The transcription factor Nerfin-1 prevents reversion of neurons into neural stem cells
|
|
|
- Zoe Norris
- 6 years ago
- Views:
Transcription
1 The transcription factor Nerfin-1 prevents reversion of neurons into neural stem cells Francesca Froldi, 1,2,6 Milan Szuperak, 1,2,6 Chen-Fang Weng, 1,2 Wei Shi, 3,4 Anthony T. Papenfuss, 1,2,3,5 and Louise Y. Cheng 1,2 1 Peter MacCallum Cancer Centre, East Melbourne, Victoria 3002, Australia; 2 Sir Peter MacCallum Department of Oncology, University of Melbourne, Parkville, Victoria 3010, Australia; 3 Bioinformatics Division, The Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria 3052, Australia; 4 Department of Computing and Information Systems, The University of Melbourne, Parkville, Victoria 3010, Australia; 5 Department of Medical Biology, University of Melbourne, Melbourne, Victoria 3010, Australia Cellular dedifferentiation is the regression of a cell from a specialized state to a more multipotent state and is implicated in cancer. However, the transcriptional network that prevents differentiated cells from reacquiring stem cell fate is so far unclear. Neuroblasts (NBs), the Drosophila neural stem cells, are a model for the regulation of stem cell self-renewal and differentiation. Here we show that the Drosophila zinc finger transcription factor Nervous fingers 1 (Nerfin-1) locks neurons into differentiation, preventing their reversion into NBs. Following Prospero-dependent neuronal specification in the ganglion mother cell (GMC), a Nerfin-1-specific transcriptional program maintains differentiation in the post-mitotic neurons. The loss of Nerfin-1 causes reversion to multipotency and results in tumors in several neural lineages. Both the onset and rate of neuronal dedifferentiation in nerfin-1 mutant lineages are dependent on Myc- and target of rapamycin (Tor)-mediated cellular growth. In addition, Nerfin-1 is required for NB differentiation at the end of neurogenesis. RNA sequencing (RNA-seq) and chromatin immunoprecipitation (ChIP) analysis show that Nerfin-1 administers its function by repression of selfrenewing-specific and activation of differentiation-specific genes. Our findings support the model of bidirectional interconvertibility between neural stem cells and their post-mitotic progeny and highlight the importance of the Nerfin-1-regulated transcriptional program in neuronal maintenance. [Keywords: stem cells; neuroblasts; cancer; dedifferentiation; Drosophila; central nervous system] Supplemental material is available for this article. Received August 11, 2014; revised version accepted December 1, Dedifferentiation is a cellular process by which a partially or terminally differentiated cell reverts to a less differentiated, more multipotent state. The bidirectional conversion between differentiated cells and cancer stem cells often underlies carcinogenesis. For example, it was recently demonstrated that glioblastoma multiforme can originate from terminally differentiated cortical astrocytes and neurons via the combined loss of NF1 and P53 (Friedmann-Morvinski et al. 2012). A combination of oncogenic hits, such as activated Kras and activation of NF-kB, has also been shown to lead to the conversion of nonstem intestinal epithelial cells into cells with stem cell properties in intestinal tumors (Schwitalla et al. 2013). Therefore, it appears that dedifferentiated cells could be the cellular origin of certain cancers, but the transcriptional programs that regulate the initiation of this event remain unclear. Furthermore, in a wild-type developmental context, although many 6 These authors contributed equally to this work. Corresponding author: louise.cheng@petermac.org Article is online at factors have been implicated in promoting stem cell differentiation, less is known about the mechanisms that render differentiated cells incapable of re-entering the cell cycle. In the Drosophila CNS, type I stem cell-like neural progenitors, the neuroblasts (NBs) (Fig. 1A, red), divide asymmetrically, producing one large daughter cell that self-renews and a second, smaller daughter cell the ganglion mother cell (GMC) that divides only once to give rise to two terminally differentiating neuronal or glial cells (Fig. 1B; for review, see Homem and Knoblich 2012). In addition to the type I NBs, eight bilateral type II NBs are located in the central brain (Fig. 1A, blue) that divide to selfrenew and generate smaller intermediate neural progenitors (INPs), which in turn undergo asymmetric divisions to Ó 2015 Froldi et al. This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see After six months, it is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at creativecommons.org/licenses/by-nc/4.0/. GENES & DEVELOPMENT 29: Published by Cold Spring Harbor Laboratory Press; ISSN /15; 129
2 Froldi et al. Figure 1. Nerfin-1 expression during postembryonic neurogenesis. (A) Larval CNS is generated by type I (red) and type II (blue) neuroblasts throughout post-embryonic neurogenesis. The earliest born neurons are located deep in the CNS, and the most recently born neurons are located superficially. (B) Schematics depicting neurogenesis in type I and type II lineages. Type I NBs express Mira and segregate Pros to the GMC. GMCs divide only once to produce two post-mitotic neurons that express Nerfin-1. Type II NBs divide to self-renew and give rise to transit-amplifying INPs. Type II NBs express Mira but not Pros. INPs divide four to eight times, generating another INP and a GMC that divides only once to give rise to two neurons, both of which express Nerfin-1. (C D09) Type I MARCM clone marked by RFP. (C C09) Superficial section: Nerfin-1 (green) is absent in the NB (yellow arrow) and in the latest born GMCs adjacent to the NB (white arrow) and is expressed at low levels in mitotic GMCs (positive for the mitotic marker ph3 + ; yellow arrowhead). (D D09) In a deep section of the clone, Nerfin-1 colocalizes with Pros (white) and Elav (blue) in differentiated neurons. (E E09) Type II lineage marked by RFP. Nerfin-1-GFP (green) is expressed in post-mitotic neurons (yellow arrowhead) but not in type II NBs (large cell; yellow arrow) or INPs (identified by cortical Pros; white in E09; white arrowhead) and at very low levels in GMCs (identified by nuclear Pros; white arrow). Bars, 10 mm. See also Supplemental Figures S1 S3. self-renew and give rise to GMCs and neurons (Fig. 1B; Bello et al. 2008; Boone and Doe 2008; Bowman et al. 2008). At each NB division, cell fate determinants such as Prospero (Pros) (Doe et al. 1991; Vaessin et al. 1991), Brain tumor (Brat) (Bello et al. 2006; Betschinger et al. 2006), and Numb (Rhyu et al. 1994) are segregated from the NB into the GMC. These fate determinants inhibit selfrenewal, direct cell cycle exit, promote neuronal differentiation, and prevent tumor formation. Pros has been shown to act as a transcriptional switch primarily in the GMCs to repress genes required for self-renewal such as Miranda (Mira), Asense (Ase), and Deadpan (Dpn) and activate genes for neuronal and glial differentiation (Fig. 1B, blue; Choksi et al. 2006). In the absence of Pros, GMCs fail to differentiate, revert to NBs, and form brain tumors in both type I and type II NB lineages (Caussinus and Gonzalez 2005; Bello et al. 2006; Betschinger et al. 2006; Choksi et al. 2006; Lee et al. 2006b; Maurange et al. 2008). However, while these studies have led to a good understanding of how neural progenitors give rise to neurons, it is not yet clear what factors are required to maintain neuronal differentiation. Recently, the RNA splicing factor midlife crisis (mlc) (Carney et al. 2013) and the BTB-zinc finger transcription factor Lola (Southall et al. 2014) have been implicated in neuronal maintenance. mlc regulates Pros expression, and its loss causes neuron-to-nb reversion. However, loss of Pros alone was not sufficient to induce neuronal dedifferentiation, and, furthermore, the reverted NBs in mlc mutants are stalled in the cell cycle and do not form tumors. Lola, on the other hand, is a Pros cofactor and acts redundantly with Pros. Loss of Lola was sufficient to cause neuron-to-nb reversion only in the optic lobe regions of the CNS where Pros is absent (Southall et al. 2014). To date, no general factor that regulates neuronal maintenance preventing dedifferentation and tumor formation in type I and II NB lineages has been found. Here we show that Nervous fingers 1 (Nerfin-1), a zinc finger transcription factor previously implicated in embryonic axon guidance in Drosophila (Stivers et al. 2000; Kuzin et al. 2005), is expressed in the post-mitotic larval neurons and maintains their differentiated status. In the absence of Nerfin-1, neurons dedifferentiate, rapidly increase in size via Myc- and target of rapamycin (Tor)-dependent mechanisms, and undergo reversion to acquire a NB cell fate, ultimately resulting in proliferative neural tumors. Using complementary approaches of RNA sequencing (RNA-seq) and chromatin immunoprecipitation (ChIP), we found that Nerfin-1, like Pros, promotes differentiation and represses self-renewal. Unlike Pros, however, Nerfin-1, rather than acting in the GMC, primarily activates a transcriptional program in neurons to maintain their differentiated state. Furthermore, we demonstrate that Nerfin-1 is required for 130 GENES & DEVELOPMENT
