SUPPLEMENTAL MATERIALS AND METHODS
|
|
|
- Lisa Cole
- 6 years ago
- Views:
Transcription
1 SUPPLEMENTAL MATERIALS AND METHODS ChIP-Seq analysis ChIPed DNA sample libraries were analyzed using Illumina parallel sequencing. Peak detection of enriched binding regions was performed using FindPeaks 1 with an estimated false discovery rate < 0.05 as the selection criterion for enriched regions. The identified peaks were mapped onto mouse genome using UCSC mus musculus reference genome version 8 (build 2006). Any peaks that are overlapping ( 1 base pair) with repetitive genomic regions or controls were discarded. Peaks from biological replicates were consolidated with their genomic coordinates merged if they overlap at least one base pair on the genome. Characterization and summary of the whole-genome occupancy pattern of peaks were performed using the Cis-regulatory Element Annotation System (CEAS) annotation tool. 2,3 ChIP-chip analysis ChIP-chip data on enriched regions and selected transcription start sites was analyzed using MA2C 4 which adjusts for sequence GC content bias. Raw intensity measures were normalized using default parameter setting. Peak detection was performed using 300bp sliding window with coverage constraint of having at least 3 probes. Final set of peaks were determined if the difference in intensity values of chipped sample and input has a significance level of p-value < Probewise averaging of all peaks was empirically estimated from all data points. An object-oriented implementation for analyzing multiple ChIP-chip experiments based on MA2C analysis is available at author s website: Analysis of Hoxa9-ER gene expression profiles Gene expression values were calculated using a robust multi-array average (RMA). 5 Temporal differential gene expression analysis was performed using EDGE. 6 The significance measures (p-values) of individual genes were adjusted for multiple hypothesis testing using Benjamini-Hochberg method. 7 A gene was deemed significant if its temporal expression change satis.es a composite criterion: 1) with adjusted p-value < 0.05; and 2) having 1.5-fold change (median expression of three replicates) compared to controls at any of the three time points (namely 72, 96, 120 hours). For genes with multiple probesets, we selected only one probeset with the highest significance. In consistency with the use of mouse genome mm8 in ChIP-Seq, genes were annotated with NetAffx Mouse430_2 annotation matrix (release 25, UCSC mm8 / NCBI build 36). When no information of official gene symbol is available, the sequence identifiers (e.g., GenBank accession numbers) are used. Clustering of Hoxa9-ER significant genes All significant genes were clustered into four groups based on their temporal patterns using Self- Organizing Maps 8 in a similar way as Huang, et al. 9 Prior to clustering, the expression of each gene was normalized to have zero mean and unit variance. A hexagonal topology of 2 by 2 prototypes was initiated and each gene was subject to the network 400 iterations. The L 2distance measure was used as distance metric. The centroid and standard deviation (SEM) of each cluster was computed and plotted in Fig. S2E. The significant genes were mapped to closest H/M peaks using CisGenome. 10 De novo motif discovery analysis For de novo motif discovery, enriched regions were distilled to a centered peak core sequence of 200 bp,
2 and overlapping peaks were collapsed into a single non-redundant core sequence prior to analysis. Subsequently, this dataset included 800 independent sequences subjected to the GADEM algorithm, an extension of MEME. 11 Three duplicate discovery runs were performed and combined into one de novo motif set; STAMP 12 was used to annotate identified motifs with the TRANSFAC 11.3 database, using E=1E-5 threshold and information content trimming >0.4; this resulted in 15/31 matches. Motifs were validated independently using multiple alternative approaches including Weeder, MEME, and CisGenome, 10,13 15 which yielded comparable results to de novo motif discovery using GADEM. Motif enrichment analysis MEME suite (Find Individual Motif Occurrences) was used to analyze enrichment of the experimentally determined Hoxa9/Meis1 consensus motif ATGATTTATGGC (Hoxa9-Meis1-Pbx1) and Meis1 consensus TGTC motif. Comprehensive search of known transcription factor binding motifs was performed for transcription factors included in Genomatix proprietary Mat Base Matrix Family Library (Version 8.2, January 2010) that includes a total of 727 motifs (170 motif families) and their corresponding position weight matrices. The DNA sequences of length 200bp from the center of each H/M peak were scanned for presence of any known transcription factor binding motif. A transcription factor binding motif is considered to be statistically significantly enriched in the H/M peaks if the number of sequences in which the motif is found to be present is significantly higher than its expected whole-genome occurrences according to standard z-test (z-score >2.81; p-value < 0.005). Evaluation of regulatory potentials of H/M peaks The base pair-wise average regulatory potential (RP) scores 16 of all H/M binding sites and their surrounding ±4 kb regions (extended H/M regions) were computed and shown as a function of distance from the center of the regions. More specifically, we compute the average RP score on a base pair basis across all extended H/M binding region. Comparison of H/M peaks and evolutionarily conserved elements The evolutionary conservation scores were obtained from UCSC phastcons17way 17 database for each enriched region and its adjacent regions extending up to 6 in width. A virtual peak is constructed by combining the left and right side of enriched region subject to the same width of enriched region. In a sliding widow fashion, six virtual peaks are constructed contiguously outwards from the center of the enriched region with no overlap with each other. A two samplet-test is performed comparing the average conservation score within the enriched region against that of each virtual peak. Script for running this analysis (using R) is available upon request. Mass spectroscopy To identify proteins co-immunoprecipitating with HA-tagged Hoxa9 and FLAG-tagged Meis1, immunoprecipitates were resolved on SDS-PAGE gels and stained with Colloidal Blue (Invitrogen) followed by destaining and cysteine reduction/carbamidomethylation (10 mm DTT+50 mmiodoacetamide). Macerated and dried gel slices were re-swollen and digested in ammonium bicarbonate buffer with trypsin (Promega). Peptides were extracted sequentially using acetonitrile/tfa gradient; extracts were pooled and concentrated prior to reverse phase chromatography (Aquasil C18,
3 Picofrit column, New Objectives). Eluted peptides were directly introduced into an ion-trap mass spectrometer (LTQ-XL, ThermoFisher) with a nano-spray (in MS/MS mode). Data were converted to mzxml format and searched against mouse IPI (v 3.50) + reverse database using X!Tandem with k-score plug-in (Global Proteome Machine). Proteins with >0.9 probability were selected (ProteinProphet29/30). Outputs were subjected to PeptideProphet and ProteinProphet analysis; proteins with a ProteinProphet probability of >0.9 were considered for further analysis. MS/MS spectra corresponding to proteins that were unique to the experimental sample were manually verified (Table S6). 18 REFERENCES 1. Robertson G, Hirst M, Bainbridge M, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4: Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system.bioinformatics. 2009;25: Song JS, Johnson WE, Zhu X, et al. Model-based analysis of two-color arrays (MA2C).Genome Biol. 2007;8:R Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci U S A. 2005;102: Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57: Kohonen T. Self-Organizing Maps. Series in Information Sciences. 1995; Huang J, Jennings NR, Fox J. An Agent-based Approach to Health Care Management. Int Journal of Applied Artificial Intelligence. 1995;9: Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26: Li L. GADEM: a genetic algorithm guided formation of spaced dyads coupled with an EM algorithm for motif discovery. J Comput Biol. 2009;16: Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities.nucleic Acids
4 Res. 2007;35:W Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching.nucleic Acids Res. 2009;37:W Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W Taylor J, Tyekucheva S, King DC, Hardison RC, Miller W, Chiaromonte F. ESPERR: learning strong and weak signals in genomic sequence alignments to identify functional elements.genome Res. 2006;16: Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15: Nesvizhskii AI, Vitek O, Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat Methods. 2007;4:
5 Figure S1. Endogenous Hoxa9 and Meis1 protein levels in transformed mouse hematopoietic progenitor (MHP) cells Hoxa9 and Meis1 protein levels from whole cell lysates of MLL-AF9 transduced (MLL) and HA- Hoxa9/FLAG-Meis1 transduced (HM2) MHP cells were detected by western blot using anti-hoxa9 (Millipore) and anti-meis1 (Abcam) antibodies. Hoxa9 and Meis1 protein levels are shown to be comparable. β-actin, used as a loading control, was detected by an anti β-actin mouse monoclonal antibody (Sigma). Figure S2. Comparison of conservation scores in H/M peaks and the extended regions outside of H/M peaks, related to Fig. 3A For each extended region, an equal length of sequence segment was chosen successively outside of H/M sequences and their associated conservation was computed. Error bars are 2 SEM.
