Supplementary Information
|
|
|
- Jesse Kelley
- 5 years ago
- Views:
Transcription
1 Supplementary Information Genome-wide profiling of DNA methylation provides insights into epigenetic regulation of fungal development in a plant pathogenic fungus, Magnaporthe oryzae Junhyun Jeon, Jaeyoung Choi, Gir-Won Lee, Sook-Young Park, Aram Huh, Ralph A. Dean, Yong-Hwan Lee
2 Table S1. Data statistics after reads alignment of BS-seq Sample Raw reads (M) Raw data (Gb) Mapped reads (M) Mapped data (Gb) Average map rate (%) Whole genome average coverage depth ( ) Mycelia Conidia Appressoria ΔModim-2 mycelia ΔMorid mycelia
3 Table S2. List of loci that are validated for DNA methylation in conidia-derived mycelia Chr. a Locus position Feature B b M c B M d upstream of MGG_ upstream of MGG_ Intergenic region ORF (MGG16885) ORF (MGG889) e 7 a Chromosome Multiple positions including Supercontig 7: a retrotransposon, MAGGY e b Number of mc sites in BS-PCR experiment c Number of mc sites in methylc-seq d Number of mc sites that overlap between BS-PCR and methylc-seq e Sequences that are predicted to have no methylation or methylation by Southern blot analysis and used as negative and positive control of the experiment *Note: BS-PCR and Sanger sequencing for loci listed above were also carried out for conidia, appressoria, and ΔModim-2, confirming little or absence of mc sites predicted in methylcseq.
4 Table S3. List of primers used in this study Name Sequences (5 3 ) Use MGG889 UF GTGCAGGTTCGTTCCTACTT KO a MGG889 UR GCACAGGTACACTTGTTTAGAGATCTGATGGTCAAGGTGAGAA KO MGG889 DF CCTTCAATATCATCTTCTGTCGACATCATTGTCCTGGAAAACA KO MGG889 DR GGTTGGCTGGCTAAACTAGA KO MGG889 nested F AATGGATCATCAGCGAAGG KO MGG889 nested R TGATGGGTTATTCGTAATGGC KO MGG889 RT F AAGGTAAATCCGACTGAACA RT MGG889 RT R AGGCTTGCGAGAAATGAC RT MGG2795 UF GCAGGACCGCATCTCGCTCA KO MGG2795 UR GCACAGGTACACTTGTTTAGAGACCCTCGCGGCATCCCTCAG KO >MGG2795 DF CCTTCAATATCATCTTCTGTCGAGTACCATGCGCCTTCCACA KO MGG2795 DR AAGCAGGCTCCGAAGAAGAA KO MGG2795 nested F CATCGAGCGGCGCAAGAAG KO MGG2795 nested R GCCTTTTCGCCACCATTCT KO MoDIM-2 RT F AAGGTAAATCCGACTGAACA RT b MoDIM-2 RT R AGGCTTGCGAGAAATGAC RT MoRID RT F CAGCTCTATTGTCATTTGTGGC RT MoRID RT R TCCGAGTCTGCAAAGTTGTC RT MGG_889 BS F TTAAAAAAGGTAAATTTGATTGAAT BS-PCR c MGG_889 BS R CAAACCACCTTATATTTCCATA BS-PCR MAGGY BS F GAATATAATTAATGATTTGATTTGA BS-PCR MAGGY BS R TCATACTACTTTAACCAAAACATT BS-PCR MGG_151 P_F AAAAAATAGGATTATATAAGGAA BS-PCR MGG_151 P_R TTAAAATATCAAATAATTTATTTTT BS-PCR MGG_15334 P_F AAGGTTGAGTAATTATTGGTAA BS-PCR MGG_15334 P_R CCACAACTTTATAATAATTTTTC BS-PCR MGG_1673 ORF F ATGTTAATAATAATGGTGTTAATAG BS-PCR MGG_1673 ORF R CTACCCTATACCAAATTTAATAC BS-PCR MGG_16885 ORF F TATTAAATTTGGTATAGGGTAGT BS-PCR MGG_16885 ORF R TTATTTTACATATAATAAACCCAC BS-PCR Chr1_Ig F GTTATTATGTTTGATAGTGATTATT BS-PCR Chr1_Ig R TACATACTCACAATTTACAAAAA BS-PCR a Gene knockout b Real-time PCR c Sanger sequencing of PCR product using bisulphite-treated genomic DNA as template
5 Figure S1 A P LTR ORF1 5.4kb ORF2 P LTR 133 hits including Supercontig 7: A T C A C A C A T T T G C C G C A A A G C A A C G G C C A C G A C G C C A T C C T A G T G G T C G T A G A C C G B Exon1 4.65kb Exon2 Supercontig 5: A A A A T C C G A C G G A A C A A C T T A C C C T G G C T G G G A C G G G G A A G G A G G C A G G A A G C G T T C C G A A T Figure S1. Representative regions in which DNA methylation predicted in bisulfite sequencing (BS -seq) were validated in our study. DNA methylation from Sanger sequencing of bisulfite-pcr (line s and circles) and BS-seq (bar graph) in one MAGGY locus (A) and MGG889 (B). Each line rep resents a Sanger sequencing result for loci indicated by the red bar below the ORF diagram. Empty and filled circles indicate non-methylated and methylated cytosines, respectively. Triangles indicate the positions of cytosines in a CG context.
6 Figure S2 A Number of mc sites 1, 2, 3, 4, 5, Proportion (%) % 2% 4% 6% 8% 1% Mycelia Conidia Appressoria mcg mchg mchh ΔModim-2 Δdmt1 ΔMorid Δdmt2 B Mycelia Conidia Appressoria ΔModim-2 ΔMorid Figure S2. Summary of DNA methylation in the genome of Magnaporthe oryzae. (A) Number (left ) and proportion (right) of methylcytosine (mc) sites in different sequence contexts identified in ea ch sample subjected to bisulfite sequencing (BS-seq). (B) Distribution of the average methylation l evel of individual mc sites. The x-axis represents the methylation level of mc divided into 1 inter vals. The y-axis represents the percentage of mcs that fall into methylation-level bins.
