GUGAUAAUGGAGCGAGAUUUUCUGUUGUGCUUGAUCUAACCAUGUGCUUGCGAGGUAUGA GAAAAACAUGGUUCCGUCAAGCACCAUGGAACGUCACGCAGCUUUCUACA
|
|
|
- Jennifer McBride
- 6 years ago
- Views:
Transcription
1 Precursor mmu-mir-8- GUGAUAAUGGAGCGAGAUUUUCUGUUGUGCUUGAUCUAACCAUGUGCUUGCGAGGUAUGA GAAAAACAUGGUUCCGUCAAGCACCAUGGAACGUCACGCAGCUUUCUACA Precursor mmu-mir-8- GACCAGUUGCCGCGGGGCUUUCCUUUGUGCUUGAUCUAACCAUGUGGUGGAACGAUGGAA ACGGAACAUGGUUCUGUCAAGCACCGCGGAAAGCAUCGCUCUCUCCUGCA Mature mir 8 UUGUGCUUGAUCUAACCAUGU Supplemental Figure S: The RNA sequence of primary and mature mir-8- and - are illustrated. Both mir-8- and - yield a single species of mature microrna, mir-8. The seed sequence is marked by a box.
2 Supplemental Figure. S A. MC3T3 Dkk elative transcript levels Days Tob Relative transcript levels Days Sfrp Sost B. BMSCs Rela7ve transcript levels.5.5 Days Dkk Tob Sfrp Supplemental Figure. S: Expression of mir- 8 target genes during osteoblast differen7a7on. Total RNA was analyzed for expression of Dkk, Tob, Sfrp and Sost by real Dme PCR at the indicated Dme points using MC3T3 cells (A) and BMSCs (B). Sost is not detected in BMSCs. Expression was normalized with GAPDH RNA.
3
4 Supplemental Figure S4: Axin CyclinD Fig S4. Stimulated Wnt signaling by mir-8 MC3T3 cells were treated by Control, mir-8 or anti-mir-8 Lentivirus. Expression of Wnt target genes, Axin and CyclinD, was analyzed by qpcr.
5 Supplemental Table S: Nucleotide sequences of primers and probes used for qrt-pcr and northern blot Bone Marker Genes - Murine Real Time qpcr Primers (5-3 ) Runx F: CGGCCCTCCCTGAACTCT R: TGCCTGCCTGGGATCTGTA Alp F: CCAACTCTTTTGTGCCAGAGA R: GGCTACATTGGTGTTGAGCTTTT Ocn F: CTGACAAAGCCTTCATGTCCAA R: GCGCCGGAGTCTGTTCACTA Tob F: ATGGCGTCCTCTCTGCTTG R: TGAAAGGTCAGCGTATGGCTT Sost F: AGCCTTCAGGAATGATGCCAC R: CTTTGGCGTCATAGGGATGGT Dkk F: TCAGTCAGCCAACCGATCTG R: TCTCTGTGGCATCGTTTCTTTT Sfrp F: CGTGGGCTCTTCCTCTTG R: ATGTTCTGGTACTCGATCCG Tcf F: AGCTTTCTCCACTCTACGAACA R: AATCCAGAGAGATCGGGGGTC Lef F: TGTTTATCCCATCACGGGTGG R: CATGGAAGTGTCGCCTGACAG Axin F: TGACTCTCCTTCCAGATCCCA R: TGCCCACACTAGGCTGACA CyclinD F: GCGTACCCTGACACCAATCTC R: ACTTGAAGTAAGATACGGAGGGC mir-8- F: CCATGGAACGTCACGCAGC mir-8- F: GCGGAAAGCACCGTGCTC Mature mir-8 F: TTGTGCTTGATCTAACCATGT hsost F: ACTTCAGAGGAGGCAGAAATGG R: CAAGGGGGAATCTTATCCAACTTTC hdkk F: AGGAGTGTGAAGTTGGGAGGTA R: GGTTTGAGTAATGACCGTGGTT hsfrp F: ATGACAACGACATAATGGAA R: GGAGATGCGCTTGAACTCTC htcf4 F: CCTTAGGGACGGACAAAGAG R: GAAGAGTTGCCCAACATTCC hlef F: CTCTACAACAAGGGACCCTC R: CTTGTTTGGAGTTGACATCTG Control Control Primers (5 3 ) GAPDH F: AGGTCGGTGTGAACGGATTTG R: TGTAGACCATGTAGTTGAGGTCA U6 F: CGCTTCGGCAGCACATATAC R: AAAATATGGAACGCTTCACGA Northern Probes Primers (5-3 ) Mouse mir-8 GGAAATCCCTGGCAATGTGAT
6 SUPPLEMENTAL TABLE S Target UTR Fragment Sequences Cloned in pmir-report-luc Vector (Ambion Inc) DKK WT 5 CTAGTCGTGAAAATGCTATTATTAAAAGAAAGCACACCATGGTTAATCATGTGCAATACAAGCACAGGGCAGCA 3 DKK MT 5 CGATGTG CGATGTG 3 SFRP WT 5 CTAGTCCGCGACCTCATTTCCGGTTTCCCAAGCACAGTCCCTACAGAACTTCAGTTTATAAGCACACAGTAACCCA3 SFRP MT 5 CGATGTG CGATGTG 3 TOB WT 5 CTAGTCGCATCATTTTTCATTTGCCAACCAAGCACAAAGCGCATCATTTTTCATTTGCCAACCAAGCACAAAGA3 TOB MT 5 GATGTG CGATGTG 3 SOST WT 5 CTAGTGGCAGTGTTAATATCGCTTTGTGAAGCACAAGTGGGCAGTGTTAATATCGCTTTGTGAAGCACAAGTGA3 SOST MT 5 CGATGTG CGATGTG 3 Supplemental Table S: Schematic illustration of the 3 UTR of TOB, DKK, SFRP and SOST mrna target sequences with mir-8 participating nucleotides. Three putative target sites of different targets bound to mir-8 predicted by TargetScan ( MICRORNA.ORG ( and MICROCOSM ( programs. Last 7 nucleotides of the respective mrna targets (AAGCACA) were the seed sequence of mir-8. In the mutant reporter, these sequences were mutated (CGATGTG).