6 Figure S3. Hoxa9/ Meis1 binding regions show enhancer activity in luciferase assays (A) Twenty-two novel Hoxa9/ Meis1 binding regions of ~1,000 bp in length were cloned into ptal-luc luciferase reporter and electroporated into K-562 cells. Enhancer activity of the Hoxa9/ Meis1 binding
7 regions was compared to a known Runx1 conserved noncoding element (Runx1_chr16(+23)) (Nottingham et al. 2007) and two control regions randomly selected in the genome as a negative control. Data is presented as the Mean + SD in triplicate normalized to ptal-luc. (B) Comparison of ChIP-chip signal intensity in Hoxa9 / Mesi1 binding regions that were scored positively and negatively for enhancer activity in the presence and absence of 4-OHT. (C) Granular level details of ChIP-Chip signal intensity at 8 Hoxa9 / Meis1 binding regions that show enhancer activity.
8 Figure S4. Conditional transformation by Hoxa9-ER and identification of Hoxa9 regulated target genes, related to Fig. 4 (A) Hoxa9-ER cells cease dividing within 96 hours of 4-OHT withdrawal and show increased expression of the myeloid/monocytic differentiation markers Gr-1 and Mac-1 by flow cytometry (B). By day 6, the
9 majority of cells showed macrophage morphology, while myeloblast morphology was maintained in cells that were cultured in continuous 4-OHT (C). (D) Cascade of significant changes in gene expression secondary effects following 4-OHT withdrawal. (E) Cluster centroids of significant genes in Hoxa9-ER profiling. Four clusters of genes with significant changes in expression level after 4-OHT withdrawal. Genes are clustered into one of the four clusters based on their temporal dynamics. For each cluster, the centroid and SEM are shown with the total number of genes in that cluster (blue). (F) through (K) Heatmaps of genes with proximate Hoxa9 and/or Meis1 binding sites that are significantly differentially expressed over the 120 hrs period post 4-OHT withdrawal (FDR P<0.05 and median fold change >1.5) compared to controls. Genes are organized into 5 location categories according to the relative distance from their genomic features to the nearest Hoxa9/Meis1 binding sequences based on RefSeq gene annotation: (F) Sequences labeled as TSS located within 20k bp 5 to the transcription start site (G) Sequences labeled as 5 UTRs are the regions between the transcription and coding start sites. (H) Differentially regulated genes with intronic binding sites (I) sequences labeled 3 UTRs are defined as the regions between the coding and transcription termination sites. (J) Sequences labeled as TES are located within 20k bp to the transcription end site. Within each location category, genes are grouped based on their cluster designation (Fig. S4E and Table S2) and listed in descending order in their differential statistics. Data are normalized across samples such that the expression value of each individual gene has zero mean and standard deviation of one.
10 Figure S5. De novo motif discovery results, related to Fig. 5A and Fig. 5B (A) STAMP logos of de novo identified motifs that are enriched in H/M peaks. (B) Co-occurrence matrix of de novo motifs. (C) Bubble plot of co-occurrence of de novo motifs. The size of the bubbles indicates the magnitude of co-localization between two motifs.
11 Figure S6. Spatial distribution of de novo identified motifs, Related to Fig. 5A and Fig. 5B The histogram and density of de novo motif spatial distribution was computed with respect to the center of the H/M peaks.
12 Figure S7. Related to Fig. 5, Fig. 6, and Discussion (A) Motifs of Hoxa-Meis1-Pbx, STAT, C/EBPα, CREB, and ETS show complementary spatial distribution patterns. (B) Comparative motif enrichment analysis show distinct motif enrichment profiles in sequences bound by H/M (green), H/M + C/EBPα (blue), and C/EBPα alone (red).
13 Figure S8. Related to Fig. 7 and Discussion ChIP experiments showing Meis1, Pu.1, C/ebpα, Stat5a/b, P300, CBP, histone H3 and histone H4 acetylation association with Hoxasomes is dependent on Hoxa9 as evidenced by drop in ChIP signal following 4-OHT withdrawal. See Experimental Procedures and Fig. 2 legend.
14 Figure S9. Examples of Hoxa9/Meis1 binding sites carrying epigenetic signature of enhancers, related to Fig. 7 and Discussion Four H/M bindings sites near or at promoter region of Dnajc10, Cd34, Foxp1, and Flt3 are shown with epigenetic signatures of CBP, H3 acetyl, H4 acetyl, P300, and RNA pol II. For each epigenetic mark, chipchip results of 4-OHT (+) and 4-OHT withdrawal ( ) are shown.
Gene Signal Estimates from Exon Arrays
 Gene Signal Estimates from Exon Arrays I. Introduction: With exon arrays like the GeneChip Human Exon 1.0 ST Array, researchers can examine the transcriptional profile of an entire gene (Figure 1). Being
Gene Signal Estimates from Exon Arrays I. Introduction: With exon arrays like the GeneChip Human Exon 1.0 ST Array, researchers can examine the transcriptional profile of an entire gene (Figure 1). Being
Introduction to genome biology
 Introduction to genome biology Lisa Stubbs We ve found most genes; but what about the rest of the genome? Genome size* 12 Mb 95 Mb 170 Mb 1500 Mb 2700 Mb 3200 Mb #coding genes ~7000 ~20000 ~14000 ~26000
Introduction to genome biology Lisa Stubbs We ve found most genes; but what about the rest of the genome? Genome size* 12 Mb 95 Mb 170 Mb 1500 Mb 2700 Mb 3200 Mb #coding genes ~7000 ~20000 ~14000 ~26000
Analysis of Microarray Data
 Analysis of Microarray Data Lecture 3: Visualization and Functional Analysis George Bell, Ph.D. Senior Bioinformatics Scientist Bioinformatics and Research Computing Whitehead Institute Outline Review
Analysis of Microarray Data Lecture 3: Visualization and Functional Analysis George Bell, Ph.D. Senior Bioinformatics Scientist Bioinformatics and Research Computing Whitehead Institute Outline Review
Chapter 1 Analysis of ChIP-Seq Data with Partek Genomics Suite 6.6
 Chapter 1 Analysis of ChIP-Seq Data with Partek Genomics Suite 6.6 Overview ChIP-Sequencing technology (ChIP-Seq) uses high-throughput DNA sequencing to map protein-dna interactions across the entire genome.