3 Nerfin-1 prevents dedifferentiation NB terminal differentiation at the end of neurogenesis, and its overexpression causes premature NB loss. Results Nerfin-1 is expressed in neurons derived from type I and type II NB lineages The larval CNS is generated during post-embryonic neurogenesis by both type I and type II NBs (Fig. 1A,B). We investigated the localization of Nerfin-1 protein in both lineages using a transgene expressing a Nerfin-1-GFP fusion protein under control of the nerfin-1 promoter (referred to as Nerfin-1-GFP) that was previously generated to study nerfin-1 post-transcriptional silencing (Kuzin et al. 2007). We found that Nerfin-1-GFP recapitulates the expression pattern of the Nerfin-1 antibody (Supplemental Fig. S1A A0), validating it as a bona fide tool to follow Nerfin-1 expression pattern. Nerfin-1-GFP was found to be expressed in a subset of neurons (which also express the neuronal marker Pros) (Spana and Doe 1995) at 36 h after larval hatching (ALH) (Supplemental Fig. S2A B09). By 72 h ALH (Supplemental Fig. S2C D09) and 96 h ALH (Supplemental Fig. S2E F09), most neurons expressed both Pros and Nerfin-1- GFP. The mature neurons generated by NBs lie deep within the CNS at a location several cell diameters away from the parental NB, while newly born neurons are found more superficially. In superficial sections, we found that Nerfin-1-GFP was absent from type I NBs (Fig. 1C C09, yellowarrow;supplementalfigs.s1b B0, S3A C0, white arrow; Supplemental Movie S1) and early born GMCs (Fig. 1C C09, white arrow; Supplemental Fig. S3C C0, yellow arrow). However, it was detected at low levels in mature, dividing GMCs (Fig. 1C-C09, yellow arrowhead, positive for the mitotic marker phospho-histone H3 [ph3]). In deep sections, Nerfin-1-GFP was expressed in all post-mitotic neurons marked by the expression of the neuronal markers Pros and embryonic lethal abnormal vision (Elav) (Fig. 1D D09). Type II NBs produce INPs that undergo several self-renewing divisions to amplify the number of progeny (Fig. 1B). In type II lineages, Nerfin-1 was also expressed in Pros-expressing neurons (Fig. 1E E09, yellow arrowhead) and at low levels in mature GMCs (Fig. 1E E09, white arrow, nuclear Pros + cells adjacent to the INPs) but not in NBs (Fig. 1E E09, yellow arrow, distinguished by its large size) or INPs (Fig. 1E E09, white arrowhead, cortical Pros localization). Altogether, in both type I and type II lineages, Nerfin-1 expression is associated with differentiated rather than self-renewing cells. nerfin-1 loss-of-function generates neural tumors in type I and type II lineages In order to investigate the function of Nerfin-1, we used a nerfin null mutant (Kuzin et al. 2005) to generate loss-of-function MARCM (mosaic analysis with a repressible cell marker) clones (Lee and Luo 2001). Clones were induced at 24 h ALH in the larval CNS and dissected 96 h later. While control clones in the ventral nerve cord (VNC) always contained one large NB positive for the neural progenitor-specific marker Mira (Fig. 2A A0 [white arrow], C), most type I nerfin clones (88%; n = 50) contained multiple Mira + cells per clone (Fig. 2B B0,C), resulting in an average 2.9-fold increase in total clonal volume (Fig. 2D). We detected no Nerfin-1 protein in nerfin clones (Supplemental Fig. S1C,C9). In addition, Nerfin-1-GFP, a GFP-tagged full-length genomic nerfin-1 construct (Kuzin et al. 2005), was sufficient to rescue the majority of nerfin clones to one NB per clone (Supplemental Fig. S1D,E). nerfin clones were highly proliferative compared with wild-type clones, as indicated by significant increases in incorporation of the thymidine analog and S-phase marker 5-ethynyl-29-deoxyuridine (EdU) (Supplemental Fig. S4, cf. B,B9 and A,A9, quantified in E) and cells positively labeled for the mitotic marker ph3 in nerfin in comparison with control clones (Supplemental Fig. S4, cf. D,D9 and C,C9, quantified in F). Control NBs undergo terminal differentiation at ;120 h ALH (Maurange et al. 2008; Homem et al. 2014). In contrast, the ectopic nerfin Mira + cells failed to undergo timely differentiation during pupal life and continued to be highly proliferative during adult stages (indicated by ph3 + and EdU + cells compared with control) (Fig. 2E F0; Supplemental Fig. S4G H9). The presence of nerfin tumor masses consisting mostly of undifferentiated Elav /Mira + progenitors (Supplemental Fig. S4, cf. J J0 and I I0) shortened animal life span by 40% (Fig. 2G). These tumors were capable of metastasis when transplanted into the abdomen of naive adult hosts, migrating to distant organs such as the eye (Fig. 2H, arrow). In type II NB lineages, we observed a similar but less dramatic phenotype. Loss of nerfin-1 did not alter the number of type II NBs (one per clone, Ase /Mira + ) (Supplemental Fig. S5, cf. B B0 and A A0, yellow arrows) but resulted in an expansion of the Ase + INPs (Supplemental Fig. S5, cf. D D0 and C C0, quantified in F) and a 1.3-fold increase in overall clonal volume (Supplemental Fig. S5E). The INPs, which normally reside in superficial sections of control clones, are now found in both superficial (Supplemental Fig. S5K K0,M M0) and deep sections of nerfin clones typically occupied by neurons (Supplemental Fig. S5L L0,N N0); however, we did not detect a corresponding increase in proliferation, as assessed by the number of cells positively labeled by cell cycle markers (Supplemental Fig. S5G [M phase, ph3], H [S phase, EdU], I,J [CycA and CycE, G1 phase]). To investigate why the ectopic INPs are not proliferative, we used the markers Dpn and Ase to distinguish between mature and immature INPs (Bowman et al. 2008; Weng et al. 2010). Importantly, we found a significant increase in the number of immature nonproliferative Ase +,Dpn INPs (Supplemental Fig. 5SK N0,Q). As it was previously shown that immature INPs require an average of 6.6 h to mature before they undergo divisions (Weng et al. 2010; Homem et al. 2013), we assessed nerfin clone size later in development (120 h after clone induction) and found that nerfin GENES & DEVELOPMENT 131
4 Froldi et al. Figure 2. Nerfin-1 inhibits neuroblast overgrowth in the larval and adult CNS. (A B0) Size comparison of control or nerfin MARCM clones in the larval VNC. Control clones contain one large Mira + NB (red; white arrow), and nerfin clones contain 20 Mira + cells on average (quantified in C; m = 19.55, SEM = 4.10, n = 11) of variable cell sizes. The large Mira + cells (B B0, white arrow) are Pros,andthesmallMira + cells (B B0, yellow arrow) are Pros +. The increase in Mira + cell numbers results in a 2.9-fold expansion in clone volume 4 d after clone induction (D; control: m = mm 3,SEM= mm 3, n = 57; nerfin : m = mm 3, SEM = mm 3, n = 137). (E F0). In contrast to control NBs which are absent in adult MARCM clones (E E0), nerfin mutant Mira + cells (F,F9) fail to undergo timely differentiation during pupal life and continue to be highly proliferative during adult stages (F0; indicated by ph3 +, white), resulting in large tumor clones that are detrimental to the adult hosts, shortening their average life span by 40% (G; control: median survival = 42 d, n = 56; nerfin : median survival = 25 d, n = 57). (H) Upon transplantation of nerfin tumor fragments into the abdomen of naive hosts, nerfin brain cells (GFP +, right; n = 10) but not GFP + control brain cells (left; n = 10) overgrow and metastasize to distant sites such as the eye (arrow). (I) Histogram depicting NB cell size; 100% of control NBs (n = 37) are between 9 and 12 mm in diameter. In nerfin clones (n = 82), 26.3% of Mira + cells are between 3 and 6 mm, 39.7% are between 6 and 9 mm, and 44% are between 9 and 12 mm. (J) Histogram depicting the percentage of Mira + cells in M phase (control: m = 34.4%, SEM = 2.06%, n = 5; large nerfin Mira + cells $8 mm: m = 32.12%, SEM = 5.6%, n = 10; small nerfin Mira + cell <8 mm: m = 15.6%, SEM = 3.03%, n = 10). (K) Eighty-seven percent of control NBs (n = 23), 84% of large nerfin Mira + cells (n = 24), and 46.2% of small nerfin Mira + cells (n = 24) correctly localize basal polarity marker Mira. (L T9) representative images of apical markers Insc and apkc and basal marker Mira in large and small NBs at metaphase. E F0 are maximum projections. Bars: A B0,L T9, 10mm; E F0, 50mm. (ns) P > 0.05; (*) P # 0.05; (**) P # 0.01; (***) P # See also Supplemental Figures S4 S7. clones are 1.7-fold larger than control clones (Supplemental Fig. 5SR) as a result of an increase in the number of mature INPs (data not shown). However, unlike type I lineages (Supplemental Fig. S5P,P9, white arrows) that contained multiple NBs, nerfin type II neurons (Supplemental Fig. S5P P0, yellow arrows) dedifferentiate to give rise to ectopic INPs but do not fully revert to NBs by 120 h ALH. nerfin mutant clones contain ectopic Mira + cells of variable size Upon close examination of nerfin type I tumor clones, we found that unlike wild-type NBs, which maintain a constant size of ;9 12 mm in diameter throughout neurogenesis (Maurange et al. 2008), nerfin Mira + cells varied between 3 and 12 mm in diameter (Fig. 2I). We then correlated NB size with specific marker expression. Control NBs (Fig. 2A A0) ornerfin Mira + cells that were $8 mm in diameter never expressed the neuronal differentiation marker Pros (Fig. 2B B0, white arrow). In contrast, a proportion of the smaller nerfin Mira + cells (<8 mm) expressed both NB and neuronal markers (Mira + /Pros + cells) (Fig. 2B B0, yellow arrow). Given that wild-type NBs were never <8 mm, we divided nerfin ectopic Mira + cells into two groups: Large Mira + cells, defined to be $8 mm, were comparable in size with wildtype NBs, whereas small Mira + cells, defined to be <8 mm, 132 GENES & DEVELOPMENT
