Supplementary information. Supplementary Figures
|
|
|
- Lily Powell
- 6 years ago
- Views:
Transcription
1 Supplementary information Supplementary Figures
2 Supplementary Figure 1. A. i. HA-JMY expressing U2OS cells were treated with SAHA (6h). DAPI was used to visualise nuclei. ii. U2OS cells stably expressing HA-JMY were untreated (-) or treated with AZD2014 (AZD) or grown under starvation conditions (EBSS) for 6h. JMY was detected with anti-ha. Scale bar = 10 m. B. U2OS cells expressing HA-JMY were treated with rapamycin (rapa, overnight), AZD2014 (AZD, 4h) tubastatin A (tuba; overnight) or bafilomycin A1 (baf; 4h). Inset shows enlarged region as denoted. JMY was detected with anti-ha. LC3 was detected using mouse (green) or rabbit (red) anti-lc3. Scale bar = 10 m. C. Endogenous JMY levels in various cell lines after treating with SAHA as denoted. Actin was used as a loading control. D. Endogenous JMY levels in various cell lines after treating overnight with SAHA (S), rapamycin (R) or tubastatin A (T). E. Endogenous JMY levels in various cell lines treated with bafilomycin A1 (baf) for the time points indicated.
3
4 Supplementary Figure 2. A. U2OS cells expressing JMY and derivatives were grown in the presence of EBSS for 6h. JMY was detected with anti-ha and rabbit anti-lc3 was used to detect LC3. Scale bar = 10 m. B. U2OS cells stably expressing JMYmtLIR (mtlir) were grown in EBSS for 6h. LIR was detected with anti-ha and LC3 with anti-lc3b. Scale bar = 10 m. C. Hela cells were treated with SAHA before harvesting for IP with anti-jmy antibody (JMY) or non-specific IgG control (ns). Inputs (In) represent 2% of extract. n=2 independent experiments. D. His-JMY (FL) or C-terminal JMY (C-term; ) was incubated with GST-LC3B and complexes isolated using glutathione sepharose 4B. Inputs represent 50% of total. n=2 independent experiments. Samples were run on the same gel but noncontiguous. E. In vitro transcribed and translated HA-JMY N-terminal region (1-119; WT) or mtlir proteins (as denoted) were incubated with 1 g of GST-LC3B before immunoprecipitating with glutathione sepharose beads. Inputs represent 25% of total. F. U2OS cells expressing HA-p62 with wild-type LIR (WT) or JMY LIR (p62/jmylir) sequences were grown in the presence of SAHA (6h) or EBSS (2h). p62 was detected using anti-ha antibody and LC3 with rabbit-anti-lc3. Scale bar = 10 m. Residues in p62 LIR shown to make direct contact with LC3 21 are highlighted in red. G. U2OS cells stably expressing HA-JMY (JMY), HA-JMYmtLIR (mtlir) or vector (vec) control were grown in EBSS for the time points indicated before harvesting. Actin was used as a loading control. H. U2OS cells stably expressing HA-JMY (JMY), HA-JMYmtLIR (mtlir) or vector (vec) control were treated with or without SAHA for 24h. Graph represents n=4 independent experiments, *p<0.08, Student s t-test.
5 Supplementary Figure 3. A. MCF7 cells were treated with non-targeting (NT) or JMY sirna for 72h before SAHA treatment for the times indicated. B. U2OS cells were treated with non-targeting (NT) or JMY sirna for 72h before treating with SAHA for the indicated
6 time points. Actin was used as a loading control. Graph represents n=6 independent experiments, *p <0.05, Student s t-test. C. U2OS cells were treated with non-targeting (NT) or JMY sirnas 1-3 for 72h. Six hours before harvesting, cells were treated with SAHA (+) or vehicle control (-). Actin was used as a loading control. n=3 independent experiments. D. HeLa cells were treated with non-targeting (NT) or JMY sirna for 72h before replacing with EBSS for the indicated time points. E. U2OS cells were treated with non-targeting (NT) or JMY sirna for 72h before replacing the medium with EBSS for the indicated time points. Graph represents n=5 independent experiments, *p<0.05, **p<0.001, Student s t-test. F. U2OS cells were treated with non-targeting (NT) or JMY sirna for 72h. The last 24h cells with treated with vehicle (-) or SAHA (+) with or without bafilomycin A1 (baf) for 4h. Actin was used as a loading control. Graph represents n=3 independent experiments, *p=0.05. G. U2OS cells stably overexpressing HA-JMY were treated with SAHA for 6h. JMY was detected with rabbit anti-ha and mouse anti-lamp2 was used to detect endogenous LAMP2. H. U2OS cells stably overexpressing HA-JMY were treated with SAHA for 6h. Rabbit anti-ha was used to detect JMY and mouse anti-cathepsin D was used to detect endogenous cathepsin D.