Chapter 1 Analysis of ChIP-Seq Data with Partek Genomics Suite 6.6 Overview ChIP-Sequencing technology (ChIP-Seq) uses high-throughput DNA sequencing to map protein-dna interactions across the entire genome.
Introduction to gene expression microarray data analysis
 Introduction to gene expression microarray data analysis Outline Brief introduction: Technology and data. Statistical challenges in data analysis. Preprocessing data normalization and transformation. Useful
Introduction to gene expression microarray data analysis Outline Brief introduction: Technology and data. Statistical challenges in data analysis. Preprocessing data normalization and transformation. Useful
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
WebMOTIFS: Automated discovery, filtering, and scoring of DNA sequence motifs using multiple programs and Bayesian approaches
 WebMOTIFS: Automated discovery, filtering, and scoring of DNA sequence motifs using multiple programs and Bayesian approaches Katherine A. Romer 1, Guy-Richard Kayombya 1, Ernest Fraenkel 2,3 1 Department
WebMOTIFS: Automated discovery, filtering, and scoring of DNA sequence motifs using multiple programs and Bayesian approaches Katherine A. Romer 1, Guy-Richard Kayombya 1, Ernest Fraenkel 2,3 1 Department
Regulation of eukaryotic transcription:
 Promoter definition by mass genome annotation data: in silico primer extension EMBNET course Bioinformatics of transcriptional regulation Jan 28 2008 Christoph Schmid Regulation of eukaryotic transcription:
Promoter definition by mass genome annotation data: in silico primer extension EMBNET course Bioinformatics of transcriptional regulation Jan 28 2008 Christoph Schmid Regulation of eukaryotic transcription:
Outline. Analysis of Microarray Data. Most important design question. General experimental issues
 Outline Analysis of Microarray Data Lecture 1: Experimental Design and Data Normalization Introduction to microarrays Experimental design Data normalization Other data transformation Exercises George Bell,
Outline Analysis of Microarray Data Lecture 1: Experimental Design and Data Normalization Introduction to microarrays Experimental design Data normalization Other data transformation Exercises George Bell,
Analysis of a Tiling Regulation Study in Partek Genomics Suite 6.6
 Analysis of a Tiling Regulation Study in Partek Genomics Suite 6.6 The example data set used in this tutorial consists of 6 technical replicates from the same human cell line, 3 are SP1 treated, and 3
Analysis of a Tiling Regulation Study in Partek Genomics Suite 6.6 The example data set used in this tutorial consists of 6 technical replicates from the same human cell line, 3 are SP1 treated, and 3
MulCom: a Multiple Comparison statistical test for microarray data in Bioconductor.
 MulCom: a Multiple Comparison statistical test for microarray data in Bioconductor. Claudio Isella, Tommaso Renzulli, Davide Corà and Enzo Medico May 3, 2016 Abstract Many microarray experiments compare
MulCom: a Multiple Comparison statistical test for microarray data in Bioconductor. Claudio Isella, Tommaso Renzulli, Davide Corà and Enzo Medico May 3, 2016 Abstract Many microarray experiments compare
A step-by-step guide to ChIP-seq data analysis
 A step-by-step guide to ChIP-seq data analysis December 03, 2014 Xi Chen, Ph.D. EMBL-European Bioinformatics Institute Wellcome Trust Sanger Institute Target audience Wet-lab biologists with no experience
A step-by-step guide to ChIP-seq data analysis December 03, 2014 Xi Chen, Ph.D. EMBL-European Bioinformatics Institute Wellcome Trust Sanger Institute Target audience Wet-lab biologists with no experience
Lecture #1. Introduction to microarray technology
 Lecture #1 Introduction to microarray technology Outline General purpose Microarray assay concept Basic microarray experimental process cdna/two channel arrays Oligonucleotide arrays Exon arrays Comparing
Lecture #1 Introduction to microarray technology Outline General purpose Microarray assay concept Basic microarray experimental process cdna/two channel arrays Oligonucleotide arrays Exon arrays Comparing
WORKSHOP. Transcriptional circuitry and the regulatory conformation of the genome. Ofir Hakim Faculty of Life Sciences
 WORKSHOP Transcriptional circuitry and the regulatory conformation of the genome Ofir Hakim Faculty of Life Sciences Chromosome conformation capture (3C) Most GR Binding Sites Are Distant From Regulated
WORKSHOP Transcriptional circuitry and the regulatory conformation of the genome Ofir Hakim Faculty of Life Sciences Chromosome conformation capture (3C) Most GR Binding Sites Are Distant From Regulated
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Mentor: Dr. Bino John
 MAT Shiksha Mantri Mentor: Dr. Bino John Model-based Analysis y of Tiling-arrays g y for ChIP-chip X. Shirley Liu et al., PNAS (2006) vol. 103 no. 33 12457 12462 Tiling Arrays Subtype of microarray chips
MAT Shiksha Mantri Mentor: Dr. Bino John Model-based Analysis y of Tiling-arrays g y for ChIP-chip X. Shirley Liu et al., PNAS (2006) vol. 103 no. 33 12457 12462 Tiling Arrays Subtype of microarray chips
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls
 PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls Joel Rozowsky, Ghia Euskirchen 2, Raymond K Auerbach 3, Zhengdong D Zhang, Theodore Gibson, Robert Bjornson 4, Nicholas Carriero
PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls Joel Rozowsky, Ghia Euskirchen 2, Raymond K Auerbach 3, Zhengdong D Zhang, Theodore Gibson, Robert Bjornson 4, Nicholas Carriero
Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs
 Technical Overview Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs Authors Stephen Madden, Crystal Cody, and Jungkap Park Agilent Technologies, Inc. Santa Clara, California,
Technical Overview Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs Authors Stephen Madden, Crystal Cody, and Jungkap Park Agilent Technologies, Inc. Santa Clara, California,
Intracellular receptors specify complex patterns of gene expression that are cell and gene
 SUPPLEMENTAL RESULTS AND DISCUSSION Some HPr-1AR ARE-containing Genes Are Unresponsive to Androgen Intracellular receptors specify complex patterns of gene expression that are cell and gene specific. For
SUPPLEMENTAL RESULTS AND DISCUSSION Some HPr-1AR ARE-containing Genes Are Unresponsive to Androgen Intracellular receptors specify complex patterns of gene expression that are cell and gene specific. For
Whole Transcriptome Analysis of Illumina RNA- Seq Data. Ryan Peters Field Application Specialist
 Whole Transcriptome Analysis of Illumina RNA- Seq Data Ryan Peters Field Application Specialist Partek GS in your NGS Pipeline Your Start-to-Finish Solution for Analysis of Next Generation Sequencing Data
Whole Transcriptome Analysis of Illumina RNA- Seq Data Ryan Peters Field Application Specialist Partek GS in your NGS Pipeline Your Start-to-Finish Solution for Analysis of Next Generation Sequencing Data
RNA-Sequencing analysis
 RNA-Sequencing analysis Markus Kreuz 25. 04. 2012 Institut für Medizinische Informatik, Statistik und Epidemiologie Content: Biological background Overview transcriptomics RNA-Seq RNA-Seq technology Challenges
