Neuronal Activity and CaMKII Regulate Kinesin-Mediated Transport of Synaptic AMPARs
|
|
|
- Drusilla Osborne
- 5 years ago
- Views:
Transcription
1 Neuron Supplemental Information Neuronal Activity and CaMKII Regulate Kinesin-Mediated Transport of Synaptic AMPARs Frédéric J. Hoerndli, Rui Wang, Jerry E. Mellem, Angy Kallarackal, Penelope J. Brockie, Colin Thacker, David M. Madsen, and Andres V. Maricq
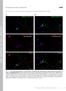
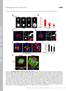
2 Figure S1. GLR-1 transport is modified by presynaptic glutamate release and postsynaptic VGCCs. Supplemental data associated with Figure 1. (A) (Left) Confocal image of GLR-1::GFP expressed in the AVA neurons. Indicated is the region where GLR-1 transport is monitored (labeled proximal processes) as well as the nerve ring that contains AMPARs activated during voltage-clamp experiments. Scale bar represents 10 m. (Right) Confocal images of GLR-1::GFP in AVA cell bodies and nerve ring processes in transgenic WT and unc-43(lf) mutants. Arrowheads indicate sites where GLR-1::GFP accumulates in unc-43(lf) mutants. Scale bar represents 5 m. (B and C) GLR-1::GFP transport in AVA neurons from eat-4(ky5) loss-of-function mutants (B) and unc-36(rnai) mutants (C), compared to wild-type worms (Control). Scale bars represent 5 m. (D) The total number of transport events normalized to WT controls. n > 1
3 8 worms per condition. *** p < 0.001, ANOVA with Tuckey s multiple comparison. Error bars indicate SEM. 2
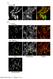
4 Figure S2. CaMKII is not required for UNC-116/KIF5 movement. Supplemental data associated with Figure 2. (A) Kymographs (left) and quantification (right) of UNC-116::GFP transport in WT and unc-43(lf) mutants normalized to WT. n = 6 worms per genotype. Scale bar represents 5 μm. (B) Quantification of UNC-116::GFP FRAP in WT and unc-43(lf) mutants. n = 6 worms per genotype. (C) Kymographs (top) and quantification (bottom) of EBP-2::GFP polymerization events in WT and unc-43(lf) mutants normalized to WT. n = 8 worms per genotype. (D) VAMP::mCherry transport in WT, unc-43(lf) mutants and unc-116(rh24) mutants (top). Quantification of the total number of VAMP::mCherry transport events 3
5 (bottom) normalized to WT. n = 11 for WT and unc-43(lf), n = 4 for unc-116(rh24). * p < Error bars indicate SEM. 4
6 Figure S3. ACh-gated currents are not disrupted in unc-43/camkii mutants. Supplemental data associated with Figure 2. Currents evoked by application of 200 M ACh (A) and the peak amplitude of AChgated current (B) in the AVA neurons of wild-type (Control) and unc-43(lf) mutants. n = 5 worms per genotype. Error bars indicate SEM. 5
7 Figure S4. UNC-43, UNC-116 and GLR-1 co-localize in the AVA cell body and processes. Supplemental data associated with Figure 4. (A and B) Confocal images of UNC-116::mTFP-1, mcherry::unc-43 and GLR-1::GFP in the AVA cell bodies (A) and AVA processes (B). (C) Confocal images of GLR- 1::GFP and UNC-43::mCherry expressed in the AVA processes of unc-116(rh24) lossof-function mutants. Scale bars represent 5 m. 6
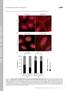
8 Figure S5. KLC-2 co-localizes with GLR-1 at synapses. Supplemental data associated with Figure 4. (A) Confocal images of GLR-1::GFP and KLC-2::TagRFP-T in the AVA processes. (B) Confocal images of wild type and mutant variants of KLC-2 fused to TagRFP-T in the AVA processes. Scale bars represent 5 m. 7
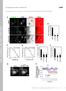
9 Figure S6. GLR-1 and GLR-2 subunit abundance are altered in unc-43/camkii mutants. Supplemental data associated with Figure 4. (A) Confocal images of SEP::GLR-1 fluorescence (left) and quantification of SEP fluorescence (right). (B) Confocal images (left) and quantification (right) of GLR- 1::mCherry and GFP::GLR-2 fluorescence in the AVA neurons of transgenic WT and unc -43(lf) mutants. (C) Glutamate-gated currents in the AVA neurons of glr-2(lf) and unc-43(lf) mutants with and without expression of a wild-type glr-2 transgene in AVA. * p < 0.05, ** p < 0.01, *** p < Scale bars represent 5 m. Error bars indicate SEM. 8
10 9
11 Figure S7. FRAP of surface GLR-1 proceeds at a different rate than FRAP of total GLR- 1. Supplemental data associated with Figures 5 and 6. (A) Confocal images of SEP and mcherry fluorescence before, immediately after, and 5, 10, and 20 min after photobleaching both SEP and mcherry. (B) The ratio of SEP to mcherry fluorescence at synaptic puncta. n = 4 worms. * p < (C) Confocal images (top) and FRAP quantification (bottom) of SEP (left) and mcherry (right) fluorescence in transgenic worms that expressed UNC-116::miniSOG in AVA. Blue bars represent light inactivation (CALI) of UNC-116::miniSOG in the AVA processes. FRAP of mcherry, but not of SEP, in transgenic worms exposed to blue light (CALI) was significantly different from transgenic worms without light exposure. n > 5 per condition. p < 0.05 (ANCOVA analysis). (D) Transport of GLR-1::GFP and mcherry::unc-43 in the AVA processes. White arrowheads indicate mcherry::unc-43 transport events. The merged kymographs show no evidence of co-transport. Scale bars represent 5 m. Error bars indicate SEM. 10