7 Density of mcgs on the chromosomes (mycelia)
8 Density of mchgs on the chromosomes (mycelia)
9 Density of mchhs on the chromosomes (mycelia)
10 Density of mcgs on the chromosomes (conidia)
11 Density of mchgs on the chromosomes (conidia)
12 Density of mchhs on the chromosomes (conidia)
13 Density of mcgs on the chromosomes (appressorium)
14 Density of mchgs on the chromosomes (appressorium)
15 Density of mchhs on the chromosomes (appressorium)
16 Density of mcgs on the chromosomes (dim-2)
17 Density of mchgs on the chromosomes (dim-2)
18 Density of mchhs on the chromosomes (dim-2)
19 Density of mcgs on the chromosomes (rid)
20 Density of mchgs on the chromosomes (rid)
21 Density of mchhs on the chromosomes (rid)
22 . Chromosomal distribution of DNA methylation in the genome of M. oryzae. The density of methylcytosines (mcs) identified on each strand throughout chromosomes of each sample depending on sequence contexts was calculated and plotted in 1kb bin (middle bar). Blue and red tick marks indicate methylation density in Watson and Crick strand, respectively. Dark grey tick marks at the top and bottom of plot indicate density of genes and transposable elements, respectively.
23 Figure S Chr 1 Chr 2 Chr 3 Chr 4 Chr 5 Chr 6 Chr 7 mc density density Figure S4. Density of methylcytosine (mc) sites and transposable elements (s) over chromoso mes. The blue bar represents the number of mc sites per 1 kb in each chromosome, and the red b ar represents the density of s in each chromosome ((total length of s/length of the correspon ding chromosome) 1). The dotted line indicates the expected mc density across chromosom es. Unassigned genome sequences are not included.
24 Figure S5 A DIM-2 CMT3 DNMT1 DNMT2 DNMT3 RID/ Masc1 B DNMT1 (Hs) MoDIM-2 (Mo) DIM-2 (Nc) PMT1 (Sp) MoRID (Mo) RID (Nc) DNMT3a (Hs) 2 a.a Figure S5. DNA methyltransferases in diverse organisms. (A) Kingdom-wide distribution of DNA methyltransferases. The cladogram was constructed for representative species of each phylum by C Vtree with the following options: K-tuple length = 7 and sequence type = amino acid ( dan.edu.cn/cvtree/). The presence (colored box) or absence (box with no color) of each family of D NA methyltransferases in different taxa was determined using the BLAST matrix function embedde d in CFGP ( To remove spurious hits, an e-value cutoff of 1e 1 5 and a score cutoff of 1 were initially used; the remaining hits were examined manually. ( B) Domain architecture of representative DNA methyltransferases. Light green, BAH domain; blue, PWWP domain; red, DNA methyltransferase domain.
25 Figure S6 A 3.4kb KpnI MoDIM-2 KpnI KpnI B KJ21 Δdim-2 Δdim-2::MoDIM-2 DC37 DC43 5kb M H M H M H M H 3kb KpnI KpnI HPH 5kb 3.1kb PstI PstI PstI PstI PstI PstI 3.4kb MoRID 5kb 4kb 3kb PstI HPH PstI PstI C D E KJ KJ21 6 ΔModim-2 ΔMorid 4 2 KJ21 Δdim-2 Δdmt1 Δdmt2 Δrid KJ21 Δdim-2 Δdmt1 Δdmt2 Δrid KJ21 Δdim-2 Δdmt1 Δdmt2 Δrid ΔModim-2 4hr 8hr 12hr Germination rate Appressorium formation rate ΔMorid Figure S6. Deletion of genes encoding DNA methyltransferases in Magnaporthe oryzae. (A) So uthern blot analysis of gene deletion mutants for MoDIM-2 (top) and MoRID (bottom). A schema tic diagram depicting gene deletion via double homologous recombination is shown on the left a nd the result of Southern hybridization using sequences indicated as a black bar as a probe is sho wn on the right. (B) Complementation of methylation defects in Modim-2 by the introduction o f a native MoDIM-2 copy (H, HpaII; M, MspI). Four separate parts of a single blot (delineated by black vertical lines) were vertically sliced and juxtaposed for qualitative comparison for clarity. (C) Conidiophore development in the mutants (8 h post-surface scraping). (D) Germination and a ppressorium formation at 4, 8, and 12 h postinoculation (hpi). (E) Pathogenicity towards rice pla nts (6 days postinoculation).
26 Figure S Mycelia Conidia Appressoria Wild-type ΔModim-2 ΔMorid Figure S7. Density (blue bar) and average methylation level (purple bar) of methylcytosine (mc) sites in different transposable elements (s). s are arranged on the x-axis in asce nding order of length.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09861 & &' -(' ()*+ ')(+,,(','-*+,&,,+ ',+' ' 23,45/0*6787*9:./09 ;78?4?@*+A786?B- &' )*+*(,-* -(' ()*+ ')(+,,(','-*+,&,,+ ',+'./)*+*(,-*..)*+*(,-*./)*+*(,-*.0)*+*(,-*..)*+*(,-*
doi:10.1038/nature09861 & &' -(' ()*+ ')(+,,(','-*+,&,,+ ',+' ' 23,45/0*6787*9:./09 ;78?4?@*+A786?B- &' )*+*(,-* -(' ()*+ ')(+,,(','-*+,&,,+ ',+'./)*+*(,-*..)*+*(,-*./)*+*(,-*.0)*+*(,-*..)*+*(,-*
Epigenetic control of tomato fruit development. Key Laboratories of Molecular Epigenetics of MOE, Northeast Normal University, Changchun, China
 Epigenetic control of tomato fruit development Silin Zhong 1,2*,, Zhangjun Fei 1,3,*,, Yun-Ru Chen 1, Yi Zheng 1, Mingyun Huang 1, Julia Vrebalov 1, Ryan McQuinn 1, Nigel Gapper 1, Bao Liu 2, Jenny Xiang
Epigenetic control of tomato fruit development Silin Zhong 1,2*,, Zhangjun Fei 1,3,*,, Yun-Ru Chen 1, Yi Zheng 1, Mingyun Huang 1, Julia Vrebalov 1, Ryan McQuinn 1, Nigel Gapper 1, Bao Liu 2, Jenny Xiang
Chemical and/or enzymatic deamination across various sequencing methods to localize modified cytosines.