7 Supplementary Figure 4. A. U2OS cells expressing HA-JMY and derivatives. JMY was detected with anti-ha antibody and phalloidin was used to visualise F-actin. Arrows denote areas of JMY and phalloidin colocalisation. Inset denotes enlarged region. Scale bar = 10 m. B. U2OS cells expressing HA-JMY and HA-JMYmtLIR (mtlir) were treated with SAHA for 6h. JMY was detected with rabbit anti-ha antibody and actin was detected with mouse anti-actin antibody. Inset denotes enlarged region. Scale bar = 10 m. C. U2OS cells
8 expressing HA-JMY and derivatives were grown in EBSS for 6h. JMY was detected with rabbit anti-ha antibody and actin was detected with mouse anti-actin antibody. Inset denotes enlarged region. Scale bar = 10 m. D. U2OS cells expressing HA-JMY were treated with SAHA for 6h. JMY was detected with anti-ha antibody and anti-lc3 was used to visualise autophagosomes. Inset denotes enlarged region. Scale bar = 10 m. E. U2OS cells stably expressing HA-JMY grown on coverslips before performing G-actin-incorporation assays using 0.4 M AlexaFluor 488-labelled G-actin. Assays were carried out for 2 min at room temperature before fixation and processing for IF. JMY was detected with anti-ha antibody.
9 Supplementary Figure 5. A. Schematic illustrates the mcherry-gfp-lc3 (LC3) and mcherry-gfp-wh2-lc3 (WH2-LC3) constructs. B. U2OS cells expressing mcherry-gfp- LC3 or mcherry-gfp-wh2-lc3 were treated with rapamycin A. Actin was visualised with mouse-anti-actin antibody. C. Enlarged area of mcherry-gfp-lc3 (left) compared to mcherry-gfp-wh2-lc3 (right) as denoted by hatched boxes in B. Arrows denote actin foci that do not colocalise with mcherry-gfp-lc3 (left) compared to the significant colocalisation of actin with mcherry-gfp-wh2-lc3 (arrows, right). Scale bar = 10 m. D. U2OS cells expressing HA-JMY WH2 were treated with SAHA 6h before processing for IF. Anti-HA and anti-lc3 antibodies were used. Arrows denote enlarged perinuclear autophagosomes. DAPI was used to visualise nuclei. Scale bar = 10 m.
10 Supplementary Figure 6. A. U2OS cells were treated with control (NT) or JMY sirna for 72h. Six hours before harvesting cells were treated with SAHA, with or without Arp2/3 inhibitor (CK-869; 20 M) before harvesting. B. U2OS cells were treated with control (NT) or JMY sirna for 72h. Two hours before harvesting the medium was replaced with EBSS with or without Arp2/3 inhibitor (CK-666 or CK-869; 40 M) as denoted before harvesting. C.
11 U2OS cells were treated with control (NT) or JMY sirna for 72h. Six hours before harvesting cells were treated with SAHA with or without the Arp2/3 inhibitor (CK-666; 20 M) as denoted before harvesting. Graph represents n=3 independent experiments, *p<0.05, Student s t-test. D. U2OS cells were treated with control (NT) or JMY sirna for 72h. The cells were treated with vehicle control (-) or AZD2014 for 24h before harvesting and analysed by FACS. Graph represents percentage sub-g1 of a representative experiment. n=3 independent experiments, *p<0.001, Student s t-test. Blots underneath represent input protein levels. E. MTT assays were performed with U2OS cells treated with control (NT) or JMY sirna for 72h before replating in DMEM (-) or EBSS (+) for 24h. Graph represents mean +/- s.e.m with DMEM (control) values normalised to 100% and EBSS values expressed as a percentage of control. Representative experiment is shown, n=2 independent experiments. *p<0.005, Student s t-test. F. MTT assays were performed with U2OS cells stably expressing HA-JMY (JMY) or vector. Cells were grown for 24h in DMEM (-) or EBSS (+) before performing MTT assay. Graph represents mean +/- s.e.m with DMEM (control) values normalised to 100% and EBSS values expressed as a percentage of control. Representative experiment is shown, n=2 independent experiments. *p=0.01, Student s t- test.