12 Figure S8. NMDA-gated currents are not appreciably disrupted by CALI inactivation of minisog::unc-43 in AVA. Supplemental data associated with Figure 8. Two examples of NMDA-gated currents in response to 0.5 second pressure applications of 1 mm NMDA at 1 minute intervals both before (black) and during (blue) CALI inactivation of minisog::unc
13 Figure S9. UNC-43/CaMKII is required for GLR-1 transport in adult animals. Supplemental data associated with Figure 9. (A and B) Kymographs (A) and quantification (B) of GLR-1::GFP transport in AVA before (Control) and after heat shock in WT and unc-43(lf) mutants, and transgenic mutants with a Phsp::unc-43 transgene. Control worms were kept at 15 before imaging. Heat shocked worms were transferred from 15 to 31 for 1 hour before imaging. Transport events were normalized to wild-type control worms (WT). n > 8 per condition and genotype. * p < Error bars indicate SEM. 12
14 Supplemental Experimental Procedures Plasmids. The following plasmids were used to generate transgenic animals: pjm23, lin- 15(+); pdm1437, Prig-3::HA::glr-1::gfp; pdm1556, Prig-3::HA::glr-1::mCherry; pdm1633, Pflp-18::unc-116::gfp; pdm1494, Prig-3::mCherry; pdm1550, Prig- 3::HA::glr-1::Dendra2; pdm1983, Prig-3::sep::mCherry::glr-1; pdm1973, Pflp- 18::gfp::glr-2; pdm1772, Pflp-18::mCherry::unc-43; pdm1475, Peat- 4::ChR2::mCherry; pct61, Pegl-20::nls::dsRed; pdm2033, Pflp-18::glr-2; pdm2032, Prig-3::stg-2; pdm1698, Prig-3::sol-1; pdm1509, Prig-3::sol-2; pdm2234, Pflp- 18::miniSOG::unc-43; pdm2235, Pflp-18::unc-116::miniSOG; pdm2241, Pflp- 18::TagRFP-T::mCaMKII; pdm2245, Pflp-18::TagRFP-T::unc-43(K42R); pdm2246, Pflp-18::TagRFP-T::unc-43(K42R; T286D); pdm2162, Pflp- 18::mCherry::miniSOG::unc-43; pdm2166, Pflp-18::unc-116:: minisog::mcherry; pdm2200, Pflp-18::unc-43; pdm2303, Peat-4::HisCl1; pdm2299, Pflp-18::HisCl1; pdm2314, Pflp-18::unc-36(cDNA); pdm2208, Pflp-18::klc-2::TagRFP-T; pdm2326, Pflp-18::klc-2(T208A)::TagRFP-T; pdm2328, Pflp-18::klc-2(S240A)::TagRFP-T; pdm2330, Pflp-18::klc-2(S276A)::TagRFP-T; pdm2336, Pflp-18::klc-2(T208A; S240A; S276A)::TagRFP-T; pdm2337, Pflp-18::klc-2(T208D; S240D; S276D)::TagRFP-T; pdm1466, Pmec-3::VAMP::mCherry; pdm1562, Prig-3::VAMP::mCherry, pct91, Pmec-3::ChIEF; pdm1501, Prig-3::ebp-2::gfp; pdm2307, Prig-3::unc-116::mTFP-1; pdm2198, Phsp 16-2::unc-43, pdam36, Prig-3::ChIEF. All fusion proteins were functional as they rescued the mutant phenotype when expressed in the relevant mutant background. 13
15 The flp-18 and rig-3 promoter sequences were based on that published in Feinberg et al., unc-116(rnai) and unc-36(rnai) were expressed in the AVA neurons using the flp-18 promoter sequences and generated using published protocols (Esposito et al., 2007). Primer sequences are available upon request. To identify potential CaMKII phosphorylation sites in KLC-2, amino acid sequences (isoforms A and B) from WormBase ( were analyzed using algorithms such as NetPhosK and GPS 2.1. Sites predicted by both NetPhosK and GPS 2.1 were chosen for mutational analysis. Transgenic arrays. In most cases, transgenic worms were generated by germline transformation of lin-15(n765ts) mutants by microinjecting the lin-15(+) rescuing plasmid (pjm23). In cases where lin-15 mutants were not used, transgenic worms were identified by expression of soluble mcherry or dsred under the regulation of a cell specific promoter encoded by the co-injected plasmid. Integrated and extra-chromosomal arrays were: akis141, Prig-3::HA::glr-1::gfp; akis154, Prig-3::HA::glr-1::Dendra2; akis201, Prig-3::sep::mCherry::glr-1; akis222, Prig-3::HA::glr-1::mCherry + Pflp- 18::gfp::glr-2; akex3707, Pflp-18::TagRFP-T::unc-43(K42R); akex3708, Pflp- 18::TagRFP-T::unc-43(K42R); akex3730, Pflp-18::TagRFP-T::unc-43(K42R; T286D); akex2952, Pflp-18::unc-116(RNAi) + Prig-3::mCherry; akex3698, Pflp-18::TagRFP- T::mCaMKII; akex3673, Pflp-18::glr-2 + Prig-3::sol-1 + Prig-3::sol-2 + Prig-3::stg- 2::gfp + Prig-3::mCherry; akex2703, Pflp-18::mCherry::unc-43 + Peat-4::::mCherry; akex3536, Pflp-18::mCherry::miniSOG::unc-43; akex3559, Pflp-18::unc- 116::mCherry::miniSOG; akex1913, Prig-3::HA::glr-1::mCherry + Pflp-18::unc- 14