5 Nerfin-1 prevents dedifferentiation were significantly smaller than wild-type NBs but larger than control neurons (Supplemental Fig. S6A). Staining for atypical protein kinase C (apkc), Inscuteable (Insc), and Mira as representative members of asymmetric division polarity complexes during M phase (Homem and Knoblich 2012) revealed no difference in the localization patterns of these proteins between wild-type and nerfin large ectopic Mira + cells (Fig. 2, cf. L,L9 and M,M9, N,N9 and O,O9, and P,P9 and Q,Q9). However, in around half of the nerfin small ectopic Mira + cells, we observed cortical mislocalization of polarity proteins (representative Mira localization is quantified in Fig. 2K, representative Insc, apkc, and Mira staining is shown in R T9; Supplemental Fig. S6B). In addition, 100% of nerfin large Mira + cells underwent asymmetric self-renewal (Supplemental Fig. S7A B09; Supplemental Movie S3) at a cell cycle speed comparable with that of control NBs (Fig. 2J; Supplemental Fig. S7E E09; Supplemental Movies S2, S6; Homem et al. 2013). In contrast, small Mira + cells sometimes exhibited cell cycle defects (Fig. 2J) and atypical size-symmetric divisions (Supplemental Fig. S7C D09; Supplemental Movies S4, S5). As these nerfin ectopic Mira + cells were capable of selfrenewing and giving rise to mature neurons, we concluded that these cells were ectopic NBs. nerfin ectopic NBs arise by dedifferentiation In order to trace the cell of origin of the ectopic NBs, we carried out a time-course experiment to establish the earliest time point at which they first arise in nerfin clones. We found no ectopic NBs 24 h after clone induction (Fig. 3A B0). By 48 h, Mira + cells with size (;5 mm) and position characteristics of post-mitotic neurons (;8 mm from the parental NB) were identified in nerfin clones (78%; n = 14) (Fig. 3C D0; Supplemental Fig. S8A D9), indicating that these ectopic NBs were likely dedifferentiated neurons. To test this hypothesis, we fed larvae with EdU + food for 4 h at 24 h after clone induction and then chased for 24 h with EdU-free food (Fig. 3I). In control animals, EdU was first incorporated by the dividing NBs during feeding and inherited by neurons generated during this time window. During EdU-free chase, additional NB divisions gave rise to further neurons, thus displacing EdU-labeled cells into deeper locations within the CNS (;7.5 mm from the parental NB) (Fig. 3I K0). Strikingly, 48 h after clone induction (the earliest time point at which ectopic NBs were first identified), we observed Mira + cells in the deep sections of nerfin clones that were also EdU +, indicating that they were in fact NBs newly derived from differentiated neurons (Fig. 3, cf. L M0, yellow arrow, and J K0). Between 48 and 72 h after clone induction, the rate of neuron-to- NB reversion rapidly increased (Fig. 3E F0,G; Supplemental Fig. S8E H0), accompanied by an enlargement in average NB cell size (Fig. 3H) and enhanced cell cycle speed (Supplemental Fig. S8I). To further support the model that nerfin ectopic NBs are derived from neurons, we carried out live-cell imaging of dissociated type I NB clones (Supplemental Fig. S9H I0; Supplemental Movies S7, S8). While control neurons did not significantly increase their cellular volume or undergo divisions during the 11 h of live imaging (Supplemental Fig. S9H H0,J), nerfin cells <7 mm (Fig. 2I; typical of small ectopic NBs or neurons and significantly smaller than wild-type NBs, as quantified in Supplemental Fig. S6A) increased their cellular volume by an average of 5.3-fold and underwent asymmetric cell divisions (Supplemental Fig. S9I I0,J). Finally, to investigate whether Nerfin-1 is continuously required for neuronal maintenance, we induced nerfin clones later in development, at 48 and 72 h ALH. We found that Nerfin-1 is indeed required to maintain neuronal differentiation throughout larval neurogenesis, as ectopic NBs arose upon removal of Nerfin-1 at both early (24 h ALH) (Fig. 3) and late time points (Supplemental Fig. S9A F9). Together, these analyses indicate that both the small and large NB populations were derived from post-mitotic neurons and that these newly dedifferentiated cells undergo rapid growth to give rise to fastproliferating large NBs, thus contributing to aggressive tumor growth. Myc- and Tor-mediated cellular growth regulates neuron-to-nb reversion in nerfin clones The increase in the size of ectopic NBs over time (Fig. 3H; Supplemental Fig. S9H J; Supplemental Movies S7, S8) and the fact that nerfin neurons were significantly larger than control neurons (Supplemental Fig. S6A) suggested that cell growth-dependent mechanisms might facilitate the neuron-to-nb reversions in nerfin clones. To test this hypothesis, we first examined the ratio of nucleolar to nuclear volume, which is an indicator of ribosomal biogenesis and cellular growth in both NBs and neurons (Fig. 4A C9, nucleolus is marked with fibrillarin [Fib]). Strikingly, we found that this ratio was significantly increased in not only nerfin NBs (Fig. 4B,B9, marked by Dpn +, quantified in D, cf. with control NBs, A,A9, white arrows) but also nerfin Dpn neurons (Fig. 4C,C9, quantified in D, cf. with control neurons, A,A9, yellow arrows), suggesting that cellular growth precedes the neuron-to-nb reversion. In addition, we found that Myc, a global regulator of ribosome biogenesis (de la Cova and Johnston 2006) was not only expressed in Mira + NBs (Fig. 4F F0, yellow arrow) but now up-regulated in Mira /Elav + neurons (Fig. 4H H0, yellow arrow) in nerfin clones compared with control clones (Fig. 4E E0,G-G0, yellow arrow). To test whether Myc is required for nerfin clonal growth, we knocked it down with RNAi (specificity verified in Supplemental Fig. S10B,B9) and found that it was sufficient to reduce nerfin clonal volume by 50% (Fig. 4I,J, quantified in K). Since Myc is implicated in cell cycle progression (Johnston et al. 1999), we next examined whether its knockdown decreased clonal growth via inhibition of the cell cycle. Interestingly, inhibition of Myc did not alter the cell cycle speed of nerfin mutant NBs (Supplemental Fig. S10A). On the other hand, Myc knockdown did cause a reduction in the number of both large and small NBs in nerfin clones (Fig. 4L), GENES & DEVELOPMENT 133
6 Figure 3. nerfin ectopic NBs arise from dedifferentiation of neurons. (A F0) Control and nerfin clones were examined at 24 h (A B9), 48 h (C D9), and 72 h (E F9) after clone induction and stained with the NB marker Mira (red). The parental NBs, identified by their large size and location in superficial sections of the brain (0 mm), are marked with white arrows, and ectopic Mira + cells are marked with yellow arrows. (A0,B0,C0,D0,E0,F0) The results are summarized in the schematics. (A B0) No ectopic NBs were identified in nerfin and control clones at 24 h after clone induction. (C F0) Ectopic NBs were detected in nerfin clones at 48 and 72 h after clone induction in the deepest sections of the clones. (G) Histogram depicting the frequency of ectopic NBs at 24, 48, and 72 h after clone induction in nerfin clones (24 h: m = 0, SEM = 0, n = 22; 48 h: m = 1.36, SEM = 0.36, n = 25; 72 h: m = 5.83, SEM = 1.23, n = 23). (H) Histogram depicting the average ectopic NB diameter at 48 and 72 h after clone induction in nerfin clones (48 h: m = 5.03, SEM = 0.17, n = 24; 72 h: m = 6.82, SEM = 0.19, n = 129). (I) Schematic depicting the EdU pulse-chase experiment in control and nerfin clones. Animals were fed EdU-containing food for 4 h at 24 h after clone induction and then chased with EdU-free food for 24 h. (J K0) In control clones, no EdU + /Mira + cells were recovered (0%; n = 18). (L M0) Innerfin clones, EdU + /Mira + ectopic NBs located in deep sections normally occupied by post-mitotic neurons were recovered in 33% of the clones (n = 18; yellow arrow). (J L0) Parental NBs are marked with white arrows. Bars, 10 mm. (***) P # See also Supplemental Figures S8 S GENES & DEVELOPMENT
7 Figure 4. Myc- and Tor- mediated cellular growth regulate neuron-to-nb reversion in nerfin clones. (A C9) The ratio of nucleolus (marked by Fib, white) to nuclear (marked by ngfp, green) volume is significantly increased in nerfin NBs (Dpn +, red; white arrow) and neurons (Dpn, yellow arrow), as quantified in D (control NBs: m = 1.89, SEM = 0.47, n = 14; nerfin NBs: m = 3.55, SEM = 0.45, n = 20; control neurons: m = 1.01, SEM = 0.13, n = 22; nerfin neurons: m = 2.08, SEM = 0.22, n = 27). Myc and Tor inhibition does not alter the nucleolus to nuclear volume ratio (mycrnai: m = 1.68, SEM = 0.47, n = 9; Tor DN : m = 1.14, SEM = 0.21, n = 9)butissufficienttorestore nerfin nucleolus to nuclear volume ratio to control levels in both nerfin neurons and NBs (D; nerfin ;mycrnai neuronal ratio: m = 0.97, SEM = 0.13, n = 10; nerfin ;mycrnai NB ratio: m = 2.24, SEM = 0.42, n = 24; nerfin ;Tor DN neuronal ratio: m = 0.67, SEM = 0.05, n = 26; nerfin ;Tor DN NB ratio: m = 1.78, SEM = 0.3, n = 20). (E H0) Myc expression (white) is up-regulated in control Mira + (red) NBs (E E0, yellow arrow; G G0, yellow arrow) and both nerfin Mira + NBs (F F0, yellow arrow), and Mira,Elav + (red) neurons (H H0, yellow arrow). (I,J) RNAi knockdown of myc significantly reduces nerfin tumor growth. (K) Histogram depicting clonal volume under Myc and Tor manipulations (control: m = mm 3,SEM= mm 3, n = 57; mycrnai: m = mm 3,SEM= mm 3, n = 46; Tor DN : m = mm 3,SEM= mm 3, n = 9; nerfin : m = mm 3,SEM= mm 3, n = 180; nerfin ;mycrnai: m = mm 3,SEM= mm 3, n = 77; nerfin ;Tor DN : m = mm 3,SEM= mm 3, n = 63). (L) The decrease in nerfin tumors under Myc and Tor manipulations is due to a reduction in both the number of large ($8 mm) and small (<8 mm) NBs (nerfin large NBs: m = 4.3, SEM = 0.44, n = 23; nerfin ;mycrnai large NBs: m = 2.17, SEM = 0.28, n = 39; nerfin ;Tor DN large NBs: m = 0.72, SEM = 0.23, n = 18; nerfin small NBs: m = 12.9, SEM = 0.90, n = 40; nerfin ;mycrnai small NBs: m = 5.30, SEM = 0.74, n = 40; nerfin ;Tor DN small NBs: m = 5.23, SEM = 0.82, n = 17). (M) RNAi knockdown of Myc and Tor was sufficient to increase the level of differentiation in nerfin clones (control: m = 83.63%, SEM = 5.18, n = 3; mycrnai: m = 86%, SEM = 2, n = 15; Tor DN : m = 82.18%, SEM = 4.17, n = 6; nerfin : m = 41.34%, SEM = 4.66, n = 21; nerfin ;mycrnai: m = 61.67%, SEM = 4.28, n = 15; nerfin ;Tor DN : m = 55%, SEM = 4.29, n = 11). Bars: A C9,E H9,10mm; I,J,25mm. (ns) P > 0.05; (*) P # 0.05; (**) P # 0.01; (***) P # 0.001; (****) P # See also Supplemental Figures S10 S11.