12 Supplementary Figure 7. A. qpcr analysis of hlc3b mrna levels from U2OS cells treated with either vehicle (-) or 6h SAHA (+). Values were normalised to GAPDH and expressed as fold over control (NT) untreated. n=4 independent experiments. B. qrt-pcr analysis of hlc3b mrna levels from U2OS cells grown in either DMEM (-) or EBSS for 30 min (+). Values were normalised to GAPDH and expressed as fold over control (NT) untreated. n=3 independent experiments. *p<0.05, Student s t-test. C. HCT116 p53 +/+ or p53 - /- cells were treated with control (NT) or JMY sirna for 72h. Six hours prior to harvesting cells were treated with SAHA (+) or vehicle (-). A representative experiment is shown, n=3 independent experiments.
13
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
M X 500 µl. M X 1000 µl
 GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr
 Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
SUPPLEMENTARY MATERIALS AND METHODS
 SUPPLEMENTARY MATERIALS AND METHODS Chemicals. All chemicals used in supplementary experiments were the same as in the manuscript except the following. Methyl-methanesulfonate (MMS) was from Fluka. NU1025
SUPPLEMENTARY MATERIALS AND METHODS Chemicals. All chemicals used in supplementary experiments were the same as in the manuscript except the following. Methyl-methanesulfonate (MMS) was from Fluka. NU1025
HCT116 SW48 Nutlin: p53
 Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
SUPPLEMENTAL FIGURE LEGENDS. Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are
 SUPPLEMENTAL FIGURE LEGENDS Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are the amino acid sequences of human DDR2, mouse DDR2 and the closest homologs in zebrafish and C. Elegans.
SUPPLEMENTAL FIGURE LEGENDS Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are the amino acid sequences of human DDR2, mouse DDR2 and the closest homologs in zebrafish and C. Elegans.
SUPPLEMENTARY INFORMATION FIGURE 1 - 1
 SUPPLEMENTARY INFORMATION FIGURE 1-1 SUPPLEMENTARY INFORMATION FIGURE 2-2 SUPPLEMENTARY INFORMATION METHODS GST-Pull-Down. Cultures of E. Coli (BL21) were transformed with pgex (Clontech) and pgex recombinant
SUPPLEMENTARY INFORMATION FIGURE 1-1 SUPPLEMENTARY INFORMATION FIGURE 2-2 SUPPLEMENTARY INFORMATION METHODS GST-Pull-Down. Cultures of E. Coli (BL21) were transformed with pgex (Clontech) and pgex recombinant
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated
 Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Supplemental Data Supplementary Figure Legends and Scheme Figure S1.
 Supplemental Data Supplementary Figure Legends and Scheme Figure S1. UTK1 inhibits the second EGF-induced wave of lamellipodia formation in TT cells. A and B, EGF-induced lamellipodia formation in TT cells,
Supplemental Data Supplementary Figure Legends and Scheme Figure S1. UTK1 inhibits the second EGF-induced wave of lamellipodia formation in TT cells. A and B, EGF-induced lamellipodia formation in TT cells,
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Translation of HTT mrna with expanded CAG repeats is regulated by
 Supplementary Information Translation of HTT mrna with expanded CAG repeats is regulated by the MID1-PP2A protein complex Sybille Krauß 1,*, Nadine Griesche 1, Ewa Jastrzebska 2,3, Changwei Chen 4, Désiree
Supplementary Information Translation of HTT mrna with expanded CAG repeats is regulated by the MID1-PP2A protein complex Sybille Krauß 1,*, Nadine Griesche 1, Ewa Jastrzebska 2,3, Changwei Chen 4, Désiree
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing
 Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
SUPPLEMENTARY INFORMATION
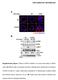 SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
(A) Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site.