16 116::gfp; akex3714, Pflp-18::miniSOG::unc-43; akex3729, Prig-3::HA::glr-1::mCherry + Pflp-18::miniSOG::unc-43; akex4217, Pflp-18::unc-36(cDNA); akex3989, Pflp- 18::HisCl1 + Prig-3::ChIEF + Prig-3::mCherry; akex3687, Pmec3::VAMP::mCherry + Pmec-3::ChIEF; akex4099, Prig-3::ebp-2::gfp; akex4100, Prig-3::ebp-2::gfp; akex4101, Prig-3::ebp-2::gfp, akex4102, Prig-3::ebp-2::gfp; akex1433, Prig- 3::VAMP::mCherry; akex3723, Pflp-18::klc-2::TagRFP-T; akex4235, Pflp-18::klc- 2(S276A)::TagRFP-T; akex4234, Pflp-18::klc-2(S240A)::TagRFP-T; akex3773, Phsp16-2::unc-43; akex3712, Pflp-18::miniSOG::unc-43; akex4047, Pflp-18::unc- 116::miniSOG; akex4033, Pflp-18::glr-2 + Prig-3::mCherry; akex4239, Peat- 4::HisCl1; akex4222, Pflp-18::unc-36(RNAi); akex 4223, Pflp-18::klc- 2(T208A)::TagRFP-T; akex 4224, Pflp-18::klc-2(T208A)::TagRFP-T; akex 4225, Pflp- 18::klc-2(T208A)::TagRFP-T; akex4226; Pflp-18::klc-2(T208D; S240D; S276S)::TagRFP-T; akex4228, Pflp-18::klc-2(T208D; S240D; S276S)::TagRFP-T; akex4231, Prig-3::unc-116::mTFP-1 + Pflp-18::mCherry::unc-43; akex4233, Pflp- 18::klc-2(T208A; S240A; S276A)::TagRFP-T; akex4236, Pflp-18::mCherry::unc-43; akex4240, Pflp-18::klc-2(T208D; S240D; S276S)::TagRFP-T; akex4241, Pflp-18::klc- 2(T208A; S240A; S276A)::TagRFP-T; akex4242, Pflp-18::klc-2(T208A; S240A; S276A)::TagRFP-T. Imaging. For UNC-116::GFP FRAP, worms were mounted as described previously for in vivo microscopy (Hoerndli et al., 2013). The bleached region included the region of interest (ROI) as well as 60 m to the left and right of the ROI. Confocal stacks of the ROI were taken 1, 2, 4, 8 and 16 minutes after photobleaching. For dual streaming 15
17 imaging of GLR-1::GFP and mcherry::unc-43, imaging was performed as described in Hoerndli et al., Heat shock. Transgenic wild-type and unc-43(lf) mutants that expressed GLR-1::GFP in AVA, and unc-43(lf) mutants that also expressed Phsp16-2::unc-43 were kept at 15 for at least one generation prior to the experiment. Control worms (no heat shock) were kept at 15 before imaging. Heat shocked worms were transferred from 15 to 31 for one hour and then kept at room temperature for one hour after heat shock and before imaging. Image analysis. For quantification of SEP puncta (Figure S6A and S6B), a linescan measurement was taken on maximal projection images and analyzed using a custom written MatLab algorithm as described in Hoerndli et al., From this analysis we extracted the total amount of SEP per m. For FRAP experiments shown in Figure 5A and 5B, the distal region of the AVA processes was chosen to minimize the effect of diffusion from the cell body. Single region of interests of 5-6 GLR-1::GFP puncta per animal, were analyzed in ImageJ, corrected for background and used to generate % FRAP shown in Figure 5B.For FRAP of GLR-2::GFP (Figure 5C 5D), the proximal region of the AVA processes was chosen because the fluorescent signal in the distal region of unc-43(lf) mutants was too low to quantify. Because the signal was more diffuse, we traced the AVA region for FRAP using the segmented line tool in ImageJ with a width of 10 pixels, extracted mean fluorescence values for this region and subtracted for background. For GLR-1::Dendra2 photoconversion experiments (Figure 5E and 5F), the 16
18 fluorescent signal was quantified by drawing ROIs on the maximal projection images of confocal stacks, corrected for background and expressed as a percentage of the signal immediately after photoconversion. This same procedure was used for experiments that combined CALI and photoconversion except that the signal was expressed as a percentage of the signal remaining after CALI (Figure 7C E). For SEP::mCherry::GLR-1 FRAP, maximal projection images were quantified by drawing ROIs around mcherry puncta, subtracting background fluorescence and calculating the percentage of the signal before photobleaching. The same regions were used to quantify SEP FRAP. 17
(A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1
 SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
SUPPLEMENTAL MATERIALS Supplemental Figure 1 Figure S1. Expression of the non-alpha nachr subunit ACR-2. (A) Schematic depicting morphology of cholinergic DA and DB motor neurons in an L1 animal. Wide-field
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
unc-33/crmp and ankyrin organize microtubules and localize kinesin to polarize
 Supplementary Information unc-33/crmp and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting Tapan A. Maniar 1, Miriam Kaplan 1, George J. Wang 2, Kang Shen 2, Li Wei
Supplementary Information unc-33/crmp and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting Tapan A. Maniar 1, Miriam Kaplan 1, George J. Wang 2, Kang Shen 2, Li Wei
Figure S1. USP-46 is expressed in several tissues including the nervous system
 Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