7 Supplemental Table S3: Pathway Ontology for mir-8 Targets Enrichment Score :.86 Gene count P value Retinol metabolism 3 4.3E- Enrichment Score :.85 Gene count P value Dilated cardiomypathy 6 4.7E- Arrhythmogenic right ventricular cardiomyopathy (ARVC) 4.E- Hypertrophic cardiomyopathy (HCM) 4.7E- Enrichment Score :.8 Gene count P value Gap junction 7.E- Vascular smooth muscle contraction 8.4E- Long-term depression 6.9E- Purine metabolism 6.5E- Enrichment Score :.78 Gene count P value Insulin signaling pathway 7 7.5E- Glioma 4.6E- Natural killer cell mediated cytotoxicity 5.7E- Chemokine signaling pathway 6 3.5E- Chronic myeloid leukemia 3 4.9E- ErbB signaling pathway 3 5.6E- Neurotrophin signaling pathway 3 7.7E- Enrichment Score :.74 Gene count P value Vascular smooth muscle contraction 8.4E- Enrichment Score :.7 Gene count P value ECM-receptor interaction 6 3.E- Enrichment Score :.46 Gene count P value Wnt signaling pathway 5 4.E- Supplemental Table S3: Pathway ontology for mir-8 targets. Using TargetScan (ver 6.), the top 5 (from approximately 8 putative targets with conserved sequences) were examined for the enrichment of gene ontology categories. The results show signaling pathways related to either cell differentiation pathways of different tissues or various cancers. Notable categories related to osteogenesis and bone metastasis include insulin, gap junctions, ECM-receptor interaction and ErbB and chemokine pathways.
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Supplementary Figure 1.
 Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1. Quantification of western blot analysis of fibroblasts (related to Figure 1) (A-F) Quantification of western blot analysis for control and IR-Mut fibroblasts. Data are expressed
Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern
 Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern blot. Northern blot analysis of mir-302b expression following infection with PAO1, PAK and Kp in (A) lung
Supplementary Figure 1 Validate the expression of mir-302b after bacterial infection by northern blot. Northern blot analysis of mir-302b expression following infection with PAO1, PAK and Kp in (A) lung
- 1 - Supplemental Data
 - 1-1 Supplemental Data 2 3 4 5 6 7 8 9 Supplemental Figure S1. Differential expression of AtPIP Genes in DC3000-inoculated plants. Gene expression in leaves was analyzed by real-time RT-PCR and expression
- 1-1 Supplemental Data 2 3 4 5 6 7 8 9 Supplemental Figure S1. Differential expression of AtPIP Genes in DC3000-inoculated plants. Gene expression in leaves was analyzed by real-time RT-PCR and expression
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Galina Gabriely, Ph.D. BWH/HMS
 Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
supplementary information
 Figure S1 ZEB1 full length mrna. (a) Analysis of the ZEB1 mrna using the UCSC genome browser (http://genome.ucsc.edu) revealed truncation of the annotated Refseq sequence (NM_030751). The probable terminus
Figure S1 ZEB1 full length mrna. (a) Analysis of the ZEB1 mrna using the UCSC genome browser (http://genome.ucsc.edu) revealed truncation of the annotated Refseq sequence (NM_030751). The probable terminus
MicroRNA Destabilization Enables. Dynamic Regulation of the mir-16 Family. In Response to Cell-Cycle Changes SUPPLEMENTAL INFORMATION
 Molecular Cell, Volume 43 SUPPLEMENTAL INFORMATION MicroRNA Destabilization Enables Dynamic Regulation of the mir-16 Family In Response to Cell-Cycle Changes Olivia S. Risland, Sue-Jean Hong, and David
Molecular Cell, Volume 43 SUPPLEMENTAL INFORMATION MicroRNA Destabilization Enables Dynamic Regulation of the mir-16 Family In Response to Cell-Cycle Changes Olivia S. Risland, Sue-Jean Hong, and David
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
HCT116 SW48 Nutlin: p53
 Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
1. Primers for PCR to amplify hairpin stem-loop precursor mir-145 plus different flanking sequence from human genomic DNA.