 Supplementary Figure 1 Chemical and/or enzymatic deamination across various sequencing methods to localize modified cytosines. (a) Upon treatment with bisulfite under acidic conditions and at elevated
Supplementary Figure 1 Chemical and/or enzymatic deamination across various sequencing methods to localize modified cytosines. (a) Upon treatment with bisulfite under acidic conditions and at elevated
SUPPLEMENTARY INFORMATION
 Supplemental Figure 1. Expression levels of cell type specific marker genes in each cell type. (a) Root meristem of ProCYCD5:GFP. GFP is specifically expressed in lower columella. (b) (e) Expression levels
Supplemental Figure 1. Expression levels of cell type specific marker genes in each cell type. (a) Root meristem of ProCYCD5:GFP. GFP is specifically expressed in lower columella. (b) (e) Expression levels
Mammalian non-cg methylations are conserved and cell-type specific and may have been involved in the evolution of transposon elements
 Mammalian non-cg methylations are conserved and cell-type specific and may have been involved in the evolution of transposon elements Weilong Guo, Michael Zhang, Hong Wu Supplementary Figures Fig. S1-S16
Mammalian non-cg methylations are conserved and cell-type specific and may have been involved in the evolution of transposon elements Weilong Guo, Michael Zhang, Hong Wu Supplementary Figures Fig. S1-S16
Erhard et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. c1-hbr allele structure. Diagram of the c1-hbr allele found in stocks segregating 1:1 for rpd1-1 and rpd1-2 homozygous mutants showing the presence of a 363 base pair (bp) Heartbreaker
Supplemental Figure 1. c1-hbr allele structure. Diagram of the c1-hbr allele found in stocks segregating 1:1 for rpd1-1 and rpd1-2 homozygous mutants showing the presence of a 363 base pair (bp) Heartbreaker
Supplemental Figure 1.
 Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Figure S1 Correlation in size of analogous introns in mouse and teleost Piccolo genes. Mouse intron size was plotted against teleost intron size for t
 Figure S1 Correlation in size of analogous introns in mouse and teleost Piccolo genes. Mouse intron size was plotted against teleost intron size for the pcloa genes of zebrafish, green spotted puffer (listed
Figure S1 Correlation in size of analogous introns in mouse and teleost Piccolo genes. Mouse intron size was plotted against teleost intron size for the pcloa genes of zebrafish, green spotted puffer (listed
Supplemental Figure legends Figure S1. (A) (B) (C) (D) Figure S2. Figure S3. (A-E) Figure S4. Figure S5. (A, C, E, G, I) (B, D, F, H, Figure S6.
 Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Identification and characterization of a pathogenicity-related gene VdCYP1 from Verticillium dahliae
 Identification and characterization of a pathogenicity-related gene VdCYP1 from Verticillium dahliae Dan-Dan Zhang*, Xin-Yan Wang*, Jie-Yin Chen*, Zhi-Qiang Kong, Yue-Jing Gui, Nan-Yang Li, Yu-Ming Bao,
Identification and characterization of a pathogenicity-related gene VdCYP1 from Verticillium dahliae Dan-Dan Zhang*, Xin-Yan Wang*, Jie-Yin Chen*, Zhi-Qiang Kong, Yue-Jing Gui, Nan-Yang Li, Yu-Ming Bao,
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for
 Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
IDN1 and IDN2: two proteins required for de novo DNA methylation in Arabidopsis thaliana.
 IDN1 and IDN2: two proteins required for de novo DNA methylation in Arabidopsis thaliana. Israel Ausin, Todd C. Mockler, Joanne Chory, and Steven E. Jacobsen Col Ler nrpd1a rdr2 dcl3-1 ago4-1 idn1-1 idn2-1
IDN1 and IDN2: two proteins required for de novo DNA methylation in Arabidopsis thaliana. Israel Ausin, Todd C. Mockler, Joanne Chory, and Steven E. Jacobsen Col Ler nrpd1a rdr2 dcl3-1 ago4-1 idn1-1 idn2-1
amplification of the 5 flanking region of CAT1 with a tail for the geneticin resistance gene cassette fusion CAT1-3F
 Supplementary materials Table S1. Primer list. Primer Sequence a (5-3 ) Description CAT1-5F CAT1-5R ATACGGATAAGGAAGCGATAGCAGCA gcacaggtacacttgtttagagagcgatccggattttaagtgaacg amplification of the 5 flanking
Supplementary materials Table S1. Primer list. Primer Sequence a (5-3 ) Description CAT1-5F CAT1-5R ATACGGATAAGGAAGCGATAGCAGCA gcacaggtacacttgtttagagagcgatccggattttaagtgaacg amplification of the 5 flanking
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Tables. Primers and probes. Target Primer / probe sequences Chemistry Human HBB F: AACTGTGTTCACTAGCAACCTCAAA
 Supplementary Tables Primers and probes Target Primer / probe sequences Chemistry H F: AACTGTGTTCACTAGCAACCTCAAA promoter R: ACAGGGCAGTAACGGCAGACT H Downstream Actb alpha (162) alpha (162.) (Pro) (16)
Supplementary Tables Primers and probes Target Primer / probe sequences Chemistry H F: AACTGTGTTCACTAGCAACCTCAAA promoter R: ACAGGGCAGTAACGGCAGACT H Downstream Actb alpha (162) alpha (162.) (Pro) (16)
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Number and length distributions of the inferred fosmids.