Supplemental Figure legends Figure S1. USP-46 is expressed in several tissues including the nervous system Transgenic animals expressing a transcriptional reporter (P::GFP) were imaged using epifluorescence
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
SUPPLEMENTARY INFORMATION
 Figure S1. lev-9 mutants are resistant to levamisole. The levamisole dose-response curves indicate that lev-9 mutants are partially resistant to levamisole similar to lev-10(kr26) mutants. unc-29(x29)
Figure S1. lev-9 mutants are resistant to levamisole. The levamisole dose-response curves indicate that lev-9 mutants are partially resistant to levamisole similar to lev-10(kr26) mutants. unc-29(x29)
mod-1::mcherry unc-47::gfp RME ser-4::gfp vm2 vm2 vm2 vm2 VNC
 A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
Supplemental Data. Wang et al. (2017). Plant Cell /tpc NIP1;2 NIP7;1
 Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat
 Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity
 Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity Supplemental figure 1. The quantification method to determine the
Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity Supplemental figure 1. The quantification method to determine the
Supplementary Figure 1. Pathologically phosphorylated Tau is localized to the presynaptic terminals of Alzheimer s disease (AD) patient brains.
 Supplementary Figure 1. Pathologically phosphorylated Tau is localized to the presynaptic terminals of Alzheimer s disease (AD) patient brains. Ultrathin (70 nm) sections of healthy control or AD patient
Supplementary Figure 1. Pathologically phosphorylated Tau is localized to the presynaptic terminals of Alzheimer s disease (AD) patient brains. Ultrathin (70 nm) sections of healthy control or AD patient
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
b number of motif occurrences
 supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Craft et al., http://www.jcb.org/cgi/content/full/jcb.201409036/dc1 Figure S1. GFP -tubulin interacts with endogenous -tubulin. Ciliary
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Craft et al., http://www.jcb.org/cgi/content/full/jcb.201409036/dc1 Figure S1. GFP -tubulin interacts with endogenous -tubulin. Ciliary
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Zebrafish activity. Zebrafish activity 4000 * WT SCN9A Muatant (I228M) Temp. Temperature. Activity. WT SCN9A Muatant (I228M) Temp.
 Temperature Temperature Zebrafish activity Zebrafish activity 4 4 4 * 3 WT SCN9 Muatant (I228M) Temp 3 3 2 2 2 1 1 1 1 2 3 SCN9 WT SCN9 I228M Figure 1. Zebrafish activity in response to temperature change.
Temperature Temperature Zebrafish activity Zebrafish activity 4 4 4 * 3 WT SCN9 Muatant (I228M) Temp 3 3 2 2 2 1 1 1 1 2 3 SCN9 WT SCN9 I228M Figure 1. Zebrafish activity in response to temperature change.
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Figure S1a. The modular dcas9 fusion system works efficiently to suppress and activate endogenous gene expression in C. elegans
 Figure S1a. The modular dcas9 fusion system works efficiently to suppress and activate endogenous gene expression in C. elegans A, a normal wild-type hermaphrodite N2 is shown. B, a dpy-5-supressed F1
Figure S1a. The modular dcas9 fusion system works efficiently to suppress and activate endogenous gene expression in C. elegans A, a normal wild-type hermaphrodite N2 is shown. B, a dpy-5-supressed F1
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Two classes of silencing RNAs move between Caenorhabditis elegans tissues.
 Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Supplementary Figure 1. Serial deletion mutants of BLITz.
 Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kanaani et al., http://www.jcb.org/cgi/content/full/jcb.200912101/dc1 Figure S1. The K2 rabbit polyclonal antibody is specific for GAD67,
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kanaani et al., http://www.jcb.org/cgi/content/full/jcb.200912101/dc1 Figure S1. The K2 rabbit polyclonal antibody is specific for GAD67,
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking
 Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7866 1.8 Fraction Bordering Fraction Aggregating.6.4.2 1.8.6.4.2 Promoter Head neurons expressing npr-1 cdna Genotype WT - - URX AQR ALN AVM gcy-35 sax-7 gcy-32+ gpa-3 RMG ILs URX AQR
doi: 1.138/nature7866 1.8 Fraction Bordering Fraction Aggregating.6.4.2 1.8.6.4.2 Promoter Head neurons expressing npr-1 cdna Genotype WT - - URX AQR ALN AVM gcy-35 sax-7 gcy-32+ gpa-3 RMG ILs URX AQR
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
Supplemental Data. Furlan et al. Plant Cell (2017) /tpc
 Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure.
Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Data. Furlan et al. Plant Cell (0) 0.0/tpc..00. Supplemental Figure.