 SUPPLEMENTRY INFORMTION SUPPLEMENTL FIGURES Figure S1. () Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site. () Western blot of ligated and unligated sciatic
SUPPLEMENTRY INFORMTION SUPPLEMENTL FIGURES Figure S1. () Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site. () Western blot of ligated and unligated sciatic
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction)
 Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Supplementary Figure Legend
 Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Wnt16 smact merge VK/AB
 A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
Center Drive, University of Michigan Health System, Ann Arbor, MI
 Leukotriene B 4 -induced reduction of SOCS1 is required for murine macrophage MyD88 expression and NFκB activation Carlos H. Serezani 1,3, Casey Lewis 1, Sonia Jancar 2 and Marc Peters-Golden 1,3 1 Division
Leukotriene B 4 -induced reduction of SOCS1 is required for murine macrophage MyD88 expression and NFκB activation Carlos H. Serezani 1,3, Casey Lewis 1, Sonia Jancar 2 and Marc Peters-Golden 1,3 1 Division
Supplementary Figure S1
 Supplementary Figure S1 Figure S1. CENP- FP constructs target to centromeres irrespective of site of tagging. FP- CENP and CENP- FP constructs were transfected into U2OS cells and counterstained with anti-
Supplementary Figure S1 Figure S1. CENP- FP constructs target to centromeres irrespective of site of tagging. FP- CENP and CENP- FP constructs were transfected into U2OS cells and counterstained with anti-
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplementary Fig. 1. Isolation and in vitro expansion of EpCAM + cholangiocytes. For collagenase perfusion, enzyme solution was injected from the
 Supplementary Fig. 1. Isolation and in vitro expansion of EpCAM + cholangiocytes. For collagenase perfusion, enzyme solution was injected from the portal vein for digesting adult livers, whereas it was
Supplementary Fig. 1. Isolation and in vitro expansion of EpCAM + cholangiocytes. For collagenase perfusion, enzyme solution was injected from the portal vein for digesting adult livers, whereas it was
Supplementary Information
 Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand
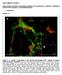 SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
Supplemental Information. HEXIM1 and NEAT1 Long Non-coding RNA Form. a Multi-subunit Complex that Regulates. DNA-Mediated Innate Immune Response
 Molecular Cell, Volume 67 Supplemental Information HEXIM1 and NEAT1 Long Non-coding RNA Form a Multi-subunit Complex that Regulates DNA-Mediated Innate Immune Response Mehdi Morchikh, Alexandra Cribier,
Molecular Cell, Volume 67 Supplemental Information HEXIM1 and NEAT1 Long Non-coding RNA Form a Multi-subunit Complex that Regulates DNA-Mediated Innate Immune Response Mehdi Morchikh, Alexandra Cribier,
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by. Regulating Occludin
 Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by Regulating Occludin Chaithanya Chelakkot 1,ǂ, Jaewang Ghim 2,3,ǂ, Nirmal Rajasekaran 4, Jong-Sun Choi 5, Jung-Hwan
Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by Regulating Occludin Chaithanya Chelakkot 1,ǂ, Jaewang Ghim 2,3,ǂ, Nirmal Rajasekaran 4, Jong-Sun Choi 5, Jung-Hwan
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Data. LMO4 Controls the Balance between Excitatory. and Inhibitory Spinal V2 Interneurons
 Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1.
 Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines
 Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Immunofluorescence images of different core histones and different histone exchange assay.