 Supplementary Figure 1 Number and length distributions of the inferred fosmids. Fosmid were inferred by mapping each pool s sequence reads to hg19. We retained only those reads that mapped to within a
Supplementary Figure 1 Number and length distributions of the inferred fosmids. Fosmid were inferred by mapping each pool s sequence reads to hg19. We retained only those reads that mapped to within a
Characterization of RNA silencing components in the plant
 1 2 3 4 5 6 Supplementary data Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum Yun Chen, Qixun Gao, Mengmeng Huang, Ye Liu, Zunyong Liu, Xin Liu, Zhonghua
1 2 3 4 5 6 Supplementary data Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum Yun Chen, Qixun Gao, Mengmeng Huang, Ye Liu, Zunyong Liu, Xin Liu, Zhonghua
% Viability. isw2 ino isw2 ino isw2 ino isw2 ino mM HU 4-NQO CPT
 a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino8 9 2.5.1.15 % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy)
a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino8 9 2.5.1.15 % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy)
Donor DNA Utilization during Gene Targeting with Zinc- finger Nucleases
 Donor DNA Utilization during Gene Targeting with Zinc- finger Nucleases Kelly J. Beumer, Jonathan K. Trautman, Kusumika Mukherjee and Dana Carroll Department of Biochemistry University of Utah School of
Donor DNA Utilization during Gene Targeting with Zinc- finger Nucleases Kelly J. Beumer, Jonathan K. Trautman, Kusumika Mukherjee and Dana Carroll Department of Biochemistry University of Utah School of
Map-Based Cloning of Qualitative Plant Genes
 Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
Supplementary Materials. Sequence-based profiling of DNA methylation: comparisons of methods and catalogue of allelic epigenetic modifications
 Supplementary Materials Sequence-based profiling of DNA methylation: comparisons of methods and catalogue of allelic epigenetic modifications Supplementary Figure 1. Analysis of biological replicates (three
Supplementary Materials Sequence-based profiling of DNA methylation: comparisons of methods and catalogue of allelic epigenetic modifications Supplementary Figure 1. Analysis of biological replicates (three
Nature Methods: doi: /nmeth Supplementary Figure 1. Construction of a sensitive TetR mediated auxotrophic off-switch.
 Supplementary Figure 1 Construction of a sensitive TetR mediated auxotrophic off-switch. A Production of the Tet repressor in yeast when conjugated to either the LexA4 or LexA8 promoter DNA binding sequences.
Supplementary Figure 1 Construction of a sensitive TetR mediated auxotrophic off-switch. A Production of the Tet repressor in yeast when conjugated to either the LexA4 or LexA8 promoter DNA binding sequences.
Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C
 CORRECTION NOTICE Nat. Genet. 47, 598 606 (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C Borbala Mifsud, Filipe Tavares-Cadete, Alice N Young, Robert Sugar,
CORRECTION NOTICE Nat. Genet. 47, 598 606 (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C Borbala Mifsud, Filipe Tavares-Cadete, Alice N Young, Robert Sugar,
C24. ros1-1. ros1-1 rdm18-1. ros1-1 rdm18-2. ros1-1 nrpe1
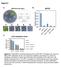 Figure S1 Methylation level 1 0.8 0.6 0.4 0.2 0 p35s methylation levels 24 ros1-1 ros1-1 rdm18-1 ros1-1 rdm18-2 ros1-1 nrpe1 mg mhg mhh Figure S1. RM18/PKL promotes silencing at the p35s-npt II transgene
Figure S1 Methylation level 1 0.8 0.6 0.4 0.2 0 p35s methylation levels 24 ros1-1 ros1-1 rdm18-1 ros1-1 rdm18-2 ros1-1 nrpe1 mg mhg mhh Figure S1. RM18/PKL promotes silencing at the p35s-npt II transgene
Supplementary Tables and Figures
 Supplementary Tables and Figures The autophagy-related genes BbATG1 and BbATG8 have different functions in differentiation, stress resistance and virulence of mycopathogen Beauveria bassiana Sheng-Hua
Supplementary Tables and Figures The autophagy-related genes BbATG1 and BbATG8 have different functions in differentiation, stress resistance and virulence of mycopathogen Beauveria bassiana Sheng-Hua
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
6/256 1/256 0/256 1/256 2/256 7/256 10/256. At3g06290 (SAC3B)
 Chr.III M 5M 1M 15M 2M 23M BAC clones F22F7 F1A16 F24F17 F24P17 T8E24 F17A9 F21O3 F17A17 F18C1 F2O1 F28L1 F5E6 F3E22 T1B9 MLP3 Number of recombinants 6/256 1/256 /256 1/256 2/256 7/256 1/256 At3g629 (SAC3B)
Chr.III M 5M 1M 15M 2M 23M BAC clones F22F7 F1A16 F24F17 F24P17 T8E24 F17A9 F21O3 F17A17 F18C1 F2O1 F28L1 F5E6 F3E22 T1B9 MLP3 Number of recombinants 6/256 1/256 /256 1/256 2/256 7/256 1/256 At3g629 (SAC3B)
Analyzing an individual sequence in the Sequence Editor
 BioNumerics Tutorial: Analyzing an individual sequence in the Sequence Editor 1 Aim The Sequence editor window is a convenient tool implemented in BioNumerics to edit and analyze nucleotide and amino acid
BioNumerics Tutorial: Analyzing an individual sequence in the Sequence Editor 1 Aim The Sequence editor window is a convenient tool implemented in BioNumerics to edit and analyze nucleotide and amino acid
SUPPLEMENTARY INFORMATION
 AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
Spermatazoa. Sertoli cells. Morc1 WT adult. Morc1 KO adult. Morc1 WT P14.5. Morc1 KO P14.5. Germ cells. Germ cells. Spermatids.