Nature Structural & Molecular Biology: doi: /nsmb.3308
 Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Methanol fixation allows better visualization of Kal7. To compare methods for
 Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Nature Methods: doi: /nmeth Supplementary Figure 1. Neither driver nor effector alone displays expression of GFP
 Supplementary Figure 1 Neither driver nor effector alone displays expression of GFP Comparison of GFP fluorescence in a driver- only strain (a) or an effector-only strain (b) to their driver + effector
Supplementary Figure 1 Neither driver nor effector alone displays expression of GFP Comparison of GFP fluorescence in a driver- only strain (a) or an effector-only strain (b) to their driver + effector
Fig. S1. Phosphorylation of eif2 by PEK-1 is increased in ire-1 mutants. Fig. S2. ire-1
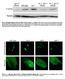 Fig. S1. Phosphorylation of eif2a by PEK-1 is increased in ire-1 mutants. Representative western blot of phosphorylated eif2a and tubulin of day-0 wild-type animals and ire-1 mutants treated with control,
Fig. S1. Phosphorylation of eif2a by PEK-1 is increased in ire-1 mutants. Representative western blot of phosphorylated eif2a and tubulin of day-0 wild-type animals and ire-1 mutants treated with control,
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Paul et al.,
 Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental Information. Glutamylation Regulates Transport, Specializes. Function, and Sculpts the Structure of Cilia
 Current Biology, Volume 27 Supplemental Information Glutamylation Regulates Transport, Specializes Function, and Sculpts the Structure of Cilia Robert O'Hagan, Malan Silva, Ken C.Q. Nguyen, Winnie Zhang,
Current Biology, Volume 27 Supplemental Information Glutamylation Regulates Transport, Specializes Function, and Sculpts the Structure of Cilia Robert O'Hagan, Malan Silva, Ken C.Q. Nguyen, Winnie Zhang,
An antibiotic selection marker for nematode transgenesis
 nature methods An antibiotic selection marker for nematode transgenesis Rosina Giordano-Santini, Stuart Milstein, Nenad Svrzikapa, Domena Tu, Robert Johnsen, David Baillie, Marc Vidal & Denis Dupuy Supplementary
nature methods An antibiotic selection marker for nematode transgenesis Rosina Giordano-Santini, Stuart Milstein, Nenad Svrzikapa, Domena Tu, Robert Johnsen, David Baillie, Marc Vidal & Denis Dupuy Supplementary
Supplementary Figure 1. The LIR motif in Kenny is necessary for its targeting to the lysosomes. (a-b) Confocal sections from fat body cells from
 Supplementary Figure 1. The LIR motif in Kenny is necessary for its targeting to the lysosomes. (a-b) Confocal sections from fat body cells from larvae expressing GFP-Kenny WT (a-a ) or GFP-Kenny F7A/L10A
Supplementary Figure 1. The LIR motif in Kenny is necessary for its targeting to the lysosomes. (a-b) Confocal sections from fat body cells from larvae expressing GFP-Kenny WT (a-a ) or GFP-Kenny F7A/L10A
DCLK-immunopositive. Bars, 100 µm for B, 50 µm for C.
 Supplementary Figure S1. Characterization of rabbit polyclonal anti-dclk antibody. (A) Immunoblotting of COS7 cells transfected with DCLK1-GFP and DCLK2-GFP expression plasmids probed with anti-dclk antibody
Supplementary Figure S1. Characterization of rabbit polyclonal anti-dclk antibody. (A) Immunoblotting of COS7 cells transfected with DCLK1-GFP and DCLK2-GFP expression plasmids probed with anti-dclk antibody
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Cell autonomous vs. cell non-autonomous gene function
 Cell autonomous vs. cell non-autonomous gene function In multicellular organisms, it is important to know in what cell(s) the activity of a gene is required. - while RNA expression can be highly informative,
Cell autonomous vs. cell non-autonomous gene function In multicellular organisms, it is important to know in what cell(s) the activity of a gene is required. - while RNA expression can be highly informative,
Supplemental Materials
 Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
Supplemental Materials Flores-Pérez et al., Supplemental Materials, page 1 of 5 Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies. (A) Controls
λ N -GFP: an RNA reporter system for live-cell imaging
 λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
Wt (Col-0) pom2-1 Wt (Ler) pom2-3 pom2-5. pom2-4 (0%) pom2-1 (0%)
 Figure 1 Wt (Col-0) Wt (Ler) pom2-3 pom2-5 pom2-2 pom2-4 B Wt (0%) (0%) Wt (4.5%) (4.5%) Wt (0%) (0%) Wt (4.5%) (4.5%) (4.5%) Figure 1. Phenotypes of pom2 and wild type (Wt) seedlings after germination
Figure 1 Wt (Col-0) Wt (Ler) pom2-3 pom2-5 pom2-2 pom2-4 B Wt (0%) (0%) Wt (4.5%) (4.5%) Wt (0%) (0%) Wt (4.5%) (4.5%) (4.5%) Figure 1. Phenotypes of pom2 and wild type (Wt) seedlings after germination
Supplemental Data. Shimoda et al. (2011). Plant Cell /tpc B C D E
 (kda) 80 58 46 30 25 B C D E (0/15) (29/29) (0/29) (0/32) Supplemental Figure 1. Immunodetection of Myc-tagged CCaMK Protein Variants on ccamk-3 Roots. () Immunodetection of myc-tag fusion CCaMK WT, 1-340
(kda) 80 58 46 30 25 B C D E (0/15) (29/29) (0/29) (0/32) Supplemental Figure 1. Immunodetection of Myc-tagged CCaMK Protein Variants on ccamk-3 Roots. () Immunodetection of myc-tag fusion CCaMK WT, 1-340
Supplementary Materials for
 www.sciencemag.org/cgi/content/full/science.1245533/dc1 Supplementary Materials for A Mechanism for Reorientation of Cortical Microtubule Arrays Driven by Microtubule Severing Jelmer J. Lindeboom, Masayoshi
www.sciencemag.org/cgi/content/full/science.1245533/dc1 Supplementary Materials for A Mechanism for Reorientation of Cortical Microtubule Arrays Driven by Microtubule Severing Jelmer J. Lindeboom, Masayoshi
Supporting Information
 Supporting Information Lee et al. 10.1073/pnas.1000866107 SI Experimental Procedures Plasmid and Strain Construction. (p)odr-3::gfp::egl-4 was constructed by excising the coding region of GFP and the first
Supporting Information Lee et al. 10.1073/pnas.1000866107 SI Experimental Procedures Plasmid and Strain Construction. (p)odr-3::gfp::egl-4 was constructed by excising the coding region of GFP and the first
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 )
 Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts
 Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Supplementary Figure 1 Activities of ABEs using extended sgrnas in HEK293T cells.