 Supplemental data: 1. Primers for PCR to amplify hairpin stem-loop precursor mir-145 plus different flanking sequence from human genomic DNA. Strategy#1: 20nt at both sides: #1_BglII-Fd primer : 5 -gga
Supplemental data: 1. Primers for PCR to amplify hairpin stem-loop precursor mir-145 plus different flanking sequence from human genomic DNA. Strategy#1: 20nt at both sides: #1_BglII-Fd primer : 5 -gga
Supporting Information
 Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
To generate the luciferase fusion to the human 3 UTRs, we sub-cloned the 3 UTR
 Plasmids To generate the luciferase fusion to the human 3 UTRs, we sub-cloned the 3 UTR fragments downstream of firefly luciferase (luc) in pgl3 control (Promega). pgl3- CDK6 was made by amplifying a 2,886
Plasmids To generate the luciferase fusion to the human 3 UTRs, we sub-cloned the 3 UTR fragments downstream of firefly luciferase (luc) in pgl3 control (Promega). pgl3- CDK6 was made by amplifying a 2,886
embryos. Asterisk represents loss of or reduced expression. Brackets represent
 Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Intron distance to transcription start (nt)*
 Schwab et al. SOM: Enhanced microrna accumulation through stemloop-adjacent introns 1 Supplemental Table 1. Characteristics of introns used in this study Intron name Length intron (nt) Length cloned fragment
Schwab et al. SOM: Enhanced microrna accumulation through stemloop-adjacent introns 1 Supplemental Table 1. Characteristics of introns used in this study Intron name Length intron (nt) Length cloned fragment
SUPPLEMENTARY INFORMATION
 SUPPLEMENTY INFORMATION doi:10.1038/nature20785 Extended data Fig 1f Extended data Fig 1h 37 KD 37 KD Extended data Fig 1i Extended data Fig 4c Extended data Fig 3b Extended data Fig 4j 37 KD 37 KD Extended
SUPPLEMENTY INFORMATION doi:10.1038/nature20785 Extended data Fig 1f Extended data Fig 1h 37 KD 37 KD Extended data Fig 1i Extended data Fig 4c Extended data Fig 3b Extended data Fig 4j 37 KD 37 KD Extended
PCR Arrays. An Advanced Real-time PCR Technology to Empower Your Pathway Analysis
 PCR Arrays An Advanced Real-time PCR Technology to Empower Your Pathway Analysis 1 Table of Contents 1. Introduction to the PCR Arrays 2. How PCR Arrays Work 3. Performance Data from PCR Arrays 4. Research
PCR Arrays An Advanced Real-time PCR Technology to Empower Your Pathway Analysis 1 Table of Contents 1. Introduction to the PCR Arrays 2. How PCR Arrays Work 3. Performance Data from PCR Arrays 4. Research
Supporting Information
 Supporting Information Lu et al. 10.1073/pnas.1106801108 Fig. S1. Analysis of spatial localization of mrnas coding for 23 zebrafish Wnt genes by whole mount in situ hybridization to identify those present
Supporting Information Lu et al. 10.1073/pnas.1106801108 Fig. S1. Analysis of spatial localization of mrnas coding for 23 zebrafish Wnt genes by whole mount in situ hybridization to identify those present
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector.
 Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Fig. S1. eif6 expression in HEK293 transfected with shrna against eif6 or pcmv-eif6 vector. (a) Western blotting analysis and (b) qpcr analysis of eif6 expression in HEK293 T cells transfected with either
Supplemental figure 1. Dys-regulated signal pathways in MSCs from TNF-Tg. mice. 965 dys-regulated genes were uploaded to IPA and David bioinformatics
 Supplemental figure 1. Dys-regulated signal pathways in MSCs from TN-Tg mice. 965 dys-regulated genes were uploaded to IPA and David bioinformatics esources software. The top 53 and top 11 dys-regulated
Supplemental figure 1. Dys-regulated signal pathways in MSCs from TN-Tg mice. 965 dys-regulated genes were uploaded to IPA and David bioinformatics esources software. The top 53 and top 11 dys-regulated
Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila
 Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
Molecular Cell, Volume 32 Supplemental Data Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila Rui Zhou, Ikuko Hotta, Ahmet M. Denli, Pengyu Hong, Norbert Perrimon, and Gregory
B. Transgenic plants with strong phenotype (%)
 A. TCTAGTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT AAATATGGTCTAAAGAAGAAGAATATGGTCTAAAGAAGAAGAATATGG 2XP35S STTM165 5 GGGGGATGAAGctaCCTGGTCCGA3 3 CCCCCUACUUC---GGACCAGGCU5 mir165 HindIII mir165 96 nt GTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT
A. TCTAGTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT AAATATGGTCTAAAGAAGAAGAATATGGTCTAAAGAAGAAGAATATGG 2XP35S STTM165 5 GGGGGATGAAGctaCCTGGTCCGA3 3 CCCCCUACUUC---GGACCAGGCU5 mir165 HindIII mir165 96 nt GTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTT
Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb
 Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
Gene Expression Analysis with Pathway-Centric DNA Microarrays
 Gene Expression Analysis with Pathway-Centric DNA Microarrays SuperArray Bioscience Corporation George J. Quellhorst, Jr. Ph.D. Manager, Customer Education Topics to be Covered Introduction to DNA Microarrays
Gene Expression Analysis with Pathway-Centric DNA Microarrays SuperArray Bioscience Corporation George J. Quellhorst, Jr. Ph.D. Manager, Customer Education Topics to be Covered Introduction to DNA Microarrays
Site-Directed Mutagenesis. Mutations in four Smad4 sites of mouse Gat1 promoter
 Supplement Supporting Materials and Methods Site-Directed Mutagenesis. Mutations in four Smad4 sites of mouse Gat1 promoter were independently generated using a two-step PCR method. The Smad4 binding site
Supplement Supporting Materials and Methods Site-Directed Mutagenesis. Mutations in four Smad4 sites of mouse Gat1 promoter were independently generated using a two-step PCR method. The Smad4 binding site
MeCP2. MeCP2/α-tubulin. GFP mir1-1 mir132
 Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Conservation Figure S1. Schematic showing 3 UTR (top; thick black line), mir132 MRE (arrow) and nucleotide sequence conservation (vertical black lines; http://genome.ucsc.edu). a GFP mir1-1 mir132 b GFP
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/362/ra12/dc1 Supplementary Materials for FOXP1 potentiates Wnt/β-catenin signaling in diffuse large B cell lymphoma Matthew P. Walker, Charles M. Stopford, Maria
www.sciencesignaling.org/cgi/content/full/8/362/ra12/dc1 Supplementary Materials for FOXP1 potentiates Wnt/β-catenin signaling in diffuse large B cell lymphoma Matthew P. Walker, Charles M. Stopford, Maria
MicroRNA-7-5p regulates osteogenic. differentiation of hmscs via targeting
 European Review for Medical and Pharmacological Sciences 2018; 22: 7826-7831 MicroRNA-7-5p regulates osteogenic differentiation of hmscs via targeting CMKLR1 B. CHEN 1,2, J. MENG 2, Y.-T. ZENG 1, Y.-X.
European Review for Medical and Pharmacological Sciences 2018; 22: 7826-7831 MicroRNA-7-5p regulates osteogenic differentiation of hmscs via targeting CMKLR1 B. CHEN 1,2, J. MENG 2, Y.-T. ZENG 1, Y.-X.
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat
 Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
Supplementary Figure 1. NORAD expression in mouse (A) and dog (B). The black boxes indicate the position of the regions alignable to the 12 repeat units in the human genome. Annotated transposable elements
mir-24-mediated down-regulation of H2AX suppresses DNA repair
 Supplemental Online Material mir-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells Ashish Lal 1,4, Yunfeng Pan 2,4, Francisco Navarro 1,4, Derek M. Dykxhoorn
Supplemental Online Material mir-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells Ashish Lal 1,4, Yunfeng Pan 2,4, Francisco Navarro 1,4, Derek M. Dykxhoorn
Table S1. Primers used in the study
 Table S1. Primers used in the study Primer name Application Sequence I1F16 Genotyping GGCAAGTGAGTGAGTGCCTA I1R11 Genotyping CCCACTCGTATTGACGCTCT V19 Genotyping GGGTCTCAAAGTCAGGGTCA D18Mit184-F Genotyping