 a. Spermatazoa Sertoli cells Morc1 WT adult c. Morc1 KO adult Morc1 WT P1.5 Morc1 KO P1.5 Morc1 WT adult Morc1 KO adult Germ cells Germ cells Spermatids Spermatozoa Supplementary Figure 1. Confirmation
a. Spermatazoa Sertoli cells Morc1 WT adult c. Morc1 KO adult Morc1 WT P1.5 Morc1 KO P1.5 Morc1 WT adult Morc1 KO adult Germ cells Germ cells Spermatids Spermatozoa Supplementary Figure 1. Confirmation
Supplementary Information Targeting fidelity of adenine and cytosine base editors in mouse embryos
 Supplementary Information ing fidelity of adenine and cytosine base s in mouse embryos Lee et al. a P = 1.012e-14 b Frequency (%) 100% 80% 60% 40% 20% 0% CB AB On-target Bystander Proximal Indels Frequency
Supplementary Information ing fidelity of adenine and cytosine base s in mouse embryos Lee et al. a P = 1.012e-14 b Frequency (%) 100% 80% 60% 40% 20% 0% CB AB On-target Bystander Proximal Indels Frequency
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Supplemental Data. Zhou et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. Confirmation of mutant mapping results. (A) Complementation assay with stably transformed genomic fragments (ComN-N) (2 kb upstream of TSS and 1.5 kb downstream of TES) and CaMV
Supplemental Figure 1. Confirmation of mutant mapping results. (A) Complementation assay with stably transformed genomic fragments (ComN-N) (2 kb upstream of TSS and 1.5 kb downstream of TES) and CaMV
SUPPLEMENTARY INFORMATION
 Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Ca 2+ /calmodulin Regulates Salicylic Acid-mediated Plant Immunity Liqun Du, Gul S. Ali, Kayla A. Simons, Jingguo Hou, Tianbao Yang, A.S.N. Reddy and B. W. Poovaiah * *To whom correspondence should be
Suppl. Table S1. Characteristics of DHS regions analyzed by bisulfite sequencing. No. CpGs analyzed in the amplicon. Genomic location specificity
 Suppl. Table S1. Characteristics of DHS regions analyzed by bisulfite sequencing. DHS/GRE Genomic location Tissue specificity DHS type CpG density (per 100 bp) No. CpGs analyzed in the amplicon CpG within
Suppl. Table S1. Characteristics of DHS regions analyzed by bisulfite sequencing. DHS/GRE Genomic location Tissue specificity DHS type CpG density (per 100 bp) No. CpGs analyzed in the amplicon CpG within
Supplemental Figure 1
 Supplemental Figure 1 Supplemental figure 1. Generation of gene targeted mice expressing an anti-id BCR. (A,B) Generation of the VDJ aid H KI mouse. (A) Targeting Construct. Top: Targeting construct for
Supplemental Figure 1 Supplemental figure 1. Generation of gene targeted mice expressing an anti-id BCR. (A,B) Generation of the VDJ aid H KI mouse. (A) Targeting Construct. Top: Targeting construct for
A) (5 points) As the starting step isolate genomic DNA from
 GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
SUPPLEMENTARY INFORMATION
 doi:.38/nature08267 Figure S: Karyotype analysis of ips cell line One hundred individual chromosomal spreads were counted for each of the ips samples. Shown here is an spread at passage 5, predominantly
doi:.38/nature08267 Figure S: Karyotype analysis of ips cell line One hundred individual chromosomal spreads were counted for each of the ips samples. Shown here is an spread at passage 5, predominantly
Supplementary Data 1.
 Supplementary Data 1. Evaluation of the effects of number of F2 progeny to be bulked (n) and average sequencing coverage (depth) of the genome (G) on the levels of false positive SNPs (SNP index = 1).
Supplementary Data 1. Evaluation of the effects of number of F2 progeny to be bulked (n) and average sequencing coverage (depth) of the genome (G) on the levels of false positive SNPs (SNP index = 1).
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and
 Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Supplementary Figure S1. Np95 interacts with de novo methyltransferases Dnmt3a and 3b. (A) Co-immunoprecipitation of endogenous Dnmt3a2 (left and right), Dnmt3b isoforms (left) and Dnmt1 (right) with GFP-Np95
Nature Methods: doi: /nmeth Supplementary Figure 1. Pilot CrY2H-seq experiments to confirm strain and plasmid functionality.
 Supplementary Figure 1 Pilot CrY2H-seq experiments to confirm strain and plasmid functionality. (a) RT-PCR on HIS3 positive diploid cell lysate containing known interaction partners AT3G62420 (bzip53)
Supplementary Figure 1 Pilot CrY2H-seq experiments to confirm strain and plasmid functionality. (a) RT-PCR on HIS3 positive diploid cell lysate containing known interaction partners AT3G62420 (bzip53)
Nature Genetics: doi: /ng.3556 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE. Supplementary Figure 1
 INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
INTEGRATED SUPPLEMENTARY FIGURE TEMPLATE Supplementary Figure 1 REF6 expression in transgenic lines. (a,b) Expression of REF6 in REF6-HA ref6 and REF6ΔZnF-HA ref6 plants detected by RT qpcr (a) and immunoblot
Supporting Information
 Supporting Information Kilian et al. 10.1073/pnas.1105861108 SI Materials and Methods Determination of the Electric Field Strength Required for Successful Electroporation. The transformation construct
Supporting Information Kilian et al. 10.1073/pnas.1105861108 SI Materials and Methods Determination of the Electric Field Strength Required for Successful Electroporation. The transformation construct
SUPPLEMENTARY INFORMATION
 Figure S1. lev-9 mutants are resistant to levamisole. The levamisole dose-response curves indicate that lev-9 mutants are partially resistant to levamisole similar to lev-10(kr26) mutants. unc-29(x29)
Figure S1. lev-9 mutants are resistant to levamisole. The levamisole dose-response curves indicate that lev-9 mutants are partially resistant to levamisole similar to lev-10(kr26) mutants. unc-29(x29)
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplemental Figure 1 A
 Supplemental Figure A prebleach postbleach 2 min 6 min 3 min mh2a.-gfp mh2a.2-gfp mh2a2-gfp GFP-H2A..9 Relative Intensity.8.7.6.5 mh2a. GFP n=8.4 mh2a.2 GFP n=4.3 mh2a2 GFP n=2.2 GFP H2A n=24. GFP n=7.