 Supplementary Figure 1 Activities of ABEs using extended sgrnas in HEK293T cells. Base editing efficiencies of ABEs with extended sgrnas at Site 18 (a), Site 19 (b), the HBB-E2 site (c), and the HBB-E3
Supplementary Figure 1 Activities of ABEs using extended sgrnas in HEK293T cells. Base editing efficiencies of ABEs with extended sgrnas at Site 18 (a), Site 19 (b), the HBB-E2 site (c), and the HBB-E3
Supplementary information:
 Supplementary information: Distinct amino acids in the C-linker domain of the plant K + subcellular localization and activity at the plasma membrane channel KAT2 determine its Nieves-Cordones et al. Part
Supplementary information: Distinct amino acids in the C-linker domain of the plant K + subcellular localization and activity at the plasma membrane channel KAT2 determine its Nieves-Cordones et al. Part
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
over time using live cell microscopy. The time post infection is indicated in the lower left corner.
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Supplementary Movie 1 Description: Fusion of NBs. BSR cells were infected
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Supplementary Movie 1 Description: Fusion of NBs. BSR cells were infected
SUPPLEMENTARY INFORMATION
 a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1.
 Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Liu et al., http :// /cgi /content /full /jcb /DC1
 Supplemental material JCB Liu et al., http ://www.jcb.org /cgi /content /full /jcb.201506081 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. Identification of sorf-1 and sorf-2. (A) RNAi of ZK563.5 increases
Supplemental material JCB Liu et al., http ://www.jcb.org /cgi /content /full /jcb.201506081 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. Identification of sorf-1 and sorf-2. (A) RNAi of ZK563.5 increases
embryos. Asterisk represents loss of or reduced expression. Brackets represent
 Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DEPRESSION is linked to a reduction in serotonin
 Copyright Ó 2008 by the Genetics Society of America DOI: 10.1534/genetics.107.079780 Regulation of Serotonin Biosynthesis by the G Proteins Ga o and Ga q Controls Serotonin Signaling in Caenorhabditis
Copyright Ó 2008 by the Genetics Society of America DOI: 10.1534/genetics.107.079780 Regulation of Serotonin Biosynthesis by the G Proteins Ga o and Ga q Controls Serotonin Signaling in Caenorhabditis
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Mos1 insertion. MosTIC protocol-11/2006. repair template - Mos1 transposase expression - Mos1 excision - DSB formation. homolog arm.
 MosTIC (Mos1 excision induced Transgene Instructed gene Conversion) Valérie Robert (vrobert@biologie.ens.fr) and Jean-Louis Bessereau (jlbesse@biologie.ens.fr) (November 2006) Introduction: MosTIC (Robert
MosTIC (Mos1 excision induced Transgene Instructed gene Conversion) Valérie Robert (vrobert@biologie.ens.fr) and Jean-Louis Bessereau (jlbesse@biologie.ens.fr) (November 2006) Introduction: MosTIC (Robert
Supplemental Information. Kinesin 1 Drives Autolysosome Tubulation
 Developmental Cell, Volume 37 Supplemental Information Kinesin 1 Drives Autolysosome Tubulation Wanqing Du, Qian Peter Su, Yang Chen, Yueyao Zhu, Dong Jiang, Yueguang Rong, Senyan Zhang, Yixiao Zhang,
Developmental Cell, Volume 37 Supplemental Information Kinesin 1 Drives Autolysosome Tubulation Wanqing Du, Qian Peter Su, Yang Chen, Yueyao Zhu, Dong Jiang, Yueguang Rong, Senyan Zhang, Yixiao Zhang,
To investigate the heredity of the WFP gene, we selected plants that were homozygous
 Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte
 Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte Carlo algorithm for one million generations to obtain
Fig. S1. Molecular phylogenetic analysis of AtHD-ZIP IV family. A phylogenetic tree was constructed using Bayesian analysis with Markov Chain Monte Carlo algorithm for one million generations to obtain
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Egan et al., http://www.jcb.org/cgi/content/full/jcb.201112101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Localization of dynein to microtubule plus ends requires
Supplemental material Egan et al., http://www.jcb.org/cgi/content/full/jcb.201112101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Localization of dynein to microtubule plus ends requires
Figure S1. MUT-16 localization in L4 hermaphrodite, adult hermaphrodite, and adult male germlines. MUT-16 DAPI. Phillips et al. S-1. male.