8 Froldi et al. suggesting that it affected both neuron-to-nb reversion (accounting for the decrease in small NBs) and NB cellular growth (which resulted in the reduction in large NB numbers). Consistent with this model, Myc knockdown was sufficient to restore the nerfin nucleolar/ nuclear volume ratio to wild-type level in both NBs and neurons (Fig. 4D). Consequently, many more neurons remained differentiated, as indicated by the percentage of Elav-positive cells per clone (Fig. 4M). Similarly, we found that slowing down cellular growth via expressing a dominant-negative form of Tor (Tor DN ) (Hennig and Neufeld 2002) also reduced the nucleolar/nuclear volume ratio (Fig. 4D) in nerfin clones. Similar to Myc knockdown, this resulted in the reduction in NB numbers (Fig. 4L), enhanced differentiation (Fig. 4M), and decreased tumor volume (Fig. 4K) independent of cell cycle progression (Supplemental Fig. S10A). We found that the Tor-dependent effects were administered by the TORC1 complex, as the Rag complex, which activates TORC1 in response to amino acids (Kim et al. 2008), and the TORC1 component Raptor (Lee and Chung 2007) were both required for nerfin clonal growth (Supplemental Fig. S11). Together, these results indicate that Myc- and Tor-dependent mechanisms are required for nerfin tumor growth by regulating both the rate of neuron-to-nb reversion and the increase in cellular volume. Self-renewal and neuronal differentiation are commonly deregulated in nerfin and pros 17 tumors The homeodomain transcription factor Pros was previously shown to promote neuronal differentiation (Bello et al. 2006; Betschinger et al. 2006; Choksi et al. 2006). In pros 17 clones, GMCs failed to give rise to neurons, resulting in tumor clones consisting almost exclusively of NBs (Fig. 5B C09). To compare the transcriptional networks required for neuronal maintenance administered by Nerfin-1 versus neuronal differentiation promoted by Pros, we conducted a genome-wide transcriptional profile analysis of nerfin and pros 17 mutant clones. GFP-labeled mutant or control cells were isolated through fluorescence-activated cell sorting (FACS) as described in Berger et al. (2012). Total RNA was extracted, sequenced (see the Materials and Methods), and compared with control to identify differentially expressed genes. We found a strong correlation between genes that were deregulated in both nerfin and pros 17 clones (Supplemental Figs. S12A D, S13), with 82% overlap between up-regulated genes in nerfin and pros 17. However, fewer genes were commonly down-regulated in nerfin and pros 17 clones, with only 44% overlap. We compared our list of differentially expressed genes with NB- and neuronal-specific genes previously identified through transcriptional profiling of FACS-sorted NBs Figure 5. Pros and Nerfin-1 act sequentially to promote neuronal specification. (A A09) Elav expression (red) is down-regulated in more cells than Pros expression (white) in nerfin clones (green). (B C09) Nerfin-1 (green, B0,C0) is absent in RFP-labeled pros 17 clones (red) that consist mostly of NBs (Mira + ; B09, white) and are almost completely devoid of differentiated cells (Elav + ; C09, white). (D G) nerfin ; prosrnai (F) phenocopies the prosrnai (E) clonal phenotype. Overexpression of Pros partially rescues nerfin clones (cf. G and D) containing zero, one (not shown) or two (G, yellow arrows) NBs per clone. Bars, 10 mm. See also Supplemental Figures S12 S GENES & DEVELOPMENT
9 Nerfin-1 prevents dedifferentiation and neurons (Berger et al. 2012) and found that 25 NB genes were up-regulated and 64 neuronal genes were downregulated in both pros 17 and nerfin mutant clones (data not shown). We validated a selected group of these differentially expressed genes by quantitative PCR (qpcr), confirming the up-regulation of the NB-specific genes mira and ase and down-regulation of the neuronal gene elav in both mutant tissues (Supplemental Fig. S14). In addition, immunostaining also confirmed the up-regulation of Ase and Dpn in nerfin type I (Supplemental Fig. S12F G0) and type II (Supplemental Fig. S5M N0,Q) clones. Interestingly, Dpn +, Mira cells were observed in nerfin mutant clones, suggesting that Dpn derepression precedes the onset of Mira expression in the process of dedifferentiation (Supplemental Fig. S12G G0, yellow arrow). Furthermore, we found up-regulation of the GMC-specific gene target of pox-n (tap) (Bush et al. 1996) and NBspecific gene zipper (zip) innerfin mutant cells. tap encodes a homolog of the mammalian protein NeuroD, which has been previously shown to be a target of the Nerfin-1 mammalian homolog INSM1 (Liu et al. 2006). Conversely, we found that the neuronal genes Sox21b, scarecrow (scro), and couch potato (cpo) (Kuzin et al. 2005; Southall et al. 2014) were down-regulated in both pros 17 and nerfin clones. Together, these results suggest that Nerfin-1 and Pros regulate similar sets of genes, enabling them to inhibit self-renewal and promote differentiation. Nerfin-1 regulates NB self-renewal and neuronal differentiation genes In order to test whether Nerfin-1 directly regulates the expression of NB and neuronal genes, we carried out a ChIP assay. We were able to show that Nerfin-1 was located in the promoter regions of the self-renewal genes dpn and ase (also targets of Pros) (Choksi et al. 2006) but not that of mira and pros (Supplemental Fig. S12E). Furthermore, we found Nerfin-1 in the proximity of the promoters of neuronal genes sox21b, cpo, andtap (Supplemental Fig. S12E). A fourfold enrichment of Nerfin-1 on the promoter of myc was also detected (Supplemental Fig. S12E), consistent with our finding that the repression of Myc-mediated cellular growth is required to block neuron-to-nb reversion (Fig. 4). Together with our genome-wide differential expression analysis, these results suggest that Nerfin-1 maintains the differentiated state of post-mitotic neurons through direct repression of NB-specific genes and activation of neuronal genes. Pros and Nerfin-1 act sequentially to promote neuronal specification Many self-renewal and differentiation genes were commonly deregulated in nerfin-1 and pros mutants; several of them were also direct targets of both Nerfin- 1 and Pros. However, loss-of-function phenotypes of nerfin-1 and pros mutants are strikingly different. In pros 17 and prosrnai clones, GMCs reverted to NBs at the expense of neuronal generation, resulting in large clones consisting almost entirely of NBs, devoid of Nerfin-1 protein and mrna (Fig. 5B C09,E; Supplemental Fig. S14). nerfin mutant clones, on the other hand, contained a mixture of NBs and neurons (Figs. 2B B0, 4F F0,H H0, 5A A09,D) resulting from failure to maintain neuronal differentiation. Consistent with this, Pros expression was reduced only in later stages of neuron-to-nb reversion in nerfin cells (Fig. 5A A09, Elav /Pros + cells, yellow arrows). These data led us to propose that Pros is required in the GMCs to promote neuronal specification prior to the requirement of Nerfin-1 in the neurons for their maintenance. To test this hypothesis, we performed an epistasis analysis between nerfin-1 and pros mutants. We found that the loss of Pros was epistatic to the loss of Nerfin-1 such that the nerfin ; prosrnai phenotype was identical to that of prosrnai alone (Fig. 5E,F; quantified in Supplemental Fig. S15C), supporting our model that Pros is required in the GMCs at an earlier stage of neurogenesis. Furthermore, Pros overexpression partially rescued the nerfin phenotype (Fig. 5G, some clones still contained multiple NBs, yellow arrows; Supplemental Fig. S15A B9, clone volume quantified in C), indicating that Pros overexpression can promote the differentiation of nerfin ectopic NBs. Nerfin-1 is required for timely NB differentiation, and ectopic Nerfin-1 promotes differentiation NBs remain proliferative until 120 h ALH, where the terminal division of most type I NBs is correlated with the nuclear accumulation of Pros that induces cell cycle exit (Maurange et al. 2008). As our results so far showed that Nerfin-1 activates differentiation-specific genes in post-mitotic neurons, we next examined whether it also plays a role in initiating NB terminal differentiation. We found that Nerfin-1 and Pros colocalized in the nucleus of 29% of the Mira + NBs examined (Fig. 6A A0; Supplemental Fig. S16), suggesting that Nerfin-1 might also play a role in promoting terminal division. Furthermore, in 24% of NBs, Nerfin-1 was detected in the absence of Pros (Fig. 6B B0, quantified in C; Supplemental Fig. S16), suggesting that Nerfin-1 may be activated in the NBs earlier than Pros at the end of neurogenesis. In addition, Nerfin-1 is also required for terminal differentiation at the end of neurogenesis, as nerfin mutant NBs do not undergo Pros-dependent cell cycle exit (Fig. 6D,D9) and remain proliferative during adult stages (Fig. 2F F0). To test whether ectopic Nerfin-1 can promote differentiation, we expressed a nerfin-1 transgene using the pan- NB driver dnab-gal4. Strikingly, we detected a gradual loss of NBs in the VNC; at 96 h ALH, we observed a 33% reduction in NBs numbers (Fig. 6E,F, quantified in M). At 110 h ALH (10 h before terminal neurogenic divisions), a 53% reduction in NB numbers was found (Fig. 6G,H, quantified in M). The disappearance of NBs was not due to an increase in cell death (data not shown) but was accounted for by an increase in premature differentiation. While larval NBs regrow to their original size after each division, terminally GENES & DEVELOPMENT 137
10 Figure 6. Nerfin-1 is required for timely NB differentiation, and ectopic nerfin-1 promotes differentiation. (A B0) At the end of neurogenesis (120 h ALH), Nerfin-1 and Pros are localized to the NB (Mira +, red) nucleus, prior to NB terminal differentiation. In 24% of NBs, both Pros (white) and Nerfin-1 (green) are expressed in the nucleus (A A0), and 29% of NBs express only Nerfin-1 but not Pros (B B0). (C) Histogram depicting the percentage of nuclear localization of Pros and Nerfin-1 in the NBs at 96 h (n = 20) and 120 h (n = 17). (D D9) nerfin NBs (Dpn +, white; yellow arrowheads) fail to undergo timely terminal differentiation at 120 h. In these NBs, nuclear Pros (red) localization does not occur. (E M) Nerfin-1 overexpression in NBs with pan-nb driver dnab-gal4 induces premature differentiation and reduction in the overall NB numbers at 96 h (E,F) and 110 h (G,H; maximum projections), as quantified in M (96 h, control: m = 122, SEM = 1.45, n = 3; dnab>nerfin-1: m = 82.4, SEM = 2.49, n = 10; 110 h, control: m = 125, SEM = 1.8, n = 3; dnab>nerfin-1: m = 58, SEM = 2.6, n = 5). (I,J) This reduction in NB numbers corresponds with a reduction in NB diameter, as quantified in L (control: m = 13.1 mm, SEM = 0.36 mm, n = 27; dnab>nerfin-1: m = 10.4 mm, SEM = 0.3 mm, n = 24), and increase in the incidence of size-symmetric divisions indicative of premature differentiation. (K) Histogram depicting the average diameter of thoracic NBs (Mira ) and their progeny (Mira + )duringtelophase(control, Mira : m = 10.5 mm, SEM = 0.3 mm, n = 10; Mira + : m = 5.3 mm, SEM = 0.4 mm, n = 10; dnab>nerfin-1,mira : m = 5.8 mm, SEM = 0.5 mm, n = 8; Mira + : m = 4.6 mm, SEM = 0.6 mm, n = 8). (N) Schematic depicting the localization of Nerfin-1 during larval and pupal neurogenesis. In larval stages (up to 96 h ALH), Nerfin-1 is expressed in neurons (which also express the differentiation marker Pros) and not in self-renewing NBs or GMCs. At the end of neurogenesis (120 h ALH), Nerfin-1 and Pros are localized to the nucleus of the NBs prior to their terminal differentiation. Bars: E H, 100mm; all others, 10 mm. (****) P # See also Supplemental Figures S16, S17.
