risk gene for mental disorders including schizophrenia Bing Lang, Lei Zhang, Guanyu Jiang, Ling Hu, Wei Lan, Lei Zhao, Irene Hunter, Michal Pruski, Ning-Ning
Control of cortex development by ULK4, a rare risk gene for mental disorders including schizophrenia Bing Lang, Lei Zhang, Guanyu Jiang, Ling Hu, Wei Lan, Lei Zhao, Irene Hunter, Michal Pruski, Ning-Ning
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplemental Information Control of apico-basal epithelial polarity by the microtubule minus-end binding protein CAMSAP3 and spectraplakin ACF7
 Supplemental Information Control of apico-basal epithelial polarity by the microtubule minus-end binding protein CAMSAP3 and spectraplakin ACF7 Ivar Noordstra, Qingyang Liu, Wilco Nijenhuis, Shasha Hua,
Supplemental Information Control of apico-basal epithelial polarity by the microtubule minus-end binding protein CAMSAP3 and spectraplakin ACF7 Ivar Noordstra, Qingyang Liu, Wilco Nijenhuis, Shasha Hua,
Figure S1 is related to Figure 1B, showing more details of outer segment of
 Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Figure S1. Sequence alignments of ATRIP and ATR TopBP1 interacting regions.
 A H. sapiens 204 TKLQTS--ERANKLAAPSVSH VSPRKNPSVVIKPEACS-PQFGKTSFPTKESFSANMS LP 259 B. taurus 201 TKLQSS--ERANKLAVPTVSH VSPRKSPSVVIKPEACS-PQFGKPSFPTKESFSANKS LP 257 M. musculus 204 TKSQSN--GRTNKPAAPSVSH
A H. sapiens 204 TKLQTS--ERANKLAAPSVSH VSPRKNPSVVIKPEACS-PQFGKTSFPTKESFSANMS LP 259 B. taurus 201 TKLQSS--ERANKLAVPTVSH VSPRKSPSVVIKPEACS-PQFGKPSFPTKESFSANKS LP 257 M. musculus 204 TKSQSN--GRTNKPAAPSVSH
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb.201506071 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. TRA IP harbors a PCNA-binding PIP box required for its recruitment
Supplemental material JCB Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb.201506071 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. TRA IP harbors a PCNA-binding PIP box required for its recruitment
Figure S1. Specificity of polyclonal anti stabilin-1 and anti stabilin-2 antibodies Lysates of 293T cells transfected with empty vector, mouse
 Figure S1. Specificity of polyclonal anti stabilin-1 and anti stabilin-2 antibodies Lysates of 293T cells transfected with empty vector, mouse stabilin-1, or mouse stabilin-2 were immunoblotted using anti
Figure S1. Specificity of polyclonal anti stabilin-1 and anti stabilin-2 antibodies Lysates of 293T cells transfected with empty vector, mouse stabilin-1, or mouse stabilin-2 were immunoblotted using anti
Supplementary information. Supplementary Figures
 Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
Supplementary information Supplementary Figures Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
No wash 2 Washes 2 Days ** ** IgG-bead phagocytosis (%)
 Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking
 Human Molecular Genetics, 2009, Vol. 18, No. 20 3926 3941 doi:10.1093/hmg/ddp335 Advance Access published on July 22, 2009 Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a
Human Molecular Genetics, 2009, Vol. 18, No. 20 3926 3941 doi:10.1093/hmg/ddp335 Advance Access published on July 22, 2009 Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