Supplemental Figure A prebleach postbleach 2 min 6 min 3 min mh2a.-gfp mh2a.2-gfp mh2a2-gfp GFP-H2A..9 Relative Intensity.8.7.6.5 mh2a. GFP n=8.4 mh2a.2 GFP n=4.3 mh2a2 GFP n=2.2 GFP H2A n=24. GFP n=7.
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
To investigate the heredity of the WFP gene, we selected plants that were homozygous
 Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Nature Genetics: doi: /ng Supplementary Figure 1. ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets.
 Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Supplementary Figure 1 ChIP-seq genome browser views of BRM occupancy at previously identified BRM targets. Gene structures are shown underneath each panel. Supplementary Figure 2 pref6::ref6-gfp complements
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Multiple choice questions (numbers in brackets indicate the number of correct answers)
 1 February 15, 2013 Multiple choice questions (numbers in brackets indicate the number of correct answers) 1. Which of the following statements are not true Transcriptomes consist of mrnas Proteomes consist
1 February 15, 2013 Multiple choice questions (numbers in brackets indicate the number of correct answers) 1. Which of the following statements are not true Transcriptomes consist of mrnas Proteomes consist
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplemental data. Li et al. (2014). Plant Cell /tpc/114/133140
 MET (CG) CMT (CHG / CHH) DRM (CHH / CG / CHG) Supplemental Figure 1. Phylogenetic Tree of DNA Methyltransferases in Maize and Arabidopsis. Maize genes are indicated by red dots. The DNA methylation sequence
MET (CG) CMT (CHG / CHH) DRM (CHH / CG / CHG) Supplemental Figure 1. Phylogenetic Tree of DNA Methyltransferases in Maize and Arabidopsis. Maize genes are indicated by red dots. The DNA methylation sequence
Supplemental Figure 1 A
 Supplemental Figure 1 A Supplemental Data. Han et al. (2016). Plant Cell 10.1105/tpc.15.00997 BamHI -1616 bp SINE repeats SalI ATG SacI +1653 bp HindIII LB HYG p35s pfwa LUC Tnos RB pfwa-bamhi-f: CGGGATCCCGCCTTTCTCTTCCTCATCTGC
Supplemental Figure 1 A Supplemental Data. Han et al. (2016). Plant Cell 10.1105/tpc.15.00997 BamHI -1616 bp SINE repeats SalI ATG SacI +1653 bp HindIII LB HYG p35s pfwa LUC Tnos RB pfwa-bamhi-f: CGGGATCCCGCCTTTCTCTTCCTCATCTGC
Result Tables The Result Table, which indicates chromosomal positions and annotated gene names, promoter regions and CpG islands, is the best way for
 Result Tables The Result Table, which indicates chromosomal positions and annotated gene names, promoter regions and CpG islands, is the best way for you to discover methylation changes at specific genomic
Result Tables The Result Table, which indicates chromosomal positions and annotated gene names, promoter regions and CpG islands, is the best way for you to discover methylation changes at specific genomic
Zhang et al., RepID facilitates replication Initiation. Supplemental Information:
 Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Nature Genetics: doi: /ng Supplementary Figure 1. Liz/Zdbf2 dynamics in vivo and in vitro.
 Supplementary igure 1 Liz/Zdbf2 dynamics in vivo and in vitro. (a) Schematic of Liz/Zdbf2 regulation during early development. Summary from Duffié et al. 18. Liz transcript initiates from the unmethylated
Supplementary igure 1 Liz/Zdbf2 dynamics in vivo and in vitro. (a) Schematic of Liz/Zdbf2 regulation during early development. Summary from Duffié et al. 18. Liz transcript initiates from the unmethylated
Supplementary Information
 Supplementary Information Supplementary Figure 1: Synthetic annealed poly(ra):poly(dt) hybrid induces type I interferon response. a) Bone marrow derived macrophages (BMDM) were transfected with or without
Supplementary Information Supplementary Figure 1: Synthetic annealed poly(ra):poly(dt) hybrid induces type I interferon response. a) Bone marrow derived macrophages (BMDM) were transfected with or without
CGAGACCTTGTCACATTGGCGTCAACTC hoga Down GATGCCTCCTGTCAAGATTGCAATGAAC
 Supplemental Table 1. PCR and qrt-pcr primers used in this study Primer designation Oligonucleotide sequence (5-3 ) PCR primers 5 SphI hoga GCATGCGAAGACTGATTCAGATAATTAGTTC 5 SalI hoga GTCGACCTGATGATCTCCACTGTCTGAAC
Supplemental Table 1. PCR and qrt-pcr primers used in this study Primer designation Oligonucleotide sequence (5-3 ) PCR primers 5 SphI hoga GCATGCGAAGACTGATTCAGATAATTAGTTC 5 SalI hoga GTCGACCTGATGATCTCCACTGTCTGAAC
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Data Retrieval from GenBank
 Data Retrieval from GenBank Peter J. Myler Bioinformatics of Intracellular Pathogens JNU, Feb 7-0, 2009 http://www.ncbi.nlm.nih.gov (January, 2007) http://ncbi.nlm.nih.gov/sitemap/resourceguide.html Accessing
Data Retrieval from GenBank Peter J. Myler Bioinformatics of Intracellular Pathogens JNU, Feb 7-0, 2009 http://www.ncbi.nlm.nih.gov (January, 2007) http://ncbi.nlm.nih.gov/sitemap/resourceguide.html Accessing
DNA METHYLATION NH 2 H 3. Solutions using bisulfite conversion and immunocapture Ideal for NGS, Sanger sequencing, Pyrosequencing, and qpcr