RNA-Sequencing analysis Markus Kreuz 25. 04. 2012 Institut für Medizinische Informatik, Statistik und Epidemiologie Content: Biological background Overview transcriptomics RNA-Seq RNA-Seq technology Challenges
Analysis of Microarray Data
 Analysis of Microarray Data Lecture 1: Experimental Design and Data Normalization George Bell, Ph.D. Senior Bioinformatics Scientist Bioinformatics and Research Computing Whitehead Institute Outline Introduction
Analysis of Microarray Data Lecture 1: Experimental Design and Data Normalization George Bell, Ph.D. Senior Bioinformatics Scientist Bioinformatics and Research Computing Whitehead Institute Outline Introduction
Gene Expression Profiling and Validation Using Agilent SurePrint G3 Gene Expression Arrays
 Gene Expression Profiling and Validation Using Agilent SurePrint G3 Gene Expression Arrays Application Note Authors Bahram Arezi, Nilanjan Guha and Anne Bergstrom Lucas Agilent Technologies Inc. Santa
Gene Expression Profiling and Validation Using Agilent SurePrint G3 Gene Expression Arrays Application Note Authors Bahram Arezi, Nilanjan Guha and Anne Bergstrom Lucas Agilent Technologies Inc. Santa
Lecture 7: April 7, 2005
 Analysis of Gene Expression Data Spring Semester, 2005 Lecture 7: April 7, 2005 Lecturer: R.Shamir and C.Linhart Scribe: A.Mosseri, E.Hirsh and Z.Bronstein 1 7.1 Promoter Analysis 7.1.1 Introduction to
Analysis of Gene Expression Data Spring Semester, 2005 Lecture 7: April 7, 2005 Lecturer: R.Shamir and C.Linhart Scribe: A.Mosseri, E.Hirsh and Z.Bronstein 1 7.1 Promoter Analysis 7.1.1 Introduction to
Identification of Microprotein-Protein Interactions via APEX Tagging
 Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian
Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian
Quality Control Assessment in Genotyping Console
 Quality Control Assessment in Genotyping Console Introduction Prior to the release of Genotyping Console (GTC) 2.1, quality control (QC) assessment of the SNP Array 6.0 assay was performed using the Dynamic
Quality Control Assessment in Genotyping Console Introduction Prior to the release of Genotyping Console (GTC) 2.1, quality control (QC) assessment of the SNP Array 6.0 assay was performed using the Dynamic
Supporting Information
 Supporting Information Ho et al. 1.173/pnas.81288816 SI Methods Sequences of shrna hairpins: Brg shrna #1: ccggcggctcaagaaggaagttgaactcgagttcaacttccttcttgacgnttttg (TRCN71383; Open Biosystems). Brg shrna
Supporting Information Ho et al. 1.173/pnas.81288816 SI Methods Sequences of shrna hairpins: Brg shrna #1: ccggcggctcaagaaggaagttgaactcgagttcaacttccttcttgacgnttttg (TRCN71383; Open Biosystems). Brg shrna
Mate-pair library data improves genome assembly
 De Novo Sequencing on the Ion Torrent PGM APPLICATION NOTE Mate-pair library data improves genome assembly Highly accurate PGM data allows for de Novo Sequencing and Assembly For a draft assembly, generate
De Novo Sequencing on the Ion Torrent PGM APPLICATION NOTE Mate-pair library data improves genome assembly Highly accurate PGM data allows for de Novo Sequencing and Assembly For a draft assembly, generate
ENCODE DCC Antibody Validation Document
 ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications
![MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications](/thumbs/74/71368870.jpg) MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
DNA Microarray Technology
 CHAPTER 1 DNA Microarray Technology All living organisms are composed of cells. As a functional unit, each cell can make copies of itself, and this process depends on a proper replication of the genetic
CHAPTER 1 DNA Microarray Technology All living organisms are composed of cells. As a functional unit, each cell can make copies of itself, and this process depends on a proper replication of the genetic
Next Gen Sequencing. Expansion of sequencing technology. Contents
 Next Gen Sequencing Contents 1 Expansion of sequencing technology 2 The Next Generation of Sequencing: High-Throughput Technologies 3 High Throughput Sequencing Applied to Genome Sequencing (TEDed CC BY-NC-ND
Next Gen Sequencing Contents 1 Expansion of sequencing technology 2 The Next Generation of Sequencing: High-Throughput Technologies 3 High Throughput Sequencing Applied to Genome Sequencing (TEDed CC BY-NC-ND
Myers Lab ChIP-seq Protocol v Modified January 10, 2014
 Myers Lab ChIP-seq Protocol V011014 1 Contact information: Dr. Florencia Pauli Behn HudsonAlpha Institute for Biotechnology 601 Genome Way Huntsville, AL 35806 Telephone: 256-327-5229 Email: fpauli@hudsonalpha.org
Myers Lab ChIP-seq Protocol V011014 1 Contact information: Dr. Florencia Pauli Behn HudsonAlpha Institute for Biotechnology 601 Genome Way Huntsville, AL 35806 Telephone: 256-327-5229 Email: fpauli@hudsonalpha.org
Perm-seq: Mapping Protein-DNA Interactions in Segmental Duplication and Highly Repetitive Regions of Genomes with Prior- Enhanced Read Mapping
 RESEARCH ARTICLE Perm-seq: Mapping Protein-DNA Interactions in Segmental Duplication and Highly Repetitive Regions of Genomes with Prior- Enhanced Read Mapping Xin Zeng 1,BoLi 2, Rene Welch 1, Constanza
RESEARCH ARTICLE Perm-seq: Mapping Protein-DNA Interactions in Segmental Duplication and Highly Repetitive Regions of Genomes with Prior- Enhanced Read Mapping Xin Zeng 1,BoLi 2, Rene Welch 1, Constanza
How to view Results with Scaffold. Proteomics Shared Resource
 How to view Results with Scaffold Proteomics Shared Resource Starting out Download Scaffold from http://www.proteomes oftware.com/proteom e_software_prod_sca ffold_download.html Follow installation instructions
How to view Results with Scaffold Proteomics Shared Resource Starting out Download Scaffold from http://www.proteomes oftware.com/proteom e_software_prod_sca ffold_download.html Follow installation instructions
Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by hemeresponsive
 Supplemental Data Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by hemeresponsive gene-2 Caiyong Chen 1, Tamika K. Samuel 1, Michael Krause 2, Harry A. Dailey 3, and Iqbal
Supplemental Data Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by hemeresponsive gene-2 Caiyong Chen 1, Tamika K. Samuel 1, Michael Krause 2, Harry A. Dailey 3, and Iqbal
Quality Measures for CytoChip Microarrays
 Quality Measures for CytoChip Microarrays How to evaluate CytoChip Oligo data quality in BlueFuse Multi software. Data quality is one of the most important aspects of any microarray experiment. This technical
Quality Measures for CytoChip Microarrays How to evaluate CytoChip Oligo data quality in BlueFuse Multi software. Data quality is one of the most important aspects of any microarray experiment. This technical
Functional Enrichment Analysis & Candidate Gene Ranking
 Functional Enrichment Analysis & Candidate Gene Ranking Anil Jegga Biomedical Informatics Contact Information: Anil Jegga Biomedical Informatics Room # 232, S Building 10th Floor CCHMC Homepage: http://anil.cchmc.org
Functional Enrichment Analysis & Candidate Gene Ranking Anil Jegga Biomedical Informatics Contact Information: Anil Jegga Biomedical Informatics Room # 232, S Building 10th Floor CCHMC Homepage: http://anil.cchmc.org
Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA.
 Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
DNA Microarray Data Oligonucleotide Arrays
 DNA Microarray Data Oligonucleotide Arrays Sandrine Dudoit, Robert Gentleman, Rafael Irizarry, and Yee Hwa Yang Bioconductor Short Course 2003 Copyright 2002, all rights reserved Biological question Experimental
DNA Microarray Data Oligonucleotide Arrays Sandrine Dudoit, Robert Gentleman, Rafael Irizarry, and Yee Hwa Yang Bioconductor Short Course 2003 Copyright 2002, all rights reserved Biological question Experimental
Probe-Level Data Normalisation: RMA and GC-RMA Sam Robson Images courtesy of Neil Ward, European Application Engineer, Agilent Technologies.
 Probe-Level Data Normalisation: RMA and GC-RMA Sam Robson Images courtesy of Neil Ward, European Application Engineer, Agilent Technologies. References Summaries of Affymetrix Genechip Probe Level Data,
Probe-Level Data Normalisation: RMA and GC-RMA Sam Robson Images courtesy of Neil Ward, European Application Engineer, Agilent Technologies. References Summaries of Affymetrix Genechip Probe Level Data,
Gene Regulation Solutions. Microarrays and Next-Generation Sequencing
 Gene Regulation Solutions Microarrays and Next-Generation Sequencing Gene Regulation Solutions The Microarrays Advantage Microarrays Lead the Industry in: Comprehensive Content SurePrint G3 Human Gene
Gene Regulation Solutions Microarrays and Next-Generation Sequencing Gene Regulation Solutions The Microarrays Advantage Microarrays Lead the Industry in: Comprehensive Content SurePrint G3 Human Gene
Bioinformatics of Transcriptional Regulation
 Bioinformatics of Transcriptional Regulation Carl Herrmann IPMB & DKFZ c.herrmann@dkfz.de Wechselwirkung von Maßnahmen und Auswirkungen Einflussmöglichkeiten in einem Dialog From genes to active compounds
Bioinformatics of Transcriptional Regulation Carl Herrmann IPMB & DKFZ c.herrmann@dkfz.de Wechselwirkung von Maßnahmen und Auswirkungen Einflussmöglichkeiten in einem Dialog From genes to active compounds
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Supplementary Data for DNA sequence+shape kernel enables alignment-free modeling of transcription factor binding.
 Supplementary Data for DNA sequence+shape kernel enables alignment-free modeling of transcription factor binding. Wenxiu Ma 1, Lin Yang 2, Remo Rohs 2, and William Stafford Noble 3 1 Department of Statistics,
Supplementary Data for DNA sequence+shape kernel enables alignment-free modeling of transcription factor binding. Wenxiu Ma 1, Lin Yang 2, Remo Rohs 2, and William Stafford Noble 3 1 Department of Statistics,
PIP-seq. Cells. Permanganate ChIP-Seq
 PIP-seq ells Formaldehyde Permanganate 5 Harvest Lyse Sonicate First dapter Ligation 3 3 5 hip Elute Reverse rosslinks Piperidine cleavage 5 3 3 5 Primer Extension Second dapter Ligation 5 3 3 5 Deep Sequencing
PIP-seq ells Formaldehyde Permanganate 5 Harvest Lyse Sonicate First dapter Ligation 3 3 5 hip Elute Reverse rosslinks Piperidine cleavage 5 3 3 5 Primer Extension Second dapter Ligation 5 3 3 5 Deep Sequencing
less sensitive than RNA-seq but more robust analysis pipelines expensive but quantitiatve standard but typically not high throughput
 Chapter 11: Gene Expression The availability of an annotated genome sequence enables massively parallel analysis of gene expression. The expression of all genes in an organism can be measured in one experiment.
Chapter 11: Gene Expression The availability of an annotated genome sequence enables massively parallel analysis of gene expression. The expression of all genes in an organism can be measured in one experiment.