11 Nerfin-1 prevents dedifferentiation differentiating pupal NBs typically do not (Homem et al. 2014). Consistent with this, we found that Nerfin-1 overexpression resulted in a reduction in the average diameter of NBs from 13.1 to 10.4 mm (Fig.6L).AsNBsbecomeprogressively smaller with each cell cycle, they typically undergo size-symmetric terminal cell divisions (Maurange et al. 2008; Homem et al. 2014) preceded by nuclear Pros localization. Indeed, we found that Nerfin-1 overexpression led to nuclear Pros accumulation (Supplemental Fig. S17A A9) and an increased incidence of size-symmetric divisions (Fig. 6I,J, quantified in K). However, the growth regulator Myc was still expressed in these NBs (Supplemental Fig. S17B,B9). These results provide evidence that Nerfin-1 not only maintains the differentiated status of neurons but also promotes differentiation during neurogenesis (Fig. 6N). Discussion Growing evidence suggests that interconversion between differentiated cells and stem cells can account for initiation of glioblastoma in the mammalian brain (Friedmann- Morvinski et al. 2012) and could be a general paradigm in cancer. Our data reveal an essential function for the zinc finger transcription factor Nerfin-1 in locking neurons into the differentiated state by preventing their reversion to NBs. This study focused mainly on type I NB lineages. We found that upon the loss of Nerfin-1, neurons undergo rapid cellular growth and reach a cell size threshold before a step-wise reversion process occurs. The reverted cells first switch on stem cell-specific genes (such as ase, dpn, andmira) while still maintaining the expression of neuronal genes (such as pros and elav). These cells with mixed neuronal/nb fates cycle slowly and occasionally undergo atypical size-symmetric divisions. As they further increase in size, neuronal genes are eventually switched off, and the reverted progenitors begin to cycle at the speed of wild-type NBs and undergo asymmetric selfrenewal. This expansion of neural progenitors results in proliferative tumorous masses that fail to differentiate and persist into adulthood (Fig. 7). With time, the homeostasis in these tumors becomes increasingly disrupted, eventually resulting in tumor masses consisting mostly of neural progenitors and few differentiated cells. A similar reversion also occurs in type II mutant NB lineages. In the absence of Nerfin-1, neurons undergo stepwise reversion to give rise to ectopic Ase +,Dpn immature and then Ase +,Dpn + mature INPs by 120 h ALH. We found that, as reported (Weng et al. 2010; Homem et al. 2013), the transition from an immature to a mature INP (capable of divisions) takes several hours, and the delay in INP maturation could partially account for the subtle overgrowth phenotype that we observed for nerfin type II lineages. Unlike linages mutant for tumor suppressors such as brat, numb/ Notch ACT,anderm, which contain multiple NBs derived from dedifferentiated INPs and generate large Figure 7. Nerfin-1 promotes differentiation and prevents neuronal reversion into neuroblasts. (A) Type I NBs express Mira (red) and Myc (orange) and segregate the homeodomain transcription factor Pros (blue) to the GMC. GMCs divide only once to produce two post-mitotic neurons that express Nerfin-1 (green). (B) Upon the loss of Nerfin-1 in neurons, Myc-dependent cellular growth ensures that neurons reach a cell size threshold before they undergo a stepwise reversion. The reverted cells first switch on stem cell-specific genes such as Mira while maintaining the expression of neuronal genes such as Pros. As these reverted NBs further increase in size, Pros is eventually switched off, and the reverted progenitors begin to cycle at the speed of wild-type neuroblasts and undergo asymmetric selfrenewal. This expansion of neural progenitors results in proliferative tumorous masses that consist of a mixture of neurons and NBs undergoing various stages of reversion. (C) When ectopically expressed earlier during development, Nerfin-1 promotes premature neuroblast cell cycle exit via size-symmetric cell divisions. type II tumors (Lee et al. 2006a,b; Wang et al. 2006; Bowman et al. 2008; Weng et al. 2010; San-Juan and Baonza 2011; Xiao et al. 2012; Zacharioudaki et al. 2012), nerfin type II mutant clones, similar to pros (Supplemental Fig. S5O; Bowman et al. 2008; Weng et al. 2010), contain only one Ase,Dpn + NB despite an expansion of INPs. As Nerfin-1 is only expressed in differentiated neurons and not in the INPs, we think it is unlikely to play a role in INP specification, accounting for why its loss failed to result in INP-to-NB reversion. However, it remains possible that full reversion from neuron to type II NB takes longer than our experimental period. Nerfin-1 acts independently of Pros to promote differentiation in neurons In contrast to other factors that have been shown to regulate neuronal differentiation through modulation of the prodifferentiation transcription factor Pros (Carney et al. 2013) or act redundantly with Pros itself (Southall et al. 2014), we show that Nerfin-1 is sufficient to cause complete neuron-to-nb conversion in type I lineages independently of Pros. Nerfin-1 and Pros act in different GENES & DEVELOPMENT 139
12 Froldi et al. cell types (neurons and GMCs, respectively). In the neurons, it is the loss of Nerfin-1 (this study) and not Pros (Carney et al. 2013) that is sufficient to induce reversion to NBs. However, many genes are commonly deregulated in nerfin and pros 17 tumors, and they share several direct targets to promote differentiation and repress self-renewal. Consistent with this, our rescue experiment suggests that Pros has the ability to partially compensate for loss of Nerfin-1, forcing ectopic stem cells to differentiate. Furthermore, Nerfin-1, like Pros, acts as both an activator and a repressor; thus, cofactors may be required to provide specificity to Nerfin-1 function. Nerfin-1 is required for timely NB cell cycle exit, and ectopic nerfin-1 promotes premature differentiation Besides maintaining the differentiated status of neurons, Nerfin-1 is also involved in promoting terminal differentiation at the end of neurogenesis. Its expression in the NB nucleus at 120 h ALH not only colocalizes with that of Pros, previously shown to be required in this process (Choksi et al. 2006; Maurange et al. 2008), but seems to precede it, with a significant proportion of NBs expressing nuclear Nerfin-1 but not Pros. Moreover, in the absence of Nerfin-1, NBs fail to undergo Pros-dependent terminal divisions and persist into adulthood. When Nerfin-1 is ectopically expressed, NBs fail to maintain their cellular volume and undergo premature cell cycle exit via a size-symmetric division (Fig. 7C). In mice, medaka, zebrafish, and Caenorhabditis elegans (Wu et al. 2001; Candal et al. 2007; Farkas et al. 2008; Forbes- Osborne et al. 2013), it has been shown that Nerfin-1 homologs are expressed in the CNS and play a role in neuronal differentiation, confirming that Nerfin-1 function is evolutionarily conserved. nerfin-1 tumor growth is facilitated by Mycand Tor-mediated mechanisms Both our immunostaining and live-imaging analysis of nerfin type I lineages demonstrated that dedifferentiated nerfin neurons undergo cellular volume increase, and this process is mediated by the global growth regulators Myc (Johnston et al. 1999) and Tor (Hennig and Neufeld 2002). At early stages of nerfin-1 tumor initiation, the up-regulation of Myc preceded the diminished expression of differentiation markers and increased expression of stem cell markers. In our ChIP analysis, we identified Myc as a direct target of Nerfin-1, confirming that the repression of Myc is likely to be key in maintaining differentiation. These results are also in line with a previously published study in which the investigators showed that in type II NBs, dedifferentiation of INPs to NBs is dependent on translational elongation factor Eif4e and Myc-mediated ribosomal cellular growth (Song and Lu 2011). Myc is indeed an important transcriptional regulator of stemness in the generation of induced pluripotent stem cells (Takahashi and Yamanaka 2006) and has been directly implicated in initiation of dedifferentiation in pancreatic cancers (Ischenko et al. 2013). Our data illustrate that deregulation of a single transcription factor, Nerfin-1, is sufficient to induce brain tumor formation in post-mitotic neurons, and overexpression of Nerfin-1 results in premature differentiation. Our results strongly support the model of bidirectional interconvertibility between stem cells and differentiated cells rather than the strict and irreversible stem cell differentiation hierarchy. Materials and methods Fly strains Larvae were raised on standard medium at 25 C. We used the following strains for generating CNS clones using the MARCM system (Lee and Luo 2001): (1) (3L) w, tub-gal4, UAS-nlsGFPT 6xmycTNLS, hs-flp; FRT2A, tubp-gal80ll9/tm6b, (2) (3L) yw, hs-flp; tub-gal4, UASmCD8GFP/CyO; FRT2A, tub-gal80/ TM6b, (3) (3R) w, tub-gal4, UAS-nlsGFPT6xmycTNLS, hs-flp; FRT82b, tubp-gal80 LL3/TM6b, and (4) (3R) yw, hs-flp;; UAS-myrRFP, tub-gal4, FRT82B, tub-gal80/tm6b. w;; FRT2A was used to generate control MARCM clones, w;; FRT2A, Df(3L)nerfin /TM6b (Kuzin et al. 2005) was used to generate nerfin clones, and w;; FRT82B, pros 17 /TM3 to generate pros 17 clones (Bloomington Stock Center). w, UAS-Dcr2; Wor-Gal4, Ase-Gal80/CyO; UAS-myrRFP was used to mark wild-type type II lineages. Flp-out clones were generated using yw,hs-flp;;act5>cd2>gal4, UAS-GFP. Other genetic elements used were UAS-nerfin-1 and P[nerfin-1.GFP-NLS.SV-40] ia (Nerfin-1-GFP protein reporter), all from Kuzin et al. (2005); UAS-myc RNAi (KK106066, Vienna Drosophila RNAi Center [VDRC]); UAS-Tor DN (Hennig and Neufeld 2002); UAS-rag DN (Kim et al. 2008); UAS-raptorRNAi (TRiP 41912, Bloomington Stock Center); UAS-prosRNAi (TRiP 42538, Bloomington Stock Center); and w; nerfin-1-gfp/cyo; HisRFP/TM6b. Pan-neuroblast (UAS-Dcr2; dnab-gal4, Bloomington Stock Center) overexpression of UAS-nerfin-1 (two copies, one located on chromosome II, and one located on chromosome III; kind gift of A. Kuzin) was performed at 29 C. Animals were dissected at either 96 h or 110 h ALH. Immunostaining Larval tissues were fixed for 20 min in 4% formaldehyde/phosphate-buffered saline (PBS) and immunostained as described (Bello et al. 2003). The primary antibodies used were rabbit anti-apkc (1:500; Santa Cruz Biotechnology), mouse anti-mira (1:50; gift of A. Gould), rat anti-ph3 (1:500; Abcam), chick anti-gfp (1:2000; Abcam), rabbit anti-dpn (1:100; gift of Y.N. Jan), guinea pig anti- Dpn (1:1000; gift of James Skeath), rat anti-pros (1:50; gift of F. Matsuzaki), rabbit anti-ase (1:50; gift of F. Matsuzaki), rat anti- CycE (1:500; gift of Helena Richardson), mouse anti-cyca (1:50; Developmental Studies Hybridoma Bank), mouse or rat anti-elav (1:100; Developmental Studies Hybridoma Bank), rabbit anti-insc (1:1000; gift of W. Chia), rabbit anti-myc (1:100; Santa Cruz Biotechnology), guinea pig anti-nerfin-1 (1:5000; gift of A. Kuzin), rabbit anti-rfp (1:100; Abcam), and mouse anti-fib (1:200; Abcam). Secondary goat antibodies conjugated to Alexa488, Alexa568, Alexa650, and Alexa505 (Molecular Probes) were used at 1:200. Images were collected on a Leica SP5 confocal microscope, and all images shown are single sections unless otherwise stated. Clone induction To induce MARCM clones for both the CNS and wing discs, larvae were heat-shocked 24 h ALH for 12 or 15 min at 37 C and 140 GENES & DEVELOPMENT
Cosa sono le cellule staminali?