 Molecular Cell, Volume 51 Supplemental Information Enhanced Chromatin Dynamics by FACT Promotes Transcriptional Restart after UV-Induced DNA Damage Christoffel Dinant, Giannis Ampatziadis-Michailidis,
Molecular Cell, Volume 51 Supplemental Information Enhanced Chromatin Dynamics by FACT Promotes Transcriptional Restart after UV-Induced DNA Damage Christoffel Dinant, Giannis Ampatziadis-Michailidis,
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Supplemental Material
 Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
TE5 KYSE510 TE7 KYSE70 KYSE140
 TE5 KYSE5 TT KYSE7 KYSE4 Supplementary Figure. Hockey stick plots showing input normalized, rank ordered H3K7ac signals for the candidate SE-associated lncrnas in this study. Rpm Rpm Rpm Chip-seq H3K7ac
TE5 KYSE5 TT KYSE7 KYSE4 Supplementary Figure. Hockey stick plots showing input normalized, rank ordered H3K7ac signals for the candidate SE-associated lncrnas in this study. Rpm Rpm Rpm Chip-seq H3K7ac
FCN1 (M-ficolin), which directly associates with immunoglobulin G1, is a molecular target of intravenous immunoglobulin therapy for Kawasaki disease
 FCN1 (M-ficolin), which directly associates with immunoglobulin G1, is a molecular target of intravenous immunoglobulin therapy for Kawasaki disease Daisuke Okuzaki, Kaori Ota, Shin-ichi Takatsuki, Yukari
FCN1 (M-ficolin), which directly associates with immunoglobulin G1, is a molecular target of intravenous immunoglobulin therapy for Kawasaki disease Daisuke Okuzaki, Kaori Ota, Shin-ichi Takatsuki, Yukari
Supporting Information
 Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Chemically defined conditions for human ipsc derivation and culture
 Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Hossain_Supplemental Figure 1
 Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Supplemental Data. Steiner et al. Plant Cell. (2012) /tpc
 Supplemental Figure 1. SPY does not interact with free GST. Invitro pull-down assay using E. coli-expressed MBP-SPY and GST, GST-TCP14 and GST-TCP15. MBP-SPY was used as bait and incubated with equal amount
Supplemental Figure 1. SPY does not interact with free GST. Invitro pull-down assay using E. coli-expressed MBP-SPY and GST, GST-TCP14 and GST-TCP15. MBP-SPY was used as bait and incubated with equal amount
Supplemental Information. Lysine-5 Acetylation Negatively Regulates. Lactate Dehydrogenase A and Is Decreased. in Pancreatic Cancer
 Cancer Cell, Volume 23 Supplemental Information Lysine-5 Acetylation Negatively Regulates Lactate Dehydrogenase A and Is Decreased in Pancreatic Cancer Di Zhao, Shao-Wu Zou, Ying Liu, Xin Zhou, Yan Mo,
Cancer Cell, Volume 23 Supplemental Information Lysine-5 Acetylation Negatively Regulates Lactate Dehydrogenase A and Is Decreased in Pancreatic Cancer Di Zhao, Shao-Wu Zou, Ying Liu, Xin Zhou, Yan Mo,
* ** ** * IB: p-p90rsk. p90rsk (Ser380) (arbitrary units) (Ser380) p90rsk. IB: p90rsk. Tubulin. IB: Tubulin. Ang II (200 nm) Ang II (200 nm)
 I: p-p9rsk I: p9rsk I: C I: p-p9rsk I: p9rsk 5 (ka) 5 5 (min) Ang II ( nm) p-p9rsk (Ser8) p9rsk p-p9rsk (Ser8) p9rsk (h) Mannitol 5 mm -Glucose 5 mm p9rsk (Ser8) (arbitrary units) p-p9rsk (Ser8) (arbitrary
I: p-p9rsk I: p9rsk I: C I: p-p9rsk I: p9rsk 5 (ka) 5 5 (min) Ang II ( nm) p-p9rsk (Ser8) p9rsk p-p9rsk (Ser8) p9rsk (h) Mannitol 5 mm -Glucose 5 mm p9rsk (Ser8) (arbitrary units) p-p9rsk (Ser8) (arbitrary
Supplemental Information. PARP1 Represses PAP and Inhibits Polyadenylation during Heat Shock
 Molecular Cell, Volume 49 Supplemental Information PARP1 Represses PAP and Inhibits Polyadenylation during Heat Shock Dafne Campigli Di Giammartino, Yongsheng Shi, and James L. Manley Supplemental Information
Molecular Cell, Volume 49 Supplemental Information PARP1 Represses PAP and Inhibits Polyadenylation during Heat Shock Dafne Campigli Di Giammartino, Yongsheng Shi, and James L. Manley Supplemental Information
Expanded View Figures
 Vasco Meneghini et al Effective lentiviral GT in NHP brains EMO Molecular Medicine Expanded View Figures Figure EV1. VN and GFP distribution in LV.GFPinjected NHPs. Vector copy number (VN) cartography
Vasco Meneghini et al Effective lentiviral GT in NHP brains EMO Molecular Medicine Expanded View Figures Figure EV1. VN and GFP distribution in LV.GFPinjected NHPs. Vector copy number (VN) cartography
by Neurobasal medium (supplemented with B27, 0.5mM glutamine, and 100 U/mL
 Supplementary Materials and methods Neuronal cultures and transfection The hippocampus was dissected from E8 rat embryos, dissociated, and neurons plated onto glass coverslips coated with poly-ornithine