Table S1. Primers used in the study Primer name Application Sequence I1F16 Genotyping GGCAAGTGAGTGAGTGCCTA I1R11 Genotyping CCCACTCGTATTGACGCTCT V19 Genotyping GGGTCTCAAAGTCAGGGTCA D18Mit184-F Genotyping
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
Summary MicroRNAs (mirnas) are genomically encoded small RNAs used by organisms to regulate the expression of proteins generated from messenger RNA
 Summary MicroRNAs (mirnas) are genomically encoded small RNAs used by organisms to regulate the expression of proteins generated from messenger RNA transcripts. The in vivo requirement of specific mirnas
Summary MicroRNAs (mirnas) are genomically encoded small RNAs used by organisms to regulate the expression of proteins generated from messenger RNA transcripts. The in vivo requirement of specific mirnas
sherwood - UltramiR shrna Collections
 sherwood - UltramiR shrna Collections Incorporating advances in shrna design and processing for superior potency and specificity sherwood - UltramiR shrna Collections Enabling Discovery Across the Genome
sherwood - UltramiR shrna Collections Incorporating advances in shrna design and processing for superior potency and specificity sherwood - UltramiR shrna Collections Enabling Discovery Across the Genome
UTR Reporter Vectors and Viruses
 UTR Reporter Vectors and Viruses 3 UTR, 5 UTR, Promoter Reporter (Version 1) Applied Biological Materials Inc. #1-3671 Viking Way Richmond, BC V6V 2J5 Canada Notice to Purchaser All abm products are for
UTR Reporter Vectors and Viruses 3 UTR, 5 UTR, Promoter Reporter (Version 1) Applied Biological Materials Inc. #1-3671 Viking Way Richmond, BC V6V 2J5 Canada Notice to Purchaser All abm products are for
QPCR ASSAYS FOR MIRNA EXPRESSION PROFILING
 TECH NOTE 4320 Forest Park Ave Suite 303 Saint Louis, MO 63108 +1 (314) 833-9764 mirna qpcr ASSAYS - powered by NAWGEN Our mirna qpcr Assays were developed by mirna experts at Nawgen to improve upon previously
TECH NOTE 4320 Forest Park Ave Suite 303 Saint Louis, MO 63108 +1 (314) 833-9764 mirna qpcr ASSAYS - powered by NAWGEN Our mirna qpcr Assays were developed by mirna experts at Nawgen to improve upon previously
Supplemental Table/Figure Legends
 MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
MiR-26a is required for skeletal muscle differentiation and regeneration in mice Bijan K. Dey, Jeffrey Gagan, Zhen Yan #, Anindya Dutta Supplemental Table/Figure Legends Suppl. Table 1: Effect of overexpression
Supporting Information
 Supporting Information Lujambio et al. 10.1073/pnas.0803055105 SI Materials and Methods RNA Isolation and mirna Expression Analysis. Total RNA was isolated from SW620,, and SIHN-0 cells, before and after
Supporting Information Lujambio et al. 10.1073/pnas.0803055105 SI Materials and Methods RNA Isolation and mirna Expression Analysis. Total RNA was isolated from SW620,, and SIHN-0 cells, before and after
Supporting Information
 Supporting Information Horie et al. 10.1073/pnas.1008499107 SI Materials and Methods ell ulture and Reagents. THP-1 cells were obtained from the American Type ell ollection. THP-1 cells were transformed
Supporting Information Horie et al. 10.1073/pnas.1008499107 SI Materials and Methods ell ulture and Reagents. THP-1 cells were obtained from the American Type ell ollection. THP-1 cells were transformed
MicroRNAs Modulate Hematopoietic Lineage Differentiation
 Chen et al., page 1 MicroRNAs Modulate Hematopoietic Lineage Differentiation Chang-Zheng Chen, Ling Li, Harvey F. Lodish, David. Bartel Supplemental Online Material Methods Cell isolation Murine bone marrow
Chen et al., page 1 MicroRNAs Modulate Hematopoietic Lineage Differentiation Chang-Zheng Chen, Ling Li, Harvey F. Lodish, David. Bartel Supplemental Online Material Methods Cell isolation Murine bone marrow
Fatchiyah
 Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
Table I. Primers used in quantitative real-time PCR for detecting gene expressions in mouse liver tissues
 Table I. Primers used in quantitative real-time PCR for detecting gene expressions in mouse liver tissues Gene name Forward primer 5' to 3' Gene name Reverse primer 5' to 3' CC_F TGCCCTGTTGGGGTTG CC_R
Table I. Primers used in quantitative real-time PCR for detecting gene expressions in mouse liver tissues Gene name Forward primer 5' to 3' Gene name Reverse primer 5' to 3' CC_F TGCCCTGTTGGGGTTG CC_R
SUPPLEMENTAL MATERIAL. Supplemental Methods. Cumate solution was from System Biosciences. Human complement C1q and complement
 SUPPLEMENTAL MATERIAL Supplemental Methods Reagents Cumate solution was from System Biosciences. Human complement Cq and complement C-esterase inhibitor (C-INH) were from Calbiochem. C-INH (Berinert) for
SUPPLEMENTAL MATERIAL Supplemental Methods Reagents Cumate solution was from System Biosciences. Human complement Cq and complement C-esterase inhibitor (C-INH) were from Calbiochem. C-INH (Berinert) for
Supplementary figures