 NH 2 DNA METHYLATION H 3 C Solutions using bisulfite conversion and immunocapture Ideal for NGS, Sanger sequencing, Pyrosequencing, and qpcr NH 2 N N H PAGE 3 Understanding DNA Methylation DNA methylation
NH 2 DNA METHYLATION H 3 C Solutions using bisulfite conversion and immunocapture Ideal for NGS, Sanger sequencing, Pyrosequencing, and qpcr NH 2 N N H PAGE 3 Understanding DNA Methylation DNA methylation
Supplementary Figure S1 Supplementary Figure S2 Supplementary Figure S3. Supplementary Figure S4
 Supplementary Figure S1 Supplementary Figure S2 Supplementary Figure S3 Supplementary Figure S4 Supplementary Figure S5 Supplementary Figure S6 Supplementary Figure S7 Supplementary Figure S8 Supplementary
Supplementary Figure S1 Supplementary Figure S2 Supplementary Figure S3 Supplementary Figure S4 Supplementary Figure S5 Supplementary Figure S6 Supplementary Figure S7 Supplementary Figure S8 Supplementary
Supplementary Information. Microfluidic high-throughput selection of microalgal strains with
 Supplementary Information Microfluidic high-throughput selection of microalgal strains with superior photosynthetic productivity using competitive phototaxis Jaoon Young Hwan Kim 1,, Ho Seok Kwak 1,, Young
Supplementary Information Microfluidic high-throughput selection of microalgal strains with superior photosynthetic productivity using competitive phototaxis Jaoon Young Hwan Kim 1,, Ho Seok Kwak 1,, Young
Kerkel et al., Supplementary Tables and Figures
 Kerkel et al., Supplementary Tables and Figures Table S1. Summary of 16 SNP tagged loci from the combined 50K and 250K MSNP screens with independently validated ASM. PBL, total peripheral blood leukocytes;
Kerkel et al., Supplementary Tables and Figures Table S1. Summary of 16 SNP tagged loci from the combined 50K and 250K MSNP screens with independently validated ASM. PBL, total peripheral blood leukocytes;
Supporting Information
 Supporting Information Park et al. 10.1073/pnas.1410555111 5 -TCAAGTCCATCTACATGGCC-3 5 -CAGCTGCCCGGCTACTACTA-3 5 -TGCAGCTGCCCGGCTACTAC-3 5 -AAGCTGGACATCACCTCCCA-3 5 -TGACAGGAACACCTACAAGT-3 5 -AAGGCACCTTTCTGTCTCCA-3
Supporting Information Park et al. 10.1073/pnas.1410555111 5 -TCAAGTCCATCTACATGGCC-3 5 -CAGCTGCCCGGCTACTACTA-3 5 -TGCAGCTGCCCGGCTACTAC-3 5 -AAGCTGGACATCACCTCCCA-3 5 -TGACAGGAACACCTACAAGT-3 5 -AAGGCACCTTTCTGTCTCCA-3
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Before starting, write your name on the top of each page Make sure you have all pages
 Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Supporting Information
 Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
(i) A trp1 mutant cell took up a plasmid containing the wild type TRP1 gene, which allowed that cell to multiply and form a colony
 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been isolated that cannot grow
1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been isolated that cannot grow
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
BME 110 Midterm Examination
 BME 110 Midterm Examination May 10, 2011 Name: (please print) Directions: Please circle one answer for each question, unless the question specifies "circle all correct answers". You can use any resource
BME 110 Midterm Examination May 10, 2011 Name: (please print) Directions: Please circle one answer for each question, unless the question specifies "circle all correct answers". You can use any resource
GATCGTGCACGATCTCGGCAATTCGGGATGCCGGCTCGTCACCGGTCGCT
 Problem. (pts) A. (5pts) Your colleague professor Eugene Mathew Lateed generated a genome-wide DNA methylation map for normal colon cells using MRE-seq and MeDIP-seq. In an intergenic region, he found
Problem. (pts) A. (5pts) Your colleague professor Eugene Mathew Lateed generated a genome-wide DNA methylation map for normal colon cells using MRE-seq and MeDIP-seq. In an intergenic region, he found
Candidate region (0.74 Mb) ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC ATC TCT GGG ACT CAT TAG CAG GAG GCT AGC
 A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM Part I: Definitions. Homology: Reverse transcriptase. Allostery: cdna library
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
Annotating 7G24-63 Justin Richner May 4, Figure 1: Map of my sequence
 Annotating 7G24-63 Justin Richner May 4, 2005 Zfh2 exons Thd1 exons Pur-alpha exons 0 40 kb 8 = 1 kb = LINE, Penelope = DNA/Transib, Transib1 = DINE = Novel Repeat = LTR/PAO, Diver2 I = LTR/Gypsy, Invader
Annotating 7G24-63 Justin Richner May 4, 2005 Zfh2 exons Thd1 exons Pur-alpha exons 0 40 kb 8 = 1 kb = LINE, Penelope = DNA/Transib, Transib1 = DINE = Novel Repeat = LTR/PAO, Diver2 I = LTR/Gypsy, Invader
CRISPR/Cas9-induced knockout and knock-in mutations in
 Supplementary Information for Scientific Reports CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii Sung-Eun Shin a,, Jong-Min Lim b,, Hyun Gi Koh a, Eun Kyung Kim h, Nam
Supplementary Information for Scientific Reports CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii Sung-Eun Shin a,, Jong-Min Lim b,, Hyun Gi Koh a, Eun Kyung Kim h, Nam
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna
 Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplementary Figure 1 Activities of ABEs using extended sgrnas in HEK293T cells.