 Cosa sono le cellule staminali? Cellule Staminali Embrionali: totipotenti o pluripotenti Provengono dalla inner cell mass di embrioni di 4-5 giorni (blastocisti) prodotti in vitro Adulte: multipotenti
Cosa sono le cellule staminali? Cellule Staminali Embrionali: totipotenti o pluripotenti Provengono dalla inner cell mass di embrioni di 4-5 giorni (blastocisti) prodotti in vitro Adulte: multipotenti
Supplementary Methods
 Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Earmuff restricts progenitor cell potential by attenuating the competence to respond to self-renewal factors
 2014. Published by The Company of Biologists Ltd (2014) 141, 1036-1046 doi:10.1242/dev.106534 RESEARCH ARTICLE STEM CELLS AND REGENERATION Earmuff restricts progenitor cell potential by attenuating the
2014. Published by The Company of Biologists Ltd (2014) 141, 1036-1046 doi:10.1242/dev.106534 RESEARCH ARTICLE STEM CELLS AND REGENERATION Earmuff restricts progenitor cell potential by attenuating the
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts
 Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
The Super Elongation Complex Drives Neural Stem Cell Fate Commitment
 Article The Super Elongation Complex Drives Neural Stem Cell Fate Commitment Graphical Abstract Authors Kun Liu, Dan Shen, Jingwen Shen,..., Chouin Wong, Weidong Feng, Yan Song Correspondence yan.song@pku.edu.cn
Article The Super Elongation Complex Drives Neural Stem Cell Fate Commitment Graphical Abstract Authors Kun Liu, Dan Shen, Jingwen Shen,..., Chouin Wong, Weidong Feng, Yan Song Correspondence yan.song@pku.edu.cn
a) Stock 4: 252 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.
![a) Stock 4: 252 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males. a) Stock 4: 252 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.](/thumbs/91/106088469.jpg) Drosophila Problem Set 2018 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
Drosophila Problem Set 2018 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
a) Stock 4: 126 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.
![a) Stock 4: 126 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males. a) Stock 4: 126 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.](/thumbs/88/116538069.jpg) Drosophila Problem Set 2012 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
Drosophila Problem Set 2012 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supporting Information
 Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Supporting Information Ho et al. 10.1073/pnas.0808899106 SI Materials and Methods In immunostaining, antibodies from Developmental Studies Hybridoma ank were -FasII (mouse, 1:200), - (mouse, 1:100), and
Supplemental Information. The Super Elongation Complex Drives. Neural Stem Cell Fate Commitment
 evelopmental ell, Volume Supplemental Information The Super longation omplex rives Neural Stem ell ate ommitment Kun Liu, an Shen, Jingwen Shen, Shihong M. Gao, o Li, houin Wong, Weidong eng, and Yan Song
evelopmental ell, Volume Supplemental Information The Super longation omplex rives Neural Stem ell ate ommitment Kun Liu, an Shen, Jingwen Shen, Shihong M. Gao, o Li, houin Wong, Weidong eng, and Yan Song
Supplementary Methods
 Caussinus and Gonzalez Supplementary Methods Mitotic clones Clones of NBs homozygous for numb 03235, mira ZZ176, pros 17 or apkc k06403 were generated by FLP/FRT mediated mitotic recombination (1). As
Caussinus and Gonzalez Supplementary Methods Mitotic clones Clones of NBs homozygous for numb 03235, mira ZZ176, pros 17 or apkc k06403 were generated by FLP/FRT mediated mitotic recombination (1). As
Figure 1-Figure Supplement 1 Validation of knockdown efficiency for trpa1 RNAi and RyR RNAi lines.
 Figure 1-Figure Supplement 1 Validation of knockdown efficiency for trpa1 RNAi and RyR RNAi lines. (A) Cartoon depicting the four characterized trpa1 isoforms. The target regions of trpa1 RNAi lines used
Figure 1-Figure Supplement 1 Validation of knockdown efficiency for trpa1 RNAi and RyR RNAi lines. (A) Cartoon depicting the four characterized trpa1 isoforms. The target regions of trpa1 RNAi lines used
The tumor suppressor Brat controls neuronal stem cell lineages by inhibiting Deadpan and Zelda
 The tumor suppressor Brat controls neuronal stem cell lineages by inhibiting Deadpan and Zelda Ilka Reichardt, François Bonnay, Victoria Steinmann, Inga Loedige, Thomas R. Burkard, Gunter Meister and Juergen
The tumor suppressor Brat controls neuronal stem cell lineages by inhibiting Deadpan and Zelda Ilka Reichardt, François Bonnay, Victoria Steinmann, Inga Loedige, Thomas R. Burkard, Gunter Meister and Juergen
Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and
 Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Fly culture and crosses were performed according to standard procedures and were
 Supplemental Material Supplemental Experimental Procedures Fly genetics Drosophila strains Fly culture and crosses were performed according to standard procedures and were raised at the indicated temperatures.
Supplemental Material Supplemental Experimental Procedures Fly genetics Drosophila strains Fly culture and crosses were performed according to standard procedures and were raised at the indicated temperatures.
A population of Nestin expressing progenitors in the cerebellum exhibits increased tumorigenicity
 A population of Nestin expressing progenitors in the cerebellum exhibits increased tumorigenicity Peng Li 1,2, Fang Du 1, Larra W. Yuelling 1, Tiffany Lin 3, Renata E. Muradimova 1, Rossella Tricarico
A population of Nestin expressing progenitors in the cerebellum exhibits increased tumorigenicity Peng Li 1,2, Fang Du 1, Larra W. Yuelling 1, Tiffany Lin 3, Renata E. Muradimova 1, Rossella Tricarico
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
midlife crisis encodes a conserved zinc-finger protein required to maintain neuronal differentiation in Drosophila
 epress. Posted online 11 September 2013 STEM CELLS AND REGENERATION RESEARCH ARTICLE 1 140, 0000-0000 (2013) doi:10.1242/dev.093781 2013. Published by The Company of Biologists Ltd midlife crisis encodes
epress. Posted online 11 September 2013 STEM CELLS AND REGENERATION RESEARCH ARTICLE 1 140, 0000-0000 (2013) doi:10.1242/dev.093781 2013. Published by The Company of Biologists Ltd midlife crisis encodes
SUPPLEMENTARY INFORMATION
 a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
A Genetic Screen to Identify Mammalian Chromatin Modifiers In Vivo.
 Gus Frangou, Stephanie Palmer & Mark Groudine Fred Hutchinson Cancer Research Center, Seattle- USA A Genetic Screen to Identify Mammalian Chromatin Modifiers In Vivo. During mammalian development and differentiation
Gus Frangou, Stephanie Palmer & Mark Groudine Fred Hutchinson Cancer Research Center, Seattle- USA A Genetic Screen to Identify Mammalian Chromatin Modifiers In Vivo. During mammalian development and differentiation
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
doi:10.1038/nature10810 Supplementary Fig. 1: Mutation of the loqs gene leads to shortened lifespan and adult-onset brain degeneration. a. Northern blot of control and loqs f00791 mutant flies. loqs f00791
Trasposable elements: Uses of P elements Problem set B at the end
 Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Supplementary Figure 1.
 Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts
 Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts Hongyan Wang, 1,2,4 Gregory W. Somers, 1 Arash Bashirullah, 3 Ulrike Heberlein, 2 Fengwei Yu, 1 and William Chia
Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts Hongyan Wang, 1,2,4 Gregory W. Somers, 1 Arash Bashirullah, 3 Ulrike Heberlein, 2 Fengwei Yu, 1 and William Chia
Supporting Information
 Supporting Information Chen et al. 10.1073/pnas.1113882109 SI Methods Strains. The following strains were used in this study: (i) y w, hsflp;; FRT82B ubigfp;(ii) y w, eyflp;; FRT82B ubigfp;(iii) yw,ubx
Supporting Information Chen et al. 10.1073/pnas.1113882109 SI Methods Strains. The following strains were used in this study: (i) y w, hsflp;; FRT82B ubigfp;(ii) y w, eyflp;; FRT82B ubigfp;(iii) yw,ubx
Sydney Brenner: Present
 Sydney Brenner: 1927 - Present Born in the small town of Germiston, South Africa Won a scholarship to the University of Witwatersrand at the age of 15 Received his PhD from Exeter College, Oxford Made
Sydney Brenner: 1927 - Present Born in the small town of Germiston, South Africa Won a scholarship to the University of Witwatersrand at the age of 15 Received his PhD from Exeter College, Oxford Made
Supplementary Figure1: ClustalW comparison between Tll, Dsf and NR2E1.