measuring gene expression December 5, 2017
 measuring gene expression December 5, 2017 transcription a usually short-lived RNA copy of the DNA is created through transcription RNA is exported to the cytoplasm to encode proteins some types of RNA
measuring gene expression December 5, 2017 transcription a usually short-lived RNA copy of the DNA is created through transcription RNA is exported to the cytoplasm to encode proteins some types of RNA
Microarray Technique. Some background. M. Nath
 Microarray Technique Some background M. Nath Outline Introduction Spotting Array Technique GeneChip Technique Data analysis Applications Conclusion Now Blind Guess? Functional Pathway Microarray Technique
Microarray Technique Some background M. Nath Outline Introduction Spotting Array Technique GeneChip Technique Data analysis Applications Conclusion Now Blind Guess? Functional Pathway Microarray Technique
Differential Gene Expression
 Biology 4361 Developmental Biology Differential Gene Expression September 28, 2006 Chromatin Structure ~140 bp ~60 bp Transcriptional Regulation: 1. Packing prevents access CH 3 2. Acetylation ( C O )
Biology 4361 Developmental Biology Differential Gene Expression September 28, 2006 Chromatin Structure ~140 bp ~60 bp Transcriptional Regulation: 1. Packing prevents access CH 3 2. Acetylation ( C O )
Outline. Evolution. Adaptive convergence. Common similarity problems. Chapter 7: Similarity searches on sequence databases
 Chapter 7: Similarity searches on sequence databases All science is either physics or stamp collection. Ernest Rutherford Outline Why is similarity important BLAST Protein and DNA Interpreting BLAST Individualizing
Chapter 7: Similarity searches on sequence databases All science is either physics or stamp collection. Ernest Rutherford Outline Why is similarity important BLAST Protein and DNA Interpreting BLAST Individualizing
Atelier Chip-Seq. Stéphanie Le Gras, IGBMC Strasbourg Violaine Saint-André, Institut Curie Paris Morgane Thomas-Chollier, ENS Paris
 Atelier Chip-Seq Stéphanie Le Gras, IGBMC Strasbourg Violaine Saint-André, Institut Curie Paris Morgane Thomas-Chollier, ENS Paris École de bioinformatique AVIESAN-IFB 2017 Get connected to the server
Atelier Chip-Seq Stéphanie Le Gras, IGBMC Strasbourg Violaine Saint-André, Institut Curie Paris Morgane Thomas-Chollier, ENS Paris École de bioinformatique AVIESAN-IFB 2017 Get connected to the server
Green Center Computational Core ChIP- Seq Pipeline, Just a Click Away
 Green Center Computational Core ChIP- Seq Pipeline, Just a Click Away Venkat Malladi Computational Biologist Computational Core Cecil H. and Ida Green Center for Reproductive Biology Science Introduc
Green Center Computational Core ChIP- Seq Pipeline, Just a Click Away Venkat Malladi Computational Biologist Computational Core Cecil H. and Ida Green Center for Reproductive Biology Science Introduc
Introduction to Bioinformatics and Gene Expression Technologies
 Introduction to Bioinformatics and Gene Expression Technologies Utah State University Fall 2017 Statistical Bioinformatics (Biomedical Big Data) Notes 1 1 Vocabulary Gene: hereditary DNA sequence at a
Introduction to Bioinformatics and Gene Expression Technologies Utah State University Fall 2017 Statistical Bioinformatics (Biomedical Big Data) Notes 1 1 Vocabulary Gene: hereditary DNA sequence at a
Proteomics Technology Note
 Targeted Proteomics for the Identification of Nucleic Acid Binding Proteins in E. coli: Finding more low abundant proteins using the QSTAR LC/MS/MS System, Multi- Dimensional Liquid Chromatography, Pro
Targeted Proteomics for the Identification of Nucleic Acid Binding Proteins in E. coli: Finding more low abundant proteins using the QSTAR LC/MS/MS System, Multi- Dimensional Liquid Chromatography, Pro
Recent technology allow production of microarrays composed of 70-mers (essentially a hybrid of the two techniques)
 Microarrays and Transcript Profiling Gene expression patterns are traditionally studied using Northern blots (DNA-RNA hybridization assays). This approach involves separation of total or polya + RNA on
Microarrays and Transcript Profiling Gene expression patterns are traditionally studied using Northern blots (DNA-RNA hybridization assays). This approach involves separation of total or polya + RNA on
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
CS 5984: Application of Basic Clustering Algorithms to Find Expression Modules in Cancer
 CS 5984: Application of Basic Clustering Algorithms to Find Expression Modules in Cancer T. M. Murali January 31, 2006 Innovative Application of Hierarchical Clustering A module map showing conditional
CS 5984: Application of Basic Clustering Algorithms to Find Expression Modules in Cancer T. M. Murali January 31, 2006 Innovative Application of Hierarchical Clustering A module map showing conditional
Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis
 Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis SWATH Acquisition on the TripleTOF 5600+ System Samuel L. Bader, Robert L. Moritz Institute of Systems Biology,
Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis SWATH Acquisition on the TripleTOF 5600+ System Samuel L. Bader, Robert L. Moritz Institute of Systems Biology,
6. GENE EXPRESSION ANALYSIS MICROARRAYS
 6. GENE EXPRESSION ANALYSIS MICROARRAYS BIOINFORMATICS COURSE MTAT.03.239 16.10.2013 GENE EXPRESSION ANALYSIS MICROARRAYS Slides adapted from Konstantin Tretyakov s 2011/2012 and Priit Adlers 2010/2011
6. GENE EXPRESSION ANALYSIS MICROARRAYS BIOINFORMATICS COURSE MTAT.03.239 16.10.2013 GENE EXPRESSION ANALYSIS MICROARRAYS Slides adapted from Konstantin Tretyakov s 2011/2012 and Priit Adlers 2010/2011
Identifying Candidate Informative Genes for Biomarker Prediction of Liver Cancer
 Identifying Candidate Informative Genes for Biomarker Prediction of Liver Cancer Nagwan M. Abdel Samee 1, Nahed H. Solouma 2, Mahmoud Elhefnawy 3, Abdalla S. Ahmed 4, Yasser M. Kadah 5 1 Computer Engineering
Identifying Candidate Informative Genes for Biomarker Prediction of Liver Cancer Nagwan M. Abdel Samee 1, Nahed H. Solouma 2, Mahmoud Elhefnawy 3, Abdalla S. Ahmed 4, Yasser M. Kadah 5 1 Computer Engineering
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11112 Figure 1 Salient features of the m 6 A methylome. a, m 6 A punctuates RNA molecules mainly around the stop codon and in unusually long internal exons.
SUPPLEMENTARY INFORMATION doi:10.1038/nature11112 Figure 1 Salient features of the m 6 A methylome. a, m 6 A punctuates RNA molecules mainly around the stop codon and in unusually long internal exons.
Mapping strategies for sequence reads
 Mapping strategies for sequence reads Ernest Turro University of Cambridge 21 Oct 2013 Quantification A basic aim in genomics is working out the contents of a biological sample. 1. What distinct elements
Mapping strategies for sequence reads Ernest Turro University of Cambridge 21 Oct 2013 Quantification A basic aim in genomics is working out the contents of a biological sample. 1. What distinct elements
Synthetic Biology. Sustainable Energy. Therapeutics Industrial Enzymes. Agriculture. Accelerating Discoveries, Expanding Possibilities. Design.