 Supplementary Figure 1 Activities of ABEs using extended sgrnas in HEK293T cells. Base editing efficiencies of ABEs with extended sgrnas at Site 18 (a), Site 19 (b), the HBB-E2 site (c), and the HBB-E3
Supplementary Figure 1 Activities of ABEs using extended sgrnas in HEK293T cells. Base editing efficiencies of ABEs with extended sgrnas at Site 18 (a), Site 19 (b), the HBB-E2 site (c), and the HBB-E3
A Naturally Occurring Epiallele associates with Leaf Senescence and Local Climate Adaptation in Arabidopsis accessions He et al.
 A Naturally Occurring Epiallele associates with Leaf Senescence and Local Climate Adaptation in Arabidopsis accessions He et al. Supplementary Notes Origin of NMR19 elements Because there are two copies
A Naturally Occurring Epiallele associates with Leaf Senescence and Local Climate Adaptation in Arabidopsis accessions He et al. Supplementary Notes Origin of NMR19 elements Because there are two copies
Supplementary Figures
 Supplementary Figures A B Supplementary Figure 1. Examples of discrepancies in predicted and validated breakpoint coordinates. A) Most frequently, predicted breakpoints were shifted relative to those derived
Supplementary Figures A B Supplementary Figure 1. Examples of discrepancies in predicted and validated breakpoint coordinates. A) Most frequently, predicted breakpoints were shifted relative to those derived
0.5% Sucrose. Supplemental Data. Chao et al. (2011). Plant Cell /tpc Col-0 tsc10a-2. Root length (mm)
 A 0% 0.5% 1% Col-0 tsc10a-2 B Root length (mm) 35 30 25 20 15 10 5 0 0% 0.50% 1% 2% 4% Sucrose concentration 2% 4% Sucrose Col-0 tsc10a-2 Supplemental Figure 1. The effect of sucrose on root growth of
A 0% 0.5% 1% Col-0 tsc10a-2 B Root length (mm) 35 30 25 20 15 10 5 0 0% 0.50% 1% 2% 4% Sucrose concentration 2% 4% Sucrose Col-0 tsc10a-2 Supplemental Figure 1. The effect of sucrose on root growth of
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
Practical solutions. for DNA methylation analysis
 DNA Methylation Analysis Tools Practical solutions for DNA methylation analysis Highly efficient and time-saving solutions Locus-specific and whole genome analysis Robust performance Innovative. Fast.
DNA Methylation Analysis Tools Practical solutions for DNA methylation analysis Highly efficient and time-saving solutions Locus-specific and whole genome analysis Robust performance Innovative. Fast.
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Lecture 9. Eukaryotic gene regulation: DNA METHYLATION
 Lecture 9 Eukaryotic gene regulation: DNA METHYLATION Recap.. Eukaryotic RNA polymerases Core promoter elements General transcription factors Enhancers and upstream activation sequences Transcriptional
Lecture 9 Eukaryotic gene regulation: DNA METHYLATION Recap.. Eukaryotic RNA polymerases Core promoter elements General transcription factors Enhancers and upstream activation sequences Transcriptional
PrimePCR Assay Validation Report
 Gene Information Gene Name keratin 78 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID KRT78 Human This gene is a member of the type II keratin gene family
Gene Information Gene Name keratin 78 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID KRT78 Human This gene is a member of the type II keratin gene family
Genome Sequence Assembly
 Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
Genome Sequence Assembly Learning Goals: Introduce the field of bioinformatics Familiarize the student with performing sequence alignments Understand the assembly process in genome sequencing Introduction:
Supplemental Data. Jing et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. Characterization of epp1 Mutants. (A) Cotyledon angles of 5-d-old Col wild-type (gray bars) and epp1-1 (black bars) seedlings under red (R), far-red (FR) and blue (BL) light conditions,
Supplemental Figure 1. Characterization of epp1 Mutants. (A) Cotyledon angles of 5-d-old Col wild-type (gray bars) and epp1-1 (black bars) seedlings under red (R), far-red (FR) and blue (BL) light conditions,
BS 50 Genetics and Genomics Week of Oct 24
 BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
Genomes summary. Bacterial genome sizes
 Genomes summary 1. >930 bacterial genomes sequenced. 2. Circular. Genes densely packed. 3. 2-10 Mbases, 470-7,000 genes 4. Genomes of >200 eukaryotes (45 higher ) sequenced. 5. Linear chromosomes 6. On
Genomes summary 1. >930 bacterial genomes sequenced. 2. Circular. Genes densely packed. 3. 2-10 Mbases, 470-7,000 genes 4. Genomes of >200 eukaryotes (45 higher ) sequenced. 5. Linear chromosomes 6. On
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Negative selection analysis of sgrnas targeting all Brd4 exons comparing day 2 to day 10 time points. Systematic evaluation of 64 Brd4 sgrnas in negative selection experiments, targeting
Supplementary Figure 1 Negative selection analysis of sgrnas targeting all Brd4 exons comparing day 2 to day 10 time points. Systematic evaluation of 64 Brd4 sgrnas in negative selection experiments, targeting
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Nature Genetics: doi: /ng Supplementary Figure 1. Neighbor-joining tree of the 183 wild, cultivated, and weedy rice accessions.
 Supplementary Figure 1 Neighbor-joining tree of the 183 wild, cultivated, and weedy rice accessions. Relationships of cultivated and wild rice correspond to previously observed relationships 40. Wild rice
Supplementary Figure 1 Neighbor-joining tree of the 183 wild, cultivated, and weedy rice accessions. Relationships of cultivated and wild rice correspond to previously observed relationships 40. Wild rice
Loss of Native Allele Protocol for KOMP-Regeneron derived ESC lines January 9 th 2009
 Loss of Native Allele Protocol for KOMP-Regeneron derived ESC lines January 9 th 2009 I. Alkaline DNA extraction in 96 well format Solutions 50mM NaOH 1M Tris-HCl ph 8.0 1. Grow ES cells in a 96-well TC
Loss of Native Allele Protocol for KOMP-Regeneron derived ESC lines January 9 th 2009 I. Alkaline DNA extraction in 96 well format Solutions 50mM NaOH 1M Tris-HCl ph 8.0 1. Grow ES cells in a 96-well TC