 Supplementary Material for Phillips et al. Figure S1. MUT-16 localization in L4 hermaphrodite, adult hermaphrodite, and adult male germlines. Figure S2. Mutator foci and P granules localize independently
Supplementary Material for Phillips et al. Figure S1. MUT-16 localization in L4 hermaphrodite, adult hermaphrodite, and adult male germlines. Figure S2. Mutator foci and P granules localize independently
Figures and figure supplements
 RESEARCH ARTICLE Figures and figure supplements Dendritic mitochondria reach stable positions during circuit development Michelle C Faits et al Faits et al. elife 2015;5:e11583. DOI: 10.7554/eLife.11583
RESEARCH ARTICLE Figures and figure supplements Dendritic mitochondria reach stable positions during circuit development Michelle C Faits et al Faits et al. elife 2015;5:e11583. DOI: 10.7554/eLife.11583
Supplementary Figures
 Supplementary Figures Supplementary Figure S1. Inhibition kinetics of p53/hdm2 binding induced by Nutlin 3 on live cells. Reproducibility of the p53/hdm2 binding disruption kinetics induced by Nutlin 3
Supplementary Figures Supplementary Figure S1. Inhibition kinetics of p53/hdm2 binding induced by Nutlin 3 on live cells. Reproducibility of the p53/hdm2 binding disruption kinetics induced by Nutlin 3
MT minus end catastrophe
 DOI: 1.138/ncb3241 MT minus end catastrophe frequency [s -1 ].2.15.1.5 control mgfp- TPX2 * * mgfp- TPX2 mini Supplementary Figure 1. TPX2 reduces catastrophes at the microtubule minus ends. Modified box-and-whiskers
DOI: 1.138/ncb3241 MT minus end catastrophe frequency [s -1 ].2.15.1.5 control mgfp- TPX2 * * mgfp- TPX2 mini Supplementary Figure 1. TPX2 reduces catastrophes at the microtubule minus ends. Modified box-and-whiskers
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Feng et al., http://www.jcb.org/cgi/content/full/jcb.201408079/dc1 Figure S1. A modest elevation of disulfide-bonded K14 in primary mouse
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Feng et al., http://www.jcb.org/cgi/content/full/jcb.201408079/dc1 Figure S1. A modest elevation of disulfide-bonded K14 in primary mouse
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
In vitro EGFP-CALI comprehensive analysis. Liang CAI. David Humphrey. Jessica Wilson. Ken Jacobson. Dept. of Cell and Developmental Biology
 In vitro EGFP-CALI comprehensive analysis Liang CAI David Humphrey Jessica Wilson Ken Jacobson Dept. of Cell and Developmental Biology 1/15 Liang s Rotation in Ken s Lab, Sep-Nov, 2003 1. CALI background
In vitro EGFP-CALI comprehensive analysis Liang CAI David Humphrey Jessica Wilson Ken Jacobson Dept. of Cell and Developmental Biology 1/15 Liang s Rotation in Ken s Lab, Sep-Nov, 2003 1. CALI background
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Supporting Information
 Supporting Information Richard et al. 10.1073/pnas.1216154110 SI Materials and Methods General Methods and Strains. The WT strain N2 and the following mutant alleles were used:, acr-16(ok789), unc-49 (e407),
Supporting Information Richard et al. 10.1073/pnas.1216154110 SI Materials and Methods General Methods and Strains. The WT strain N2 and the following mutant alleles were used:, acr-16(ok789), unc-49 (e407),
Hoopfer et al., Supplemental Data Supplemental Figure S1. Quantification of ORN axon degeneration
 Hoopfer et al., Supplemental Data Supplemental Figure S1. Quantification of ORN axon degeneration (A) Mean GFP intensity of commissural ORN axons 1 day after antennae removal. 1 day after cutting, wt ORNs
Hoopfer et al., Supplemental Data Supplemental Figure S1. Quantification of ORN axon degeneration (A) Mean GFP intensity of commissural ORN axons 1 day after antennae removal. 1 day after cutting, wt ORNs
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Yamaguchi et al., http://www.jcb.org/cgi/content/full/jcb.201009126/dc1 S1 Figure S2. The expression levels of GFP-Akt1-PH and localization
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Yamaguchi et al., http://www.jcb.org/cgi/content/full/jcb.201009126/dc1 S1 Figure S2. The expression levels of GFP-Akt1-PH and localization
Supplementary Figures 1-6
 1 Supplementary Figures 1-6 2 3 4 5 6 7 Supplementary Fig. 1: GFP-KlpA forms a homodimer. a, Hydrodynamic analysis of the purified full-length GFP-KlpA protein. Fractions from size exclusion chromatography
1 Supplementary Figures 1-6 2 3 4 5 6 7 Supplementary Fig. 1: GFP-KlpA forms a homodimer. a, Hydrodynamic analysis of the purified full-length GFP-KlpA protein. Fractions from size exclusion chromatography
Dendritic targeting of mrna
 Dendritic targeting of mrna FMRP-dependent mglur-induced dendritic targeting of MAP1b and CaMKII mrna IMP-dependent NMDAR-induced dendritic targeting of β-action mrna Discussion (12/15) RNA and protein
Dendritic targeting of mrna FMRP-dependent mglur-induced dendritic targeting of MAP1b and CaMKII mrna IMP-dependent NMDAR-induced dendritic targeting of β-action mrna Discussion (12/15) RNA and protein
Sperm cells are passive cargo of the pollen tube in plant fertilization
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
Protein localization in electron micrographs using fluorescence nanoscopy