 Synthetic Biology Accelerating Discoveries, Expanding Possibilities Sustainable Energy Therapeutics Industrial Enzymes Agriculture Design Build Generate Solutions to Advance Synthetic Biology Research
Synthetic Biology Accelerating Discoveries, Expanding Possibilities Sustainable Energy Therapeutics Industrial Enzymes Agriculture Design Build Generate Solutions to Advance Synthetic Biology Research
How to view Results with. Proteomics Shared Resource
 How to view Results with Scaffold 3.0 Proteomics Shared Resource An overview This document is intended to walk you through Scaffold version 3.0. This is an introductory guide that goes over the basics
How to view Results with Scaffold 3.0 Proteomics Shared Resource An overview This document is intended to walk you through Scaffold version 3.0. This is an introductory guide that goes over the basics
1. Cross-linking and cell harvesting
 ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
ENCODE DCC Antibody Validation Document
 ENCODE DCC Antibody Validation Document Date of Submission 06/15/2012 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: CDP (sc6327) Target: CDP Company/ Source: Santa Cruz Catalog
ENCODE DCC Antibody Validation Document Date of Submission 06/15/2012 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: CDP (sc6327) Target: CDP Company/ Source: Santa Cruz Catalog
The human noncoding genome defined by genetic diversity
 SUPPLEMENTARY INFORMATION Letters https://doi.org/10.1038/s41588-018-0062-7 In the format provided by the authors and unedited. The human noncoding genome defined by genetic diversity Julia di Iulio 1,5,
SUPPLEMENTARY INFORMATION Letters https://doi.org/10.1038/s41588-018-0062-7 In the format provided by the authors and unedited. The human noncoding genome defined by genetic diversity Julia di Iulio 1,5,
Multi-omics in biology: integration of omics techniques
 31/07/17 Летняя школа по биоинформатике 2017 Multi-omics in biology: integration of omics techniques Konstantin Okonechnikov Division of Pediatric Neurooncology German Cancer Research Center (DKFZ) 2 Short
31/07/17 Летняя школа по биоинформатике 2017 Multi-omics in biology: integration of omics techniques Konstantin Okonechnikov Division of Pediatric Neurooncology German Cancer Research Center (DKFZ) 2 Short
Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Supplementary Material
 Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions Joshua N. Burton 1, Andrew Adey 1, Rupali P. Patwardhan 1, Ruolan Qiu 1, Jacob O. Kitzman 1, Jay Shendure 1 1 Department
Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions Joshua N. Burton 1, Andrew Adey 1, Rupali P. Patwardhan 1, Ruolan Qiu 1, Jacob O. Kitzman 1, Jay Shendure 1 1 Department
Preprocessing Affymetrix GeneChip Data. Affymetrix GeneChip Design. Terminology TGTGATGGTGGGGAATGGGTCAGAAGGCCTCCGATGCGCCGATTGAGAAT
 Preprocessing Affymetrix GeneChip Data Credit for some of today s materials: Ben Bolstad, Leslie Cope, Laurent Gautier, Terry Speed and Zhijin Wu Affymetrix GeneChip Design 5 3 Reference sequence TGTGATGGTGGGGAATGGGTCAGAAGGCCTCCGATGCGCCGATTGAGAAT
Preprocessing Affymetrix GeneChip Data Credit for some of today s materials: Ben Bolstad, Leslie Cope, Laurent Gautier, Terry Speed and Zhijin Wu Affymetrix GeneChip Design 5 3 Reference sequence TGTGATGGTGGGGAATGGGTCAGAAGGCCTCCGATGCGCCGATTGAGAAT
Predicting Microarray Signals by Physical Modeling. Josh Deutsch. University of California. Santa Cruz
 Predicting Microarray Signals by Physical Modeling Josh Deutsch University of California Santa Cruz Predicting Microarray Signals by Physical Modeling p.1/39 Collaborators Shoudan Liang NASA Ames Onuttom
Predicting Microarray Signals by Physical Modeling Josh Deutsch University of California Santa Cruz Predicting Microarray Signals by Physical Modeling p.1/39 Collaborators Shoudan Liang NASA Ames Onuttom
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Parameter Estimation for the Exponential-Normal Convolution Model
 Parameter Estimation for the Exponential-Normal Convolution Model Monnie McGee & Zhongxue Chen cgee@smu.edu, zhongxue@smu.edu. Department of Statistical Science Southern Methodist University ENAR Spring
Parameter Estimation for the Exponential-Normal Convolution Model Monnie McGee & Zhongxue Chen cgee@smu.edu, zhongxue@smu.edu. Department of Statistical Science Southern Methodist University ENAR Spring
Year III Pharm.D Dr. V. Chitra
 Year III Pharm.D Dr. V. Chitra 1 Genome entire genetic material of an individual Transcriptome set of transcribed sequences Proteome set of proteins encoded by the genome 2 Only one strand of DNA serves
Year III Pharm.D Dr. V. Chitra 1 Genome entire genetic material of an individual Transcriptome set of transcribed sequences Proteome set of proteins encoded by the genome 2 Only one strand of DNA serves
Introduction to the UCSC genome browser
 Introduction to the UCSC genome browser Dominik Beck NHMRC Peter Doherty and CINSW ECR Fellow, Senior Lecturer Lowy Cancer Research Centre, UNSW and Centre for Health Technology, UTS SYDNEY NSW AUSTRALIA
Introduction to the UCSC genome browser Dominik Beck NHMRC Peter Doherty and CINSW ECR Fellow, Senior Lecturer Lowy Cancer Research Centre, UNSW and Centre for Health Technology, UTS SYDNEY NSW AUSTRALIA
Supplementary Figure 1. Utilization of publicly available antibodies in different applications.
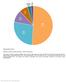 Supplementary Figure 1 Utilization of publicly available antibodies in different applications. The fraction of publicly available antibodies toward human protein targets is shown with data for the following
Supplementary Figure 1 Utilization of publicly available antibodies in different applications. The fraction of publicly available antibodies toward human protein targets is shown with data for the following
Bioinformatics Advice on Experimental Design
 Bioinformatics Advice on Experimental Design Where do I start? Please refer to the following guide to better plan your experiments for good statistical analysis, best suited for your research needs. Statistics
Bioinformatics Advice on Experimental Design Where do I start? Please refer to the following guide to better plan your experiments for good statistical analysis, best suited for your research needs. Statistics
Decoding Chromatin States with Epigenome Data Advanced Topics in Computa8onal Genomics
 Decoding Chromatin States with Epigenome Data 02-715 Advanced Topics in Computa8onal Genomics HMMs for Decoding Chromatin States Epigene8c modifica8ons of the genome have been associated with Establishing
Decoding Chromatin States with Epigenome Data 02-715 Advanced Topics in Computa8onal Genomics HMMs for Decoding Chromatin States Epigene8c modifica8ons of the genome have been associated with Establishing
Sequence assembly. Jose Blanca COMAV institute bioinf.comav.upv.es
 Sequence assembly Jose Blanca COMAV institute bioinf.comav.upv.es Sequencing project Unknown sequence { experimental evidence result read 1 read 4 read 2 read 5 read 3 read 6 read 7 Computational requirements
Sequence assembly Jose Blanca COMAV institute bioinf.comav.upv.es Sequencing project Unknown sequence { experimental evidence result read 1 read 4 read 2 read 5 read 3 read 6 read 7 Computational requirements
Ascl2 Acts as an R-spondin/Wnt-Responsive Switch to Control Stemness in Intestinal Crypts
 Cell Stem Cell Supplemental Information Ascl2 Acts as an R-spondin/Wnt-Responsive Switch to Control Stemness in Intestinal Crypts Jurian Schuijers, Jan Philipp Junker, Michal Mokry, Pantelis Hatzis, Bon-Kyoung