Supplementary Materials and methods Neuronal cultures and transfection The hippocampus was dissected from E8 rat embryos, dissociated, and neurons plated onto glass coverslips coated with poly-ornithine
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/4/157/ra4/dc1 Supplementary Materials for Genome-Wide RNAi Screen Reveals Disease-Associated Genes That Are Common to Hedgehog and Wnt Signaling Leni S. Jacob,
www.sciencesignaling.org/cgi/content/full/4/157/ra4/dc1 Supplementary Materials for Genome-Wide RNAi Screen Reveals Disease-Associated Genes That Are Common to Hedgehog and Wnt Signaling Leni S. Jacob,
Online Supplementary Information
 Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1.
 Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc RGLG Hormonal treatment H2O B RGLG µm ABA µm ACC µm GA Time (hours) µm µm MJ µm IA
 Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb.201506071 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. TRA IP harbors a PCNA-binding PIP box required for its recruitment
Supplemental material JCB Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb.201506071 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. TRA IP harbors a PCNA-binding PIP box required for its recruitment
Nature Medicine: doi: /nm.4464
 Supplementary Fig. 1. Amino acid transporters and substrates used for selectivity screening. (A) Common transporters and amino acid substrates shown. Amino acids designated by one-letter codes. Transporters
Supplementary Fig. 1. Amino acid transporters and substrates used for selectivity screening. (A) Common transporters and amino acid substrates shown. Amino acids designated by one-letter codes. Transporters
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2774 Figure S1 TRF2 dosage modulates the tumorigenicity of mouse and human tumor cells. (a) Left: immunoblotting with antibodies directed against the Myc tag of the transduced TRF2 forms
DOI: 10.1038/ncb2774 Figure S1 TRF2 dosage modulates the tumorigenicity of mouse and human tumor cells. (a) Left: immunoblotting with antibodies directed against the Myc tag of the transduced TRF2 forms
SUPPLEMENTARY INFORMATION. Small molecule activation of the TRAIL receptor DR5 in human cancer cells
 SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss
 SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
Nature Immunology: doi: /ni Supplementary Figure 1. Zranb1 gene targeting.
 Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 BALB/c LYVE1-deficient mice exhibited reduced lymphatic trafficking of all DC subsets after oxazolone-induced sensitization. (a) Schematic overview of the mouse skin oxazolone contact
Supplementary Figure 1 BALB/c LYVE1-deficient mice exhibited reduced lymphatic trafficking of all DC subsets after oxazolone-induced sensitization. (a) Schematic overview of the mouse skin oxazolone contact
GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts
 Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Supplemental Data. Regulating Gene Expression. through RNA Nuclear Retention
 Supplemental Data Regulating Gene Expression through RNA Nuclear Retention Kannanganattu V. Prasanth, Supriya G. Prasanth, Zhenyu Xuan, Stephen Hearn, Susan M. Freier, C. Frank Bennett, Michael Q. Zhang,
Supplemental Data Regulating Gene Expression through RNA Nuclear Retention Kannanganattu V. Prasanth, Supriya G. Prasanth, Zhenyu Xuan, Stephen Hearn, Susan M. Freier, C. Frank Bennett, Michael Q. Zhang,
Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous
 Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
1. Cross-linking and cell harvesting
 ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
INOS. Colorimetric Cell-Based ELISA Kit. Catalog #: OKAG00807
 INOS Colorimetric Cell-Based ELISA Kit Catalog #: OKAG00807 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research Purposes Only. Not Intended
INOS Colorimetric Cell-Based ELISA Kit Catalog #: OKAG00807 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research Purposes Only. Not Intended
Khaled_Fig. S MITF TYROSINASE R²= MITF PDE4D R²= Variance from mean mrna expression
 Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
CytoGLOW. IKK-α/β. Colorimetric Cell-Based ELISA Kit. Catalog #: CB5358
 CytoGLOW IKK-α/β Colorimetric Cell-Based ELISA Kit Catalog #: CB5358 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research Purposes Only.