 P-Box Dsf -----------------MG-TAG--DRLLD-IPCKVCGDRSSGKHYGIYSCDGCSGFFKR 39 NR2E1 -----------------MSKPAGSTSRILD-IPCKVCGDRSSGKHYGVYACDGCSGFFKR 42 Tll MQSSEGSPDMMDQKYNSVRLSPAASSRILYHVPCKVCRDHSSGKHYGIYACDGCAGFFKR
P-Box Dsf -----------------MG-TAG--DRLLD-IPCKVCGDRSSGKHYGIYSCDGCSGFFKR 39 NR2E1 -----------------MSKPAGSTSRILD-IPCKVCGDRSSGKHYGVYACDGCSGFFKR 42 Tll MQSSEGSPDMMDQKYNSVRLSPAASSRILYHVPCKVCRDHSSGKHYGIYACDGCAGFFKR
Supplemental Information. Stratum, a Homolog of the Human GEF Mss4, Partnered with Rab8, Controls the Basal Restriction
 Cell Reports, Volume 18 Supplemental Information Stratum, a Homolog of the Human GEF Mss4, Partnered with Rab8, Controls the Basal Restriction of Basement Membrane Proteins in Epithelial Cells Olivier
Cell Reports, Volume 18 Supplemental Information Stratum, a Homolog of the Human GEF Mss4, Partnered with Rab8, Controls the Basal Restriction of Basement Membrane Proteins in Epithelial Cells Olivier
embryos. Asterisk represents loss of or reduced expression. Brackets represent
 Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
New Tool for Late Stage Visualization of Oskar Protein in Drosophila melanogaster. Oocytes. Kathleen Harnois. Supervised by Paul Macdonald
 New Tool for Late Stage Visualization of Oskar Protein in Drosophila melanogaster Oocytes Kathleen Harnois Supervised by Paul Macdonald Institute of Cellular and Molecular Biology, The University of Texas
New Tool for Late Stage Visualization of Oskar Protein in Drosophila melanogaster Oocytes Kathleen Harnois Supervised by Paul Macdonald Institute of Cellular and Molecular Biology, The University of Texas
Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling
 Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling Go-Woon Kim, Jong-Hoon Won, Ok-Kyung Lee, Sang-Soo Lee, Jeong-Hoon Han, Orkhon Tsogtbaatar, Sujin Nam, Yeon Kim, and Kyung-Ok Cho* Supplementary
Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling Go-Woon Kim, Jong-Hoon Won, Ok-Kyung Lee, Sang-Soo Lee, Jeong-Hoon Han, Orkhon Tsogtbaatar, Sujin Nam, Yeon Kim, and Kyung-Ok Cho* Supplementary
fga phenotypes were also analyzed in the tracheal system, the organ responsible for
 SUPPLEMENTARY TEXT Tracheal defects fga phenotypes were also analyzed in the tracheal system, the organ responsible for oxygen delivery in the fly. In fga homozygous mutant alleles, we observed defects
SUPPLEMENTARY TEXT Tracheal defects fga phenotypes were also analyzed in the tracheal system, the organ responsible for oxygen delivery in the fly. In fga homozygous mutant alleles, we observed defects
Brain tumor specifies intermediate progenitor cell identity by attenuating β-catenin/armadillo activity
 2014. Published by The Company of Biologists Ltd (2014) 141, 51-62 doi:10.1242/dev.099382 RESEARCH ARTICLE STEM CELLS AND REGENERATION Brain tumor specifies intermediate progenitor cell identity by attenuating
2014. Published by The Company of Biologists Ltd (2014) 141, 51-62 doi:10.1242/dev.099382 RESEARCH ARTICLE STEM CELLS AND REGENERATION Brain tumor specifies intermediate progenitor cell identity by attenuating
Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and
 Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and Tin TFBS motifs and located in the flanking or intronic
Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and Tin TFBS motifs and located in the flanking or intronic
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms
 Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Otefin, a Nuclear Membrane Protein, Determines
 Developmental Cell 14 Supplemental Data Otefin, a Nuclear Membrane Protein, Determines the Fate of Germline Stem Cells in Drosophila via Interaction with Smad Complexes Xiaoyong Jiang, Laixin Xia, Dongsheng
Developmental Cell 14 Supplemental Data Otefin, a Nuclear Membrane Protein, Determines the Fate of Germline Stem Cells in Drosophila via Interaction with Smad Complexes Xiaoyong Jiang, Laixin Xia, Dongsheng
Biology Open (2015) 000, 1 16 doi: /bio
 Supplementary Material Annekatrin König and Halyna R. Shcherbata doi: 10.1242/ bio.201410553 Fig. S1. Histones are differentially modified in somatic and germline cells in the germarium. Wildtype (OregonR)
Supplementary Material Annekatrin König and Halyna R. Shcherbata doi: 10.1242/ bio.201410553 Fig. S1. Histones are differentially modified in somatic and germline cells in the germarium. Wildtype (OregonR)
Supplemental Figure 1 (Figure S1), related to Figure 1 Figure S1 provides evidence to demonstrate Nfatc1Cre is a mouse line that directed gene
 Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Name AP Biology Mrs. Laux Take home test #11 on Chapters 14, 15, and 17 DUE: MONDAY, DECEMBER 21, 2009
 MULTIPLE CHOICE QUESTIONS 1. Inducible genes are usually actively transcribed when: A. the molecule degraded by the enzyme(s) is present in the cell. B. repressor molecules bind to the promoter. C. lactose
MULTIPLE CHOICE QUESTIONS 1. Inducible genes are usually actively transcribed when: A. the molecule degraded by the enzyme(s) is present in the cell. B. repressor molecules bind to the promoter. C. lactose
ANNOUNCEMENTS. HW2 is due Thursday 2/4 by 12:00 pm. Office hours: Monday 12:50 1:20 (ECCH 134)
 ANNOUNCEMENTS HW2 is due Thursday 2/4 by 12:00 pm. Office hours: Monday 12:50 1:20 (ECCH 134) Lectures 6 8 Outline Stem Cell Division - symmetric - asymmetric Stem cell lineage potential - pluripotent
ANNOUNCEMENTS HW2 is due Thursday 2/4 by 12:00 pm. Office hours: Monday 12:50 1:20 (ECCH 134) Lectures 6 8 Outline Stem Cell Division - symmetric - asymmetric Stem cell lineage potential - pluripotent
Genetic Basis of Development & Biotechnologies
 Genetic Basis of Development & Biotechnologies 1. Steps of embryonic development: cell division, morphogenesis, differentiation Totipotency and pluripotency 2. Plant cloning 3. Animal cloning Reproductive
Genetic Basis of Development & Biotechnologies 1. Steps of embryonic development: cell division, morphogenesis, differentiation Totipotency and pluripotency 2. Plant cloning 3. Animal cloning Reproductive
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
a previous report, Lee and colleagues showed that Oct4 and Sox2 bind to DXPas34 and Xite
 SUPPLEMENTARY INFORMATION doi:1.138/nature9496 a 1 Relative RNA levels D12 female ES cells 24h Control sirna 24h sirna Oct4 5 b,4 Oct4 Xist (wt) Xist (mut) Tsix 3' (wt) Tsix 5' (wt) Beads Oct4,2 c,2 Xist
SUPPLEMENTARY INFORMATION doi:1.138/nature9496 a 1 Relative RNA levels D12 female ES cells 24h Control sirna 24h sirna Oct4 5 b,4 Oct4 Xist (wt) Xist (mut) Tsix 3' (wt) Tsix 5' (wt) Beads Oct4,2 c,2 Xist
A Survey of Genetic Methods
 IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
Nature Immunology: doi: /ni.3694
 Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
CELL BIOLOGY - CLUTCH CH. 7 - GENE EXPRESSION.
 !! www.clutchprep.com CONCEPT: CONTROL OF GENE EXPRESSION BASICS Gene expression is the process through which cells selectively to express some genes and not others Every cell in an organism is a clone
!! www.clutchprep.com CONCEPT: CONTROL OF GENE EXPRESSION BASICS Gene expression is the process through which cells selectively to express some genes and not others Every cell in an organism is a clone
Lecture 8: Transgenic Model Systems and RNAi
 Lecture 8: Transgenic Model Systems and RNAi I. Model systems 1. Caenorhabditis elegans Caenorhabditis elegans is a microscopic (~1 mm) nematode (roundworm) that normally lives in soil. It has become one
Lecture 8: Transgenic Model Systems and RNAi I. Model systems 1. Caenorhabditis elegans Caenorhabditis elegans is a microscopic (~1 mm) nematode (roundworm) that normally lives in soil. It has become one
Biology 4361 Developmental Biology Lecture 4. The Genetic Core of Development
 Biology 4361 Developmental Biology Lecture 4. The Genetic Core of Development The only way to get from genotype to phenotype is through developmental processes. - Remember the analogy that the zygote contains
Biology 4361 Developmental Biology Lecture 4. The Genetic Core of Development The only way to get from genotype to phenotype is through developmental processes. - Remember the analogy that the zygote contains
Gradients of the Drosophila Chinmo BTB-Zinc Finger Protein Govern Neuronal Temporal Identity
 Gradients of the Drosophila Chinmo BTB-Zinc Finger Protein Govern Neuronal Temporal Identity Sijun Zhu, 1,2,5 Suewei Lin, 3 Chih-Fei Kao, 3 Takeshi Awasaki, 3 Ann-Shyn Chiang, 4 and Tzumin Lee 1,3, * 1
Gradients of the Drosophila Chinmo BTB-Zinc Finger Protein Govern Neuronal Temporal Identity Sijun Zhu, 1,2,5 Suewei Lin, 3 Chih-Fei Kao, 3 Takeshi Awasaki, 3 Ann-Shyn Chiang, 4 and Tzumin Lee 1,3, * 1
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
SUPPORTING ONLINE MATERIAL
 SUPPORTING ONLINE MATERIAL MATERIAL AND METHODS Fly stocks All Drosophila stocks were grown at 25 C on yeast-containing corn meal/molasses medium. The nanos (nos) alleles nos 18 and nos 53 have been previously
SUPPORTING ONLINE MATERIAL MATERIAL AND METHODS Fly stocks All Drosophila stocks were grown at 25 C on yeast-containing corn meal/molasses medium. The nanos (nos) alleles nos 18 and nos 53 have been previously
Neural Stem Cell Characterization Kit
 Neural Stem Cell Characterization Kit Catalog No. SCR019 FOR RESEARCH USE ONLY Not for use in diagnostic procedures USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209 Europe +44 (0) 23 8026 2233
Neural Stem Cell Characterization Kit Catalog No. SCR019 FOR RESEARCH USE ONLY Not for use in diagnostic procedures USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209 Europe +44 (0) 23 8026 2233
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila
 A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila Joy S Wu & Liqun Luo Howard Hughes Medical Institute, Department of Biological Sciences, Neurosciences Program, Stanford
A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila Joy S Wu & Liqun Luo Howard Hughes Medical Institute, Department of Biological Sciences, Neurosciences Program, Stanford
Supporting Information
 Supporting Information Lukaszewicz and Anderson 1.173/pnas.1162318 SI Text 1 Modeling GOF Electroporation. We tested whether mke overexpression could lead indirectly to an increase in the size of the neurogenic
Supporting Information Lukaszewicz and Anderson 1.173/pnas.1162318 SI Text 1 Modeling GOF Electroporation. We tested whether mke overexpression could lead indirectly to an increase in the size of the neurogenic
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Linköping University Post Print. Segment-specific Neuronal Sub-type Specification by the Integration of Anteroposterior and Temporal Cues
 Linköping University Post Print Segment-specific Neuronal Sub-type Specification by the Integration of Anteroposterior and Temporal Cues Daniel Karlsson, Magnus Baumgardt and Stefan Thor N.B.: When citing
Linköping University Post Print Segment-specific Neuronal Sub-type Specification by the Integration of Anteroposterior and Temporal Cues Daniel Karlsson, Magnus Baumgardt and Stefan Thor N.B.: When citing