Cell Stem Cell Supplemental Information Ascl2 Acts as an R-spondin/Wnt-Responsive Switch to Control Stemness in Intestinal Crypts Jurian Schuijers, Jan Philipp Junker, Michal Mokry, Pantelis Hatzis, Bon-Kyoung
Bayesian Variable Selection and Data Integration for Biological Regulatory Networks
 Bayesian Variable Selection and Data Integration for Biological Regulatory Networks Shane T. Jensen Department of Statistics The Wharton School, University of Pennsylvania stjensen@wharton.upenn.edu Gary
Bayesian Variable Selection and Data Integration for Biological Regulatory Networks Shane T. Jensen Department of Statistics The Wharton School, University of Pennsylvania stjensen@wharton.upenn.edu Gary
Genome Biology and Biotechnology
 Genome Biology and Biotechnology 10. The proteome Prof. M. Zabeau Department of Plant Systems Biology Flanders Interuniversity Institute for Biotechnology (VIB) University of Gent International course
Genome Biology and Biotechnology 10. The proteome Prof. M. Zabeau Department of Plant Systems Biology Flanders Interuniversity Institute for Biotechnology (VIB) University of Gent International course
Axiom mydesign Custom Array design guide for human genotyping applications
 TECHNICAL NOTE Axiom mydesign Custom Genotyping Arrays Axiom mydesign Custom Array design guide for human genotyping applications Overview In the past, custom genotyping arrays were expensive, required
TECHNICAL NOTE Axiom mydesign Custom Genotyping Arrays Axiom mydesign Custom Array design guide for human genotyping applications Overview In the past, custom genotyping arrays were expensive, required
Microarray Analysis of Gene Expression in Huntington's Disease Peripheral Blood - a Platform Comparison. CodeLink compatible
 Microarray Analysis of Gene Expression in Huntington's Disease Peripheral Blood - a Platform Comparison CodeLink compatible Microarray Analysis of Gene Expression in Huntington's Disease Peripheral Blood
Microarray Analysis of Gene Expression in Huntington's Disease Peripheral Blood - a Platform Comparison CodeLink compatible Microarray Analysis of Gene Expression in Huntington's Disease Peripheral Blood
Discovery of Transcription Factor Binding Sites with Deep Convolutional Neural Networks
 Discovery of Transcription Factor Binding Sites with Deep Convolutional Neural Networks Reesab Pathak Dept. of Computer Science Stanford University rpathak@stanford.edu Abstract Transcription factors are
Discovery of Transcription Factor Binding Sites with Deep Convolutional Neural Networks Reesab Pathak Dept. of Computer Science Stanford University rpathak@stanford.edu Abstract Transcription factors are
Machine Learning Methods for RNA-seq-based Transcriptome Reconstruction
 Machine Learning Methods for RNA-seq-based Transcriptome Reconstruction Gunnar Rätsch Friedrich Miescher Laboratory Max Planck Society, Tübingen, Germany NGS Bioinformatics Meeting, Paris (March 24, 2010)
Machine Learning Methods for RNA-seq-based Transcriptome Reconstruction Gunnar Rätsch Friedrich Miescher Laboratory Max Planck Society, Tübingen, Germany NGS Bioinformatics Meeting, Paris (March 24, 2010)
Nature Genetics: doi: /ng Supplementary Figure 1. Analysis of DHS STARR-seq in the P5424 cell line.
 Supplementary Figure 1 Analysis of DHS STARR-seq in the P5424 cell line. (a) Luciferase enhancer assays of proximal DHSs defined as active or inactive enhancers by STARR-seq in P5424 cells. For each candidate,
Supplementary Figure 1 Analysis of DHS STARR-seq in the P5424 cell line. (a) Luciferase enhancer assays of proximal DHSs defined as active or inactive enhancers by STARR-seq in P5424 cells. For each candidate,
Ensembl Funcgen: A Database and API for Epigenomics and Gene Regulation Data.
 Ensembl Funcgen: A Database and API for Epigenomics and Gene Regulation Data. Nathan Johnson Ensembl Regulation EBI is an Outstation of the European Molecular Biology Laboratory.! Workshop Overview http://www.ebi.ac.uk/~njohnson/courses/23.05.2013-
Ensembl Funcgen: A Database and API for Epigenomics and Gene Regulation Data. Nathan Johnson Ensembl Regulation EBI is an Outstation of the European Molecular Biology Laboratory.! Workshop Overview http://www.ebi.ac.uk/~njohnson/courses/23.05.2013-
user s guide Question 3
 Question 3 During a positional cloning project aimed at finding a human disease gene, linkage data have been obtained suggesting that the gene of interest lies between two sequence-tagged site markers.
Question 3 During a positional cloning project aimed at finding a human disease gene, linkage data have been obtained suggesting that the gene of interest lies between two sequence-tagged site markers.
Gene Expression Data Analysis (I)
 Gene Expression Data Analysis (I) Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Bioinformatics tasks Biological question Experiment design Microarray experiment
Gene Expression Data Analysis (I) Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Bioinformatics tasks Biological question Experiment design Microarray experiment
Differential Gene Expression
 Biology 4361 Developmental Biology Differential Gene Expression June 19, 2008 Differential Gene Expression Overview Chromatin structure Gene anatomy RNA processing and protein production Initiating transcription:
Biology 4361 Developmental Biology Differential Gene Expression June 19, 2008 Differential Gene Expression Overview Chromatin structure Gene anatomy RNA processing and protein production Initiating transcription:
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
On the detection and refinement of transcription factor binding sites using ChIP-Seq data
 Nucleic Acids Research Advance Access published January 6, 2010 Nucleic Acids Research, 2009, 1 14 doi:10.1093/nar/gkp1180 On the detection and refinement of transcription factor binding sites using ChIP-Seq
Nucleic Acids Research Advance Access published January 6, 2010 Nucleic Acids Research, 2009, 1 14 doi:10.1093/nar/gkp1180 On the detection and refinement of transcription factor binding sites using ChIP-Seq
Supplemental Data. LMO4 Controls the Balance between Excitatory. and Inhibitory Spinal V2 Interneurons
 Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Outline. Array platform considerations: Comparison between the technologies available in microarrays
 Microarray overview Outline Array platform considerations: Comparison between the technologies available in microarrays Differences in array fabrication Differences in array organization Applications of
Microarray overview Outline Array platform considerations: Comparison between the technologies available in microarrays Differences in array fabrication Differences in array organization Applications of
ChroMoS Guide (version 1.2)
 ChroMoS Guide (version 1.2) Background Genome-wide association studies (GWAS) reveal increasing number of disease-associated SNPs. Since majority of these SNPs are located in intergenic and intronic regions
ChroMoS Guide (version 1.2) Background Genome-wide association studies (GWAS) reveal increasing number of disease-associated SNPs. Since majority of these SNPs are located in intergenic and intronic regions
EECS730: Introduction to Bioinformatics
 EECS730: Introduction to Bioinformatics Lecture 14: Microarray Some slides were adapted from Dr. Luke Huan (University of Kansas), Dr. Shaojie Zhang (University of Central Florida), and Dr. Dong Xu and
EECS730: Introduction to Bioinformatics Lecture 14: Microarray Some slides were adapted from Dr. Luke Huan (University of Kansas), Dr. Shaojie Zhang (University of Central Florida), and Dr. Dong Xu and