CytoGLOW IKK-α/β Colorimetric Cell-Based ELISA Kit Catalog #: CB5358 Please read the provided manual entirely prior to use as suggested experimental protocols may have changed. Research Purposes Only.
supplementary information
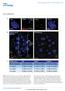 DOI: 10.1038/ncb2017 Figure S1 53BP1 sirna results in a heterochromatic DSB repair defect. Panel A: NIH3T3 cells were transfected with murine 53BP1 sirna. 48 hrs later, cells were irradiated with 3 Gy
DOI: 10.1038/ncb2017 Figure S1 53BP1 sirna results in a heterochromatic DSB repair defect. Panel A: NIH3T3 cells were transfected with murine 53BP1 sirna. 48 hrs later, cells were irradiated with 3 Gy
affects the development of newborn neurons Brain Mind Institute and School of Life Sciences, Ecole Polytechnique Fédérale de
 Shedding of neurexin 3β ectodomain by ADAM10 releases a soluble fragment that affects the development of newborn neurons Erika Borcel* a, Magda Palczynska* a, Marine Krzisch b, Mitko Dimitrov a, Giorgio
Shedding of neurexin 3β ectodomain by ADAM10 releases a soluble fragment that affects the development of newborn neurons Erika Borcel* a, Magda Palczynska* a, Marine Krzisch b, Mitko Dimitrov a, Giorgio
from α- to β-cleavage
 Supplementary Information Specific antibody binding to the APP 672-699 region shifts APP processing from α- to β-cleavage Song Li 1,6,8, Juan Deng 1,7,8, Huayan Hou 1,8, Jun Tian 1, Brian Giunta 2,3, Yanjiang
Supplementary Information Specific antibody binding to the APP 672-699 region shifts APP processing from α- to β-cleavage Song Li 1,6,8, Juan Deng 1,7,8, Huayan Hou 1,8, Jun Tian 1, Brian Giunta 2,3, Yanjiang
Supplementary Information
 Supplementary Information promotes cancer cell invasion and proliferation by receptor-mediated endocytosis-dependent and -independent mechanisms, respectively Kensaku Shojima, Akira Sato, Hideaki Hanaki,
Supplementary Information promotes cancer cell invasion and proliferation by receptor-mediated endocytosis-dependent and -independent mechanisms, respectively Kensaku Shojima, Akira Sato, Hideaki Hanaki,
Changsong Yang, Martin Pring, Martin A. Wear, Minzhou Huang, John A. Cooper, Tatyana M. Svitkina, Sally H. Zigmond
 Mammalian CARMIL Inhibits Actin Filament Capping by Capping Protein Changsong Yang, Martin Pring, Martin A. Wear, Minzhou Huang, John A. Cooper, Tatyana M. Svitkina, Sally H. Zigmond Supplemental Experimental
Mammalian CARMIL Inhibits Actin Filament Capping by Capping Protein Changsong Yang, Martin Pring, Martin A. Wear, Minzhou Huang, John A. Cooper, Tatyana M. Svitkina, Sally H. Zigmond Supplemental Experimental
Supplemental Information. A Tyrosine Phosphorylation Cycle. Regulates Fungal Activation of a Plant. Receptor Ser/Thr Kinase
 Cell Host & Microbe, Volume 23 Supplemental Information A Tyrosine Phosphorylation Cycle Regulates Fungal Activation of a Plant Receptor Ser/Thr Kinase Jun Liu, Bing Liu, Sufen Chen, Ben-Qiang Gong, Lijuan
Cell Host & Microbe, Volume 23 Supplemental Information A Tyrosine Phosphorylation Cycle Regulates Fungal Activation of a Plant Receptor Ser/Thr Kinase Jun Liu, Bing Liu, Sufen Chen, Ben-Qiang Gong, Lijuan