 Relative intensity Relative intensity Relative intensity Supplementary figures a None Caffeine None Caffeine c None Caffeine 6 6 6 ISG 6 6 6 UBE1L 6 6 6 UBCH8 6 6 6 EFP 1 1 DOX (h) 1 1 CPT (h) 1 1 UV (h)
Relative intensity Relative intensity Relative intensity Supplementary figures a None Caffeine None Caffeine c None Caffeine 6 6 6 ISG 6 6 6 UBE1L 6 6 6 UBCH8 6 6 6 EFP 1 1 DOX (h) 1 1 CPT (h) 1 1 UV (h)
Supplementary Figure 1. ips_3y+mir-302b. ips_3y+mir-372. ips_3y+mir-302b + mir-372. ips_4y. ips_4y+mir-302b. ips_4y + mir-372
 Supplementary Figure 1 ips_3y+mir-32b ips_3y+ ips_3y+mir-32b + ips_4y ips_4y+mir-32b ips_4y + Nature Biotechnology: doi:1.138/nb.t.1862 TRA-1-6 DAPI Oct3/4 Supplementary Figure 1: ips cells derived from
Supplementary Figure 1 ips_3y+mir-32b ips_3y+ ips_3y+mir-32b + ips_4y ips_4y+mir-32b ips_4y + Nature Biotechnology: doi:1.138/nb.t.1862 TRA-1-6 DAPI Oct3/4 Supplementary Figure 1: ips cells derived from
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Pennsylvania 15260, USA and 6 Department of Bioinformatics and Biosystems Technology, Technical University of Applied Sciences Wildau, Hochschulring
 Robust gene expression and mutation analyses of RNA-sequencing of formalin-fixed diagnostic tumor samples Stefan Graw 1,6, Richard Meier 1,6, Kay Minn 1, Clark Bloomer 2, Andrew K Godwin 3, Brooke Fridley
Robust gene expression and mutation analyses of RNA-sequencing of formalin-fixed diagnostic tumor samples Stefan Graw 1,6, Richard Meier 1,6, Kay Minn 1, Clark Bloomer 2, Andrew K Godwin 3, Brooke Fridley
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kawai and Amano, http://www.jcb.org/cgi/content/full/jcb.201110008/dc1 Figure S1. Regulation of mirna expression by BRCA1. (A) Confirmation
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kawai and Amano, http://www.jcb.org/cgi/content/full/jcb.201110008/dc1 Figure S1. Regulation of mirna expression by BRCA1. (A) Confirmation
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
Reviewers' Comments: Reviewer #1 (Remarks to the Author)
 Reviewers' Comments: Reviewer #1 (Remarks to the Author) The manuscript reports a novel form of microrna sponge, designated a "zipper" an LNA antisense sequence that bridges from the 3' end of one microrna
Reviewers' Comments: Reviewer #1 (Remarks to the Author) The manuscript reports a novel form of microrna sponge, designated a "zipper" an LNA antisense sequence that bridges from the 3' end of one microrna
Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice
 Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice Supplementary Material Supplementary Methods Materials Spironolactone, aldosterone and β-glycerophosphate were
Spironolactone ameliorates PIT1-dependent vascular osteoinduction in klotho-hypomorphic mice Supplementary Material Supplementary Methods Materials Spironolactone, aldosterone and β-glycerophosphate were
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
SUPPLEMENTARY INFORMATION
 Materials and Methods Transgenic Plant Materials and DNA Constructs. VEX1::H2B-GFP, ACA3::H2B-GFP, KRP6::H2B-GFP, KRP6::mock21ts-GFP, KRP6::TE21ts- GFP and KRP6::miR161ts-GFP constructs were generated
Materials and Methods Transgenic Plant Materials and DNA Constructs. VEX1::H2B-GFP, ACA3::H2B-GFP, KRP6::H2B-GFP, KRP6::mock21ts-GFP, KRP6::TE21ts- GFP and KRP6::miR161ts-GFP constructs were generated
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
Next Generation Sequencing. Target Enrichment
 Next Generation Sequencing Target Enrichment Next Generation Sequencing Your Partner in Every Step from Sample to Data NGS: Revolutionizing Genetic Analysis with Single-Molecule Resolution Next generation
Next Generation Sequencing Target Enrichment Next Generation Sequencing Your Partner in Every Step from Sample to Data NGS: Revolutionizing Genetic Analysis with Single-Molecule Resolution Next generation
LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation
 LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation Mi Young Park, 1# Raki Sudan, 1# Neetu Srivastava, 1 Sudha Neelam, 1 Christie Youngs, 1 Jia- Wang Wang, 4 Robert W. Engelman, 5,6,7
LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation Mi Young Park, 1# Raki Sudan, 1# Neetu Srivastava, 1 Sudha Neelam, 1 Christie Youngs, 1 Jia- Wang Wang, 4 Robert W. Engelman, 5,6,7
Document S1. Supplemental Experimental Procedures and Three Figures (see next page)
 Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
b alternative classical none
 Supplementary Figure. 1: Related to Figure.1 a d e b alternative classical none NIK P-IkBa Total IkBa Tubulin P52 (Lighter) P52 (Darker) RelB (Lighter) RelB (Darker) HDAC1 Control-Sh RelB-Sh NF-kB2-Sh
Supplementary Figure. 1: Related to Figure.1 a d e b alternative classical none NIK P-IkBa Total IkBa Tubulin P52 (Lighter) P52 (Darker) RelB (Lighter) RelB (Darker) HDAC1 Control-Sh RelB-Sh NF-kB2-Sh
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1. Validation of CDK9-inhibitor treatment.
 Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Khaled_Fig. S MITF TYROSINASE R²= MITF PDE4D R²= Variance from mean mrna expression
 Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Blimp-1/PRDM1 rabbit monoclonal antibody (C14A4) was purchased from Cell Signaling
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Methods Antibodies Blimp-1/PRDM1 rabbit monoclonal antibody (C14A4) was purchased from Cell Signaling (Danvers, MA) and used at 1:1000 to detect the total PRDM1 protein
1 2 3 4 5 6 7 8 9 10 11 Supplementary Methods Antibodies Blimp-1/PRDM1 rabbit monoclonal antibody (C14A4) was purchased from Cell Signaling (Danvers, MA) and used at 1:1000 to detect the total PRDM1 protein
Supplementary Figure 1. Characterization of the POP2 transcriptional and post-transcriptional regulatory elements. (A) POP2 nucleotide sequence
 1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Bootcamp: Molecular Biology Techniques and Interpretation
 Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 MOLECULAR BIOLOGY BIO-2B02 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 MOLECULAR BIOLOGY BIO-2B02 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question
Supplementary Figure 1. ERK signaling is not activated at early hypertension. a, Western blot analysis for the level of phospho-erk (perk) and total
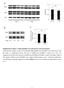 Supplementary Figure 1. ERK signaling is not activated at early hypertension. a, Western blot analysis for the level of phospho-erk (perk) and total ERK in the aortic tissue from the saline- or AngII-infused
Supplementary Figure 1. ERK signaling is not activated at early hypertension. a, Western blot analysis for the level of phospho-erk (perk) and total ERK in the aortic tissue from the saline- or AngII-infused
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
Marilyn G. Rimando, Hao-Hsiang Wu, Yu-An Liu, Chien-Wei Lee, Shu-Wen Kuo, Yin-
 Supplementary Information Glucocorticoid receptor and Histone Deacetylase 6 mediate the differential effect of dexamethasone during osteogenesis of Mesenchymal stromal cells (MSCs) Marilyn G. Rimando,
Supplementary Information Glucocorticoid receptor and Histone Deacetylase 6 mediate the differential effect of dexamethasone during osteogenesis of Mesenchymal stromal cells (MSCs) Marilyn G. Rimando,
Functional microrna targets in protein coding sequences. Merve Çakır
 Functional microrna targets in protein coding sequences Martin Reczko, Manolis Maragkakis, Panagiotis Alexiou, Ivo Grosse, Artemis G. Hatzigeorgiou Merve Çakır 27.04.2012 microrna * micrornas are small
Functional microrna targets in protein coding sequences Martin Reczko, Manolis Maragkakis, Panagiotis Alexiou, Ivo Grosse, Artemis G. Hatzigeorgiou Merve Çakır 27.04.2012 microrna * micrornas are small
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for
 Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials Molecular Biology of the Cell
 Supplemental Materials Molecular Biology of the Cell Zorbas et al. Fig S1: Positions of sirnas on transcripts targeted in this work, and sirna-mediated depletion efficiency A, The regions targeted
Supplemental Materials Molecular Biology of the Cell Zorbas et al. Fig S1: Positions of sirnas on transcripts targeted in this work, and sirna-mediated depletion efficiency A, The regions targeted
Flowcytometry-based purity analysis of peritoneal macrophage culture.
 Liao et al., KLF4 regulates macrophage polarization Revision of Manuscript 45444-RG- Supplementary Figure Legends Figure S Flowcytometry-based purity analysis of peritoneal macrophage culture. Thioglycollate
Liao et al., KLF4 regulates macrophage polarization Revision of Manuscript 45444-RG- Supplementary Figure Legends Figure S Flowcytometry-based purity analysis of peritoneal macrophage culture. Thioglycollate
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3389 Supplementary Figure 1 Hepatocyte-specific overexpression of activated Yap causes hepatomegaly and accelerates DMBA-induced liver tumor formation. (a) Schematic diagram of the construct
DOI: 10.1038/ncb3389 Supplementary Figure 1 Hepatocyte-specific overexpression of activated Yap causes hepatomegaly and accelerates DMBA-induced liver tumor formation. (a) Schematic diagram of the construct
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit
 Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
Supplemental Figure 1
 Supplemental Figure 1 Supplemental figure 1. Generation of gene targeted mice expressing an anti-id BCR. (A,B) Generation of the VDJ aid H KI mouse. (A) Targeting Construct. Top: Targeting construct for
Supplemental Figure 1 Supplemental figure 1. Generation of gene targeted mice expressing an anti-id BCR. (A,B) Generation of the VDJ aid H KI mouse. (A) Targeting Construct. Top: Targeting construct for
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Pattern Formation via Small RNA Mobility SUPPLEMENTAL FIGURE 1 SUPPLEMENTAL INFORMATION. Daniel H. Chitwood et al.