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte
 Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte Carlo algorithm for one million generations to obtain
Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte Carlo algorithm for one million generations to obtain
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl
 Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Reviewers' Comments: Reviewer #1 (Remarks to the Author)
 Reviewers' Comments: Reviewer #1 (Remarks to the Author) In this study, Rosenbluh et al reported direct comparison of two screening approaches: one is genome editing-based method using CRISPR-Cas9 (cutting,
Reviewers' Comments: Reviewer #1 (Remarks to the Author) In this study, Rosenbluh et al reported direct comparison of two screening approaches: one is genome editing-based method using CRISPR-Cas9 (cutting,
Supplement Figure S1:
 Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
SUPPLEMENTARY INFORMATION
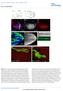 DOI: 10.1038/ncb2806 Supplemental Material: Figure S1 Wnt4 PCP phenotypes are not mediated through canonical Wnt signaling. (a-a ) Illustration of wg, dwnt4 and dwnt6 expression patterns in wing (a) and
DOI: 10.1038/ncb2806 Supplemental Material: Figure S1 Wnt4 PCP phenotypes are not mediated through canonical Wnt signaling. (a-a ) Illustration of wg, dwnt4 and dwnt6 expression patterns in wing (a) and
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Fig. 1. Multiple five micron sections of liver tissues of rats treated
 Supplementary Figure Legends Supplementary Fig. 1. Multiple five micron sections of liver tissues of rats treated with either vehicle (left; n=3) or CCl 4 (right; n=3) were co-immunostained for NRP-1 (green)
Supplementary Figure Legends Supplementary Fig. 1. Multiple five micron sections of liver tissues of rats treated with either vehicle (left; n=3) or CCl 4 (right; n=3) were co-immunostained for NRP-1 (green)
Supplementary Figure S1. Alterations in Fzr1( / );Nestin-Cre brains. (a) P10 Cdh1-deficient brains display low levels of Myelin basic protein (MBP)
 Supplementary Figure S1. Alterations in Fzr1( / );Nestin-Cre brains. (a) P10 Cdh1-deficient brains display low levels of Myelin basic protein (MBP) in the cortex (area 1 as defined in Fig. 2a), and corpus
Supplementary Figure S1. Alterations in Fzr1( / );Nestin-Cre brains. (a) P10 Cdh1-deficient brains display low levels of Myelin basic protein (MBP) in the cortex (area 1 as defined in Fig. 2a), and corpus
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
beta-galactosidase (A420) ctrl
 0.6 beta-galactosidase (A420) 0.4 0.2 ctrl 2A 22A 35B 36B 58A 64A 68E 75C 86Fa 86Fb 102F Fig. S1. Levels of induced expression at various attp sites. The induced β-galactosidase activity was measured from
0.6 beta-galactosidase (A420) 0.4 0.2 ctrl 2A 22A 35B 36B 58A 64A 68E 75C 86Fa 86Fb 102F Fig. S1. Levels of induced expression at various attp sites. The induced β-galactosidase activity was measured from
b number of motif occurrences
 supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
Aneuploidy causes premature differentiation of neural and intestinal stem cells
 Received 3 Apr 15 Accepted 14 Oct 15 Published 17 Nov 15 DOI:.38/ncomms9894 OPEN Aneuploidy causes premature differentiation of neural and intestinal stem cells Delphine Gogendeau 1, Katarzyna Siudeja
Received 3 Apr 15 Accepted 14 Oct 15 Published 17 Nov 15 DOI:.38/ncomms9894 OPEN Aneuploidy causes premature differentiation of neural and intestinal stem cells Delphine Gogendeau 1, Katarzyna Siudeja
ANNOUNCEMENTS. HW1 is due Thursday 1/28 by 12:00 pm. Office hours: Monday 12:50 1:20 (ECCH 134) Wednesday 9:45 10:15 (ECCH 134)
 ANNOUNCEMENTS HW1 is due Thursday 1/28 by 12:00 pm. Office hours: Monday 12:50 1:20 (ECCH 134) Wednesday 9:45 10:15 (ECCH 134) Lecture 5: Kinetics of Cell Growth in Culture Phenomenological mathematical
ANNOUNCEMENTS HW1 is due Thursday 1/28 by 12:00 pm. Office hours: Monday 12:50 1:20 (ECCH 134) Wednesday 9:45 10:15 (ECCH 134) Lecture 5: Kinetics of Cell Growth in Culture Phenomenological mathematical
mod-1::mcherry unc-47::gfp RME ser-4::gfp vm2 vm2 vm2 vm2 VNC
 A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
John Gurdon was testing the hypothesis of genomic equivalence or that when cells divide they retain a full genomic compliment.
 1. (15 pts) John Gurdon won the 2012 Nobel Prize in Physiology or Medicine for work he did in the 1960 s. What was the major developmental hypothesis he set out to test? What techniques did he development
1. (15 pts) John Gurdon won the 2012 Nobel Prize in Physiology or Medicine for work he did in the 1960 s. What was the major developmental hypothesis he set out to test? What techniques did he development
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Experimental genetics - 2 Partha Roy
 Partha Roy Experimental genetics - 2 Making genetically altered animal 1) Gene knock-out k from: a) the entire animal b) selected cell-type/ tissue c) selected cell-type/tissue at certain time 2) Transgenic
Partha Roy Experimental genetics - 2 Making genetically altered animal 1) Gene knock-out k from: a) the entire animal b) selected cell-type/ tissue c) selected cell-type/tissue at certain time 2) Transgenic
Supplementary Materials
 Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
Differential Gene Expression
 Biology 4361 Developmental Biology Differential Gene Expression September 28, 2006 Chromatin Structure ~140 bp ~60 bp Transcriptional Regulation: 1. Packing prevents access CH 3 2. Acetylation ( C O )
Biology 4361 Developmental Biology Differential Gene Expression September 28, 2006 Chromatin Structure ~140 bp ~60 bp Transcriptional Regulation: 1. Packing prevents access CH 3 2. Acetylation ( C O )
Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of
 Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and
 Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Chapter 11: Regulation of Gene Expression
 Chapter Review 1. It has long been known that there is probably a genetic link for alcoholism. Researchers studying rats have begun to elucidate this link. Briefly describe the genetic mechanism found
Chapter Review 1. It has long been known that there is probably a genetic link for alcoholism. Researchers studying rats have begun to elucidate this link. Briefly describe the genetic mechanism found
Histone H3 Methylation Antibody Panel Pack I - Active Genes Base Catalog # C Component Size Shipping Temperature
 Histone H3 Methylation Antibody Panel Pack I - Active Genes Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K4D Histone H3K4me2 (H3K4 Dimethyl) Polyclonal Antibody
Histone H3 Methylation Antibody Panel Pack I - Active Genes Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K4D Histone H3K4me2 (H3K4 Dimethyl) Polyclonal Antibody
Evolutionary conservation of midline repulsion by Robo family receptors in flies and mice
 University of Arkansas, Fayetteville ScholarWorks@UARK Biological Sciences Undergraduate Honors Theses Biological Sciences 5-2018 Evolutionary conservation of midline repulsion by Robo family receptors
University of Arkansas, Fayetteville ScholarWorks@UARK Biological Sciences Undergraduate Honors Theses Biological Sciences 5-2018 Evolutionary conservation of midline repulsion by Robo family receptors
Methanol fixation allows better visualization of Kal7. To compare methods for
 Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Flybow genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster
 Nature Methods Flybow genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster Dafni Hadjieconomou, Shay Rotkopf, Cyrille Alexandre, Donald M Bell, Barry J Dickson & Iris
Nature Methods Flybow genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster Dafni Hadjieconomou, Shay Rotkopf, Cyrille Alexandre, Donald M Bell, Barry J Dickson & Iris
Blocking endocycle apoptosis page 1. Table S1: Relative abundance of mrna levels in the endocycling salivary gland versus mitotic
 Blocking endocycle apoptosis page 1 SUPPLEMENTAL DATA Table S1: Relative abundance of mrna levels in the endocycling salivary gland versus mitotic cycling brain-disc. Gene E/M 1 E rr /M rr E rr /E M rr
Blocking endocycle apoptosis page 1 SUPPLEMENTAL DATA Table S1: Relative abundance of mrna levels in the endocycling salivary gland versus mitotic cycling brain-disc. Gene E/M 1 E rr /M rr E rr /E M rr
Development Supplementary information SUPPLEMENTARY METHODS
 SUPPLEMENTARY METHODS BrdU and EdU labeling For in vivo pulse/chase BrdU labeling experiments, recently hatched first-instar larvae were collected and grown in standard medium at 25 C. At the desired developmental
SUPPLEMENTARY METHODS BrdU and EdU labeling For in vivo pulse/chase BrdU labeling experiments, recently hatched first-instar larvae were collected and grown in standard medium at 25 C. At the desired developmental
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
sides of the aleurone (Al) but it is excluded from the basal endosperm transfer layer
 Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
Histone H3K27 Methylation Antibody Panel Pack Base Catalog # C Component Size Shipping Temperature
 Histone H3K27 Methylation Antibody Panel Pack Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K27M Histone H3K27me1 (H3K27 Monomethyl) Polyclonal Antibody 25 µg
Histone H3K27 Methylation Antibody Panel Pack Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K27M Histone H3K27me1 (H3K27 Monomethyl) Polyclonal Antibody 25 µg
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin
 Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
unc-33/crmp and ankyrin organize microtubules and localize kinesin to polarize
 Supplementary Information unc-33/crmp and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting Tapan A. Maniar 1, Miriam Kaplan 1, George J. Wang 2, Kang Shen 2, Li Wei
Supplementary Information unc-33/crmp and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting Tapan A. Maniar 1, Miriam Kaplan 1, George J. Wang 2, Kang Shen 2, Li Wei
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Transgenesis. Stable integration of foreign DNA into host genome Foreign DNA is passed to progeny germline transmission
 Transgenic Mice Transgenesis Stable integration of foreign DNA into host genome Foreign DNA is passed to progeny germline transmission integrates into all cells including sperm or egg Knockin mice DNA
Transgenic Mice Transgenesis Stable integration of foreign DNA into host genome Foreign DNA is passed to progeny germline transmission integrates into all cells including sperm or egg Knockin mice DNA
CS262 Lecture 12 Notes Single Cell Sequencing Jan. 11, 2016
 CS262 Lecture 12 Notes Single Cell Sequencing Jan. 11, 2016 Background A typical human cell consists of ~6 billion base pairs of DNA and ~600 million bases of mrna. It is time-consuming and expensive to
CS262 Lecture 12 Notes Single Cell Sequencing Jan. 11, 2016 Background A typical human cell consists of ~6 billion base pairs of DNA and ~600 million bases of mrna. It is time-consuming and expensive to