GTTCGGGTTCC TTTTGAGCAG
 Supplementary Figures Splice variants of the SIP1 transcripts play a role in nodule organogenesis in Lotus japonicus. Wang C, Zhu H, Jin L, Chen T, Wang L, Kang H, Hong Z, Zhang Z. 5 UTR CDS 3 UTR TCTCAACCATCCTTTGTCTGCTTCCGCCGCATGGGTGAGGTCATTTTGTCTAGATGACGTGCAATTTACAATGA
Supplementary Figures Splice variants of the SIP1 transcripts play a role in nodule organogenesis in Lotus japonicus. Wang C, Zhu H, Jin L, Chen T, Wang L, Kang H, Hong Z, Zhang Z. 5 UTR CDS 3 UTR TCTCAACCATCCTTTGTCTGCTTCCGCCGCATGGGTGAGGTCATTTTGTCTAGATGACGTGCAATTTACAATGA
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and
 Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
42 fl organelles = 34.5 fl (1) 3.5X X 0.93 = 78,000 (2)
 SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
Supplemental Information
 Molecular Cell, Volume 54 Supplemental Information BRD7, a Tumor Suppressor, Interacts with p85 and Regulates PI3K Activity Yu-Hsin Chiu, Jennifer Y. Lee, and Lewis C. Cantley Supplemental Materials Supplemental
Molecular Cell, Volume 54 Supplemental Information BRD7, a Tumor Suppressor, Interacts with p85 and Regulates PI3K Activity Yu-Hsin Chiu, Jennifer Y. Lee, and Lewis C. Cantley Supplemental Materials Supplemental
mcherry Monoclonal Antibody (16D7) Catalog Number M11217 Product data sheet
 Website: thermofisher.com Customer Service (US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 mcherry Monoclonal Antibody (16D7) Catalog Number M11217 Product data sheet Details
Website: thermofisher.com Customer Service (US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 mcherry Monoclonal Antibody (16D7) Catalog Number M11217 Product data sheet Details
EGFR (Phospho-Ser695)
 Assay Biotechnology Company www.assaybiotech.com Tel: 1-877-883-7988 Fax: 1-877-610-9758 EGFR (Phospho-Ser695) Colorimetric Cell-Based ELISA Kit Catalog #: OKAG02090 Please read the provided manual entirely
Assay Biotechnology Company www.assaybiotech.com Tel: 1-877-883-7988 Fax: 1-877-610-9758 EGFR (Phospho-Ser695) Colorimetric Cell-Based ELISA Kit Catalog #: OKAG02090 Please read the provided manual entirely
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/323/5910/124/dc1 Supporting Online Material for Regulation of Neuronal Survival Factor MEF2D by Chaperone-Mediated Autophagy Qian Yang, Hua She, Marla Gearing, Emanuela
www.sciencemag.org/cgi/content/full/323/5910/124/dc1 Supporting Online Material for Regulation of Neuronal Survival Factor MEF2D by Chaperone-Mediated Autophagy Qian Yang, Hua She, Marla Gearing, Emanuela
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
cdna by PCR with primers BglII-PacI-MAML (5 -GGCCAGATCTTTAATTAAGCCGCCAC CATGGCGCTGCCGCGGCACA-3 ) and XhoI-PmeI-GFP (5 - GGCCCTCGAGGTTTAAACT
 Supplementary Materials and Methods Generation of ptof-dnmaml1 Platinum Pfx DNA polymerase (Invitrogen) was used to amplify GFP-DNMAML1 cdna by PCR with primers BglII-PacI-MAML (5 -GGCCAGATCTTTAATTAAGCCGCCAC
Supplementary Materials and Methods Generation of ptof-dnmaml1 Platinum Pfx DNA polymerase (Invitrogen) was used to amplify GFP-DNMAML1 cdna by PCR with primers BglII-PacI-MAML (5 -GGCCAGATCTTTAATTAAGCCGCCAC