 SUPPLEMENTAL INFORMATION Pattern Formation via Small RNA Mobility Daniel H. Chitwood et al. SUPPLEMENTAL FIGURE 1 Supplemental Figure 1. The ARF3 promoter drives expression throughout leaves. (A, B) Expression
SUPPLEMENTAL INFORMATION Pattern Formation via Small RNA Mobility Daniel H. Chitwood et al. SUPPLEMENTAL FIGURE 1 Supplemental Figure 1. The ARF3 promoter drives expression throughout leaves. (A, B) Expression
Gene expression analysis. Gene expression analysis. Total RNA. Rare and abundant transcripts. Expression levels. Transcriptional output of the genome
 Gene expression analysis Gene expression analysis Biology of the transcriptome Observing the transcriptome Computational biology of gene expression sven.nelander@wlab.gu.se Recent examples Transcriptonal
Gene expression analysis Gene expression analysis Biology of the transcriptome Observing the transcriptome Computational biology of gene expression sven.nelander@wlab.gu.se Recent examples Transcriptonal
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms
 Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Alteration of microrna expression correlates to fatty acidmediated. insulin resistance in mouse myoblasts
 Alteration of microrna expression correlates to fatty acidmediated insulin resistance in mouse myoblasts Supplemental Data Correlation coefficient matrix con PA PA 0.9425 1.0000 PA+OA 0.9407 0.9626 con
Alteration of microrna expression correlates to fatty acidmediated insulin resistance in mouse myoblasts Supplemental Data Correlation coefficient matrix con PA PA 0.9425 1.0000 PA+OA 0.9407 0.9626 con
Exam 3 4/25/07. Total of 7 questions, 100 points.
 Exam 3 4/25/07 BISC 4A P. Sengupta Total of 7 questions, 100 points. QUESTION 1. Circle the correct answer. Total of 40 points 4 points each. 1. Which of the following is typically attacked by the antibody-mediated
Exam 3 4/25/07 BISC 4A P. Sengupta Total of 7 questions, 100 points. QUESTION 1. Circle the correct answer. Total of 40 points 4 points each. 1. Which of the following is typically attacked by the antibody-mediated
Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex
 POSTER PRESENTATION Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex Hai-Chon Lee *, Je-In Youn, Kyungwha Lee, Hwanyul Yong, Seung-Yong
POSTER PRESENTATION Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex Hai-Chon Lee *, Je-In Youn, Kyungwha Lee, Hwanyul Yong, Seung-Yong
Regulation of hepcidin expression by inflammation-induced activin B
 Regulation of hepcidin expression by inflammation-induced activin B Yohei Kanamori, Makoto Sugiyama, Osamu Hashimoto, Masaru Murakami, Tohru Matsui and Masayuki Funaba Supplemental methods Liver cell separation
Regulation of hepcidin expression by inflammation-induced activin B Yohei Kanamori, Makoto Sugiyama, Osamu Hashimoto, Masaru Murakami, Tohru Matsui and Masayuki Funaba Supplemental methods Liver cell separation
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/496/eaam6291/dc1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells
www.sciencesignaling.org/cgi/content/full/10/496/eaam6291/dc1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells
SUPPLEMENTARY FIGURES AND TABLES
 SUPPLEMENTARY FIGURES AND TABLES Suppl. Table 1 Oligonucleotides used in the study The table shows the structure and nucleotide position of the oligonucleotides used in the study along with their application
SUPPLEMENTARY FIGURES AND TABLES Suppl. Table 1 Oligonucleotides used in the study The table shows the structure and nucleotide position of the oligonucleotides used in the study along with their application
Sample & Assay Technologies. Challenging biological samples: stabilization and simultaneous purification of DNA and RNA
 Challenging biological samples: stabilization and simultaneous purification of DNA and RNA October 10, 2012 Stefanie Schröer Challenging biological samples Precious and nonhomogeneous samples Clinical
Challenging biological samples: stabilization and simultaneous purification of DNA and RNA October 10, 2012 Stefanie Schröer Challenging biological samples Precious and nonhomogeneous samples Clinical
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2.
 Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