 Nature Methods Protein localization in electron micrographs using fluorescence nanoscopy Shigeki Watanabe, Annedore Punge, Gunther Hollopeter, Katrin I Willig, Robert John Hobson, M Wayne Davis, Stefan
Nature Methods Protein localization in electron micrographs using fluorescence nanoscopy Shigeki Watanabe, Annedore Punge, Gunther Hollopeter, Katrin I Willig, Robert John Hobson, M Wayne Davis, Stefan
Flybow genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster
 Nature Methods Flybow genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster Dafni Hadjieconomou, Shay Rotkopf, Cyrille Alexandre, Donald M Bell, Barry J Dickson & Iris
Nature Methods Flybow genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster Dafni Hadjieconomou, Shay Rotkopf, Cyrille Alexandre, Donald M Bell, Barry J Dickson & Iris
SUPPLEMENTARY INFORMATION
 SULEMENTARY INFORMATION doi:10.1038/nature12117 Supplementary Discussion Although loss of leads to initiation of postembryonic development even under starvation conditions, ectopic overexpression of under
SULEMENTARY INFORMATION doi:10.1038/nature12117 Supplementary Discussion Although loss of leads to initiation of postembryonic development even under starvation conditions, ectopic overexpression of under
Figure S1 early log mid log late log stat.
 Figure S1 early log mid log late log stat. C D E F Fig. S1. The chromosome-expressed EI localizes to the cell poles independent of the growth phase and growth media. MG1655 (ptsi-mcherry) cells, which
Figure S1 early log mid log late log stat. C D E F Fig. S1. The chromosome-expressed EI localizes to the cell poles independent of the growth phase and growth media. MG1655 (ptsi-mcherry) cells, which
Supplementary Information. A superfolding Spinach2 reveals the dynamic nature of. trinucleotide repeat RNA
 Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
Supplementary Information A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA Rita L. Strack 1, Matthew D. Disney 2 & Samie R. Jaffrey 1 1 Department of Pharmacology, Weill Medical
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
Expanded View Figures
 The EMO Journal mediated aggrephagy reconstitution Gabriele Zaffagnini et al Expanded View Figures Figure EV1. and ubiquitinpositive proteins spontaneously cluster in solution. Representative micrographs
The EMO Journal mediated aggrephagy reconstitution Gabriele Zaffagnini et al Expanded View Figures Figure EV1. and ubiquitinpositive proteins spontaneously cluster in solution. Representative micrographs
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplemental Data. Borg et al. Plant Cell (2014) /tpc
 Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/323/5911/256/dc1 Supporting Online Material for HDAC4 Regulates Neuronal Survival in Normal and Diseased Retinas Bo Chen and Constance L. Cepko E-mail: bochen@genetics.med.harvard.edu,
www.sciencemag.org/cgi/content/full/323/5911/256/dc1 Supporting Online Material for HDAC4 Regulates Neuronal Survival in Normal and Diseased Retinas Bo Chen and Constance L. Cepko E-mail: bochen@genetics.med.harvard.edu,
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin
 Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
15 June 2011 Supplementary material Bagriantsev et al.
 Supplementary Figure S1 Characterization of K 2P 2.1 (TREK-1) GOF mutants A, Distribution of the positions of mutated nucleotides, represented by a red x, from a pool of 18 unselected K 2P 2.1 (KCNK2)
Supplementary Figure S1 Characterization of K 2P 2.1 (TREK-1) GOF mutants A, Distribution of the positions of mutated nucleotides, represented by a red x, from a pool of 18 unselected K 2P 2.1 (KCNK2)
Supplemental Data. Zhou et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. Confirmation of mutant mapping results. (A) Complementation assay with stably transformed genomic fragments (ComN-N) (2 kb upstream of TSS and 1.5 kb downstream of TES) and CaMV
Supplemental Figure 1. Confirmation of mutant mapping results. (A) Complementation assay with stably transformed genomic fragments (ComN-N) (2 kb upstream of TSS and 1.5 kb downstream of TES) and CaMV
-7 ::(teto) ::(teto) 120. GFP membranes merge. overexposed. Supplemental Figure 1 (Marquis et al.)
 P spoiie -gfp -7 ::(teto) 120-91 ::(teto) 120 GFP membranes merge overexposed Supplemental Figure 1 (Marquis et al.) Figure S1 TetR-GFP is stripped off the teto array in sporulating cells that contain
P spoiie -gfp -7 ::(teto) 120-91 ::(teto) 120 GFP membranes merge overexposed Supplemental Figure 1 (Marquis et al.) Figure S1 TetR-GFP is stripped off the teto array in sporulating cells that contain
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/323/5920/1500/dc1 Supporting Online Material for RSY-1 Is a Local Inhibitor of Presynaptic Assembly in C. elegans Maulik R. Patel and Kang Shen To whom correspondence
www.sciencemag.org/cgi/content/full/323/5920/1500/dc1 Supporting Online Material for RSY-1 Is a Local Inhibitor of Presynaptic Assembly in C. elegans Maulik R. Patel and Kang Shen To whom correspondence
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
